Genome-Wide Association Analysis for Chronic Superficial Keratitis in the Australian Racing Greyhound
Abstract
1. Introduction
2. Materials and Methods
3. Results
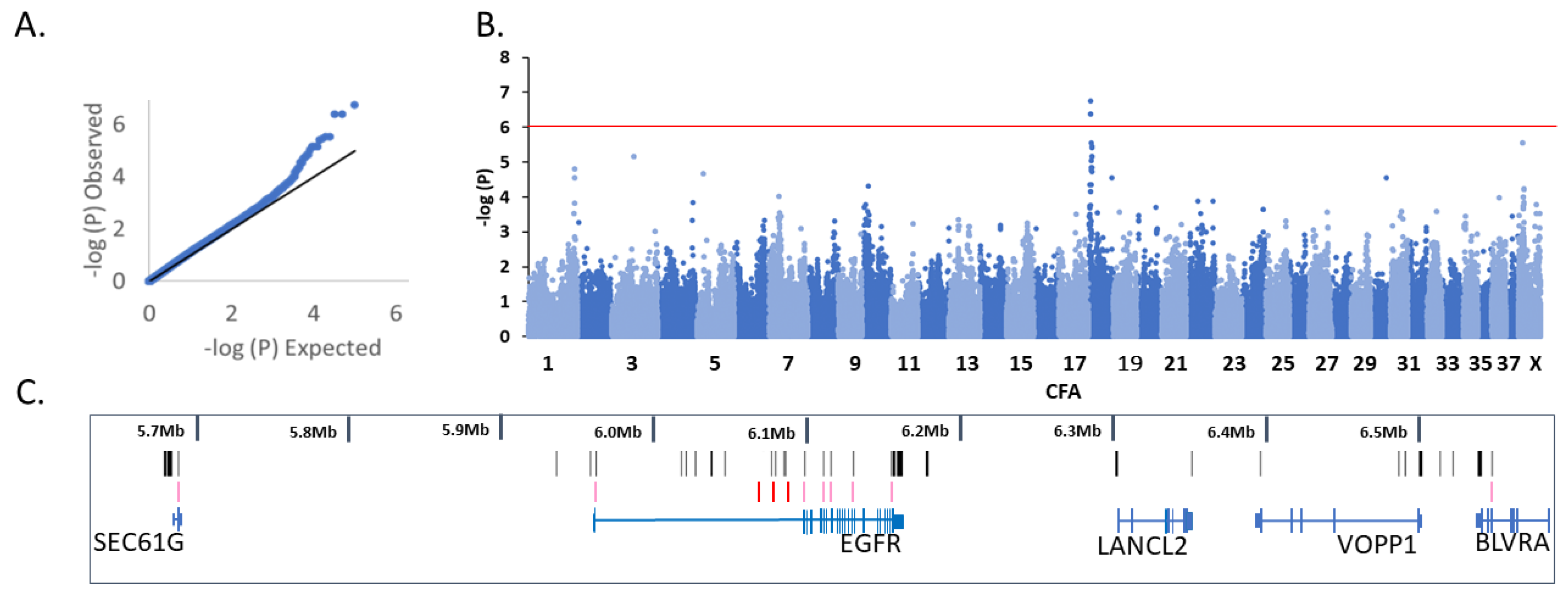
3.1. Genome-Wide Association Analysis
3.2. Variant Analysis
3.2.1. CFA18
3.2.2. CFX
3.2.3. CFA3
3.3. Haplotype Risk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slatter, D.H.; Lavach, J.D.; Severin, G.A.; Young, S. Uberreiter’s syndrome (chronic superficial keratitis) in dogs in the Rocky Mountain area—A study of 463 cases. J. Small Anim. Pract. 1977, 18, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Lynch, G.L. Ophthalmic examination findings in a group of retired racing Greyhounds. Vet. Ophthalmol. 2007, 10, 363–367. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, D.G.; Rooney, N.J.; Brock, C.; Church, D.B.; Brodbelt, D.C.; Pegram, C. Greyhounds under general veterinary care in the UK during 2016: Demography and common disorders. Canine Genet. Epidemiol. 2019, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Chavkin, M.J.; Roberts, S.M.; Salman, M.D.; Severin, G.A.; Scholten, N.J. Risk factors for development of chronic superficial keratitis in dogs. J. Am. Vet. Med. Assoc. 1994, 204, 1630–1634. [Google Scholar]
- Cheng, S.; Wigney, D.; Haase, B.; Wade, C.M. Inheritance of chronic superficial keratitis in Australian Greyhounds. Anim. Genet. 2016, 47, 629. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.L. Histological and immunohistochemical evaluation of canine chronic superficial keratitis. Res. Vet. Sci. 1999, 67, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Jokinen, P.; Rusanen, E.M.; Kennedy, L.J.; Lohi, H. MHC class II risk haplotype associated with canine chronic superficial keratitis in German Shepherd dogs. Vet. Immunol. Immunopathol. 2011, 140, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.S.; Zapata, G.; Crespi, J.A.; Posik, D.M.; Diaz, S.; It, V.; Peral-Garcia, P.; Giovambattista, G. A study of the association between chronic superficial keratitis and polymorphisms in the upstream regulatory regions of DLA-DRB1, DLA-DQB1 and DLA-DQA1. Vet. Immunol. Immunopathol. 2013, 156, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Stanley, R.G. Superficial stromal keratitis in the dog. Aust. Vet. J. 1988, 65, 321–323. [Google Scholar] [CrossRef]
- Gannon, J.R.; Craig, A.M.; Fegan, D.P. Care of the Racing and Retired Greyhound, 1st ed.; American Greyhound Council: Abilene, KS, USA, 2007. [Google Scholar]
- Yam, G.H.F.; Riau, A.K.; Funderburgh, M.L.; Mehta, J.S.; Jhanji, V. Keratocyte biology. Exp. Eye Res. 2020, 196, 108062. [Google Scholar] [CrossRef]
- Kamil, S.; Mohan, R.R. Corneal stromal wound healing: Major regulators and therapeutic targets. Ocul. Surf. 2021, 19, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.L. Major histocompatibility class II expression in the normal canine cornea and in canine chronic superficial keratitis. Vet. Ophthalmol. 2005, 8, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.H.; Okuda, H.K.; Lipton, D.E.; Reed, C. Chronic superficial keratitis in dogs: Detection of cellular hypersensitivity. Am. J. Vet. Res. 1975, 36, 669–671. [Google Scholar]
- Kaur, A.; Kumar, V.; Singh, S.; Singh, J.; Upadhyay, N.; Datta, S.; Singla, S.; Kumar, V. Toll-like receptor-associated keratitis and strategies for its management. 3 Biotech. 2015, 5, 611–619. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc. 2009, 2009, 71. [Google Scholar] [CrossRef]
- Wang, C.; Wallerman, O.; Arendt, M.L.; Sundstrom, E.; Karlsson, A.; Nordin, J.; Makelainen, S.; Pielberg, G.R.; Hanson, J.; Ohlsson, A.; et al. A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun. Biol. 2021, 4, 185. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Chew, T.; Willet, C.E.; Haase, B.; Wade, C.M. Genomic Characterization of External Morphology Traits in Kelpies Does Not Support Common Ancestry with the Australian Dingo. Genes 2019, 10, 337. [Google Scholar] [CrossRef]
- Hinrichs, A.S.; Raney, B.J.; Speir, M.L.; Rhead, B.; Casper, J.; Karolchik, D.; Kuhn, R.M.; Rosenbloom, K.R.; Zweig, A.S.; Haussler, D.; et al. UCSC Data Integrator and Variant Annotation Integrator. Bioinformatics 2016, 32, 1430–1432. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, V.; Drogemuller, C.; Leeb, T.; Dog Biomedical Variant Database Consortium. A comprehensive biomedical variant catalogue based on whole genome sequences of 582 dogs and eight wolves. Anim. Genet. 2019, 50, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.C.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Lambiase, A.; Micera, A.; Sacchetti, M.; Mantelli, F.; Bonini, S. Toll-like receptors in ocular surface diseases: Overview and new findings. Clin. Sci. 2011, 120, 441–450. [Google Scholar] [CrossRef]
- Andrew, S.E. Immune-mediated canine and feline keratitis. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 269–290. [Google Scholar] [CrossRef]
- Eichenbaum, J.D.; Lavach, J.D.; Gould, D.H.; Severin, G.A.; Paulsen, M.E.; Jones, R.L. Immunohistochemical staining patterns of canine eyes affected with chronic superficial keratitis. Am. J. Vet. Res. 1986, 47, 1952–1955. [Google Scholar]
- Farrugia, M.; Baron, B. The Role of Toll-Like Receptors in Autoimmune Diseases through Failure of the Self-Recognition Mechanism. Int. J. Inflam. 2017, 2017, 8391230. [Google Scholar] [CrossRef]

| Position on CFA18 (bp) | 5,961,614 | 6,068,508 | 6,077,388 | 6,087,347 | 6,110,158 | 6,115,276 | 6,154,817 |
| Variant identifier | rs851737129 | rs22613273 | rs22642459 | rs22653533 | . | . | rs397512405 |
| Location | EGFR | ARRAY | ARRAY | ARRAY | EGFR | EGFR | EGFR |
| Feature | splice_region_variant | intronic | intronic | intronic | missense_variant | splice_region_variant | potential stop codon |
| Reference allele a | G | A | C | T | G | A | T |
| Alternate allele (s) | C | G | T | C | A | T | C |
| USCF1455 (case) | G G | G G | T T | C C | G G | T A | C T |
| USCF3181 (control) | C G | A G | C T | T C | A G | T A | C T |
| Source of annotation | UU-GSD1 a | ARRAY c | ARRAY | ARRAY | VAI b | VAI | UU-GSD1 |
| MAF722 d | 0.552 | 0.397 | 0.432 | 0.383 | 0.0009158 | 0.063 | 0.132 |
| MAF590 e | 0.491 | 0.479 | 0.486 | 0.476 | - | 0.036 | 0.074 |
| Case | Control | Penetrance | ||
|---|---|---|---|---|
| CFA18 Haplotype a | Homozygous GAG (Risk18) | 65 | 21 | 0.76 |
| Heterozygous | 3 | 16 | 0.16 | |
| Homozygous AGA (Low-risk18) | 0 | 1 | 0.00 | |
| Uncalled | 2 | 2 | ||
| CFX:BICF2G630538106 b | G G (RiskX)(Female) G (Male) | 63 | 26 | 0.71 |
| A G (Female only) | 3 | 5 | 0.38 | |
| AA (Low-riskX) (Female) A (Male) | 2 | 8 | 0.20 | |
| Uncalled | 2 | 0 | ||
| Interaction | Risk18/RiskX | 61 | 15 | 0.80 |
| Risk18/Low-riskX | 2 | 4 | 0.33 | |
| Low-risk18/RiskX | 0 | 0 | - | |
| Low-risk18/Low-riskX | 0 | 1 | 0.00 | |
| Other | 7 | 19 | 0.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karamatic, S.; Goode, R.; Bageswaran, N.; Willet, C.E.; Samaha, G.; Ferguson, R.; Mazrier, H.; Wade, C.M. Genome-Wide Association Analysis for Chronic Superficial Keratitis in the Australian Racing Greyhound. Genes 2022, 13, 1328. https://doi.org/10.3390/genes13081328
Karamatic S, Goode R, Bageswaran N, Willet CE, Samaha G, Ferguson R, Mazrier H, Wade CM. Genome-Wide Association Analysis for Chronic Superficial Keratitis in the Australian Racing Greyhound. Genes. 2022; 13(8):1328. https://doi.org/10.3390/genes13081328
Chicago/Turabian StyleKaramatic, Steven, Rebecca Goode, Niruba Bageswaran, Cali E. Willet, Georgina Samaha, Ray Ferguson, Hamutal Mazrier, and Claire M. Wade. 2022. "Genome-Wide Association Analysis for Chronic Superficial Keratitis in the Australian Racing Greyhound" Genes 13, no. 8: 1328. https://doi.org/10.3390/genes13081328
APA StyleKaramatic, S., Goode, R., Bageswaran, N., Willet, C. E., Samaha, G., Ferguson, R., Mazrier, H., & Wade, C. M. (2022). Genome-Wide Association Analysis for Chronic Superficial Keratitis in the Australian Racing Greyhound. Genes, 13(8), 1328. https://doi.org/10.3390/genes13081328






