Molecular Characterization of TGF-Beta Gene Family in Buffalo to Identify Gene Duplication and Functional Mutations
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of TGF-β Genes in Buffalo
2.2. Characterization of Buffalo TGF-β Genes
2.3. Multiple Sequence Alignment
2.4. Structural Features Analysis
2.5. Phylogenetic Analysis
2.6. Synteny and Gene Duplications Analysis of TGF-β Superfamily Genes
2.7. Evaluation of Functional Mutation (SNPs) Effect in Buffalo TGF-β Proteins
3. Results
3.1. Genomic Identification of Buffalo TGF-β Genes
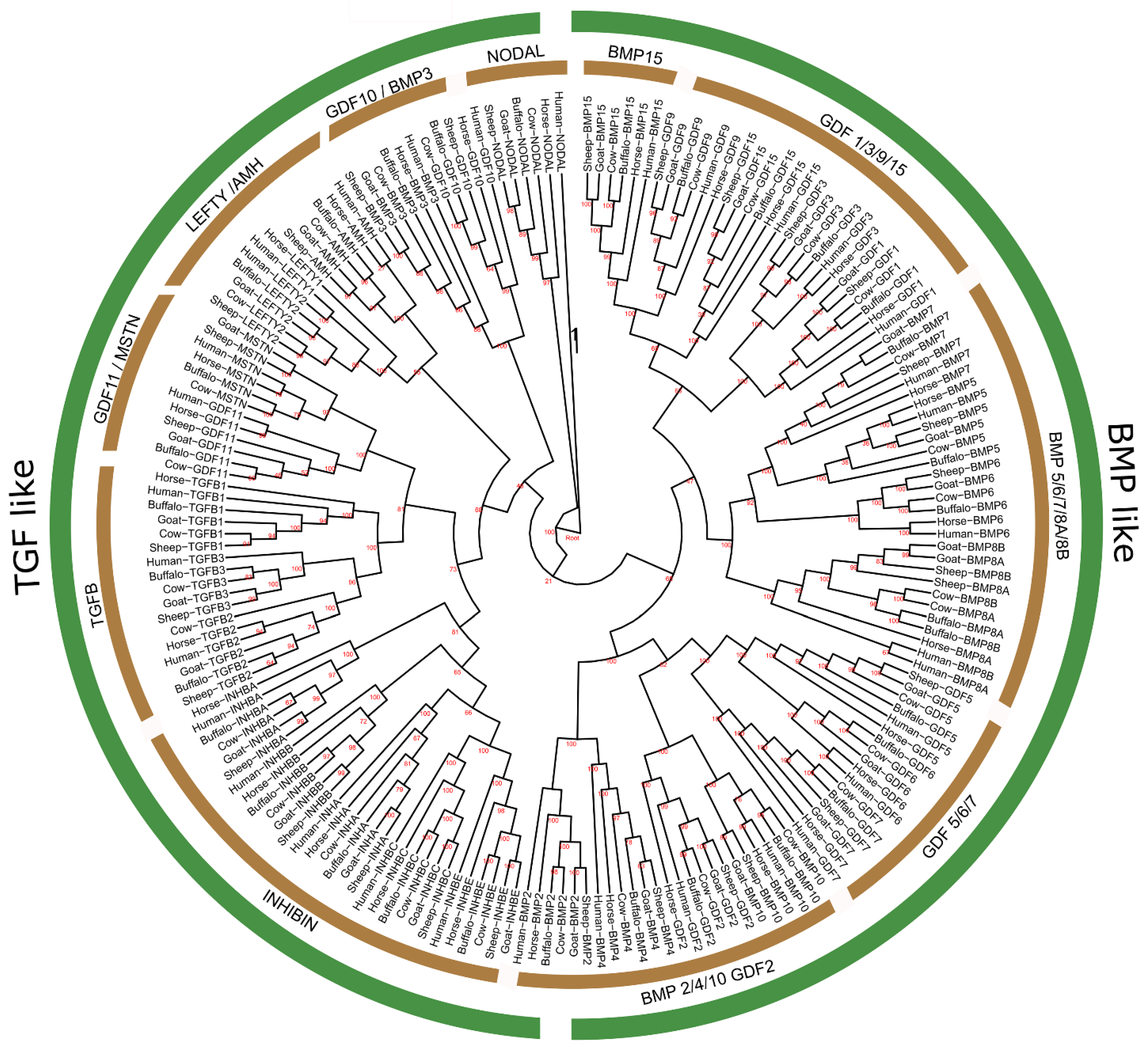
3.2. Phylogenetic Analysis of TGF-β Gene Family
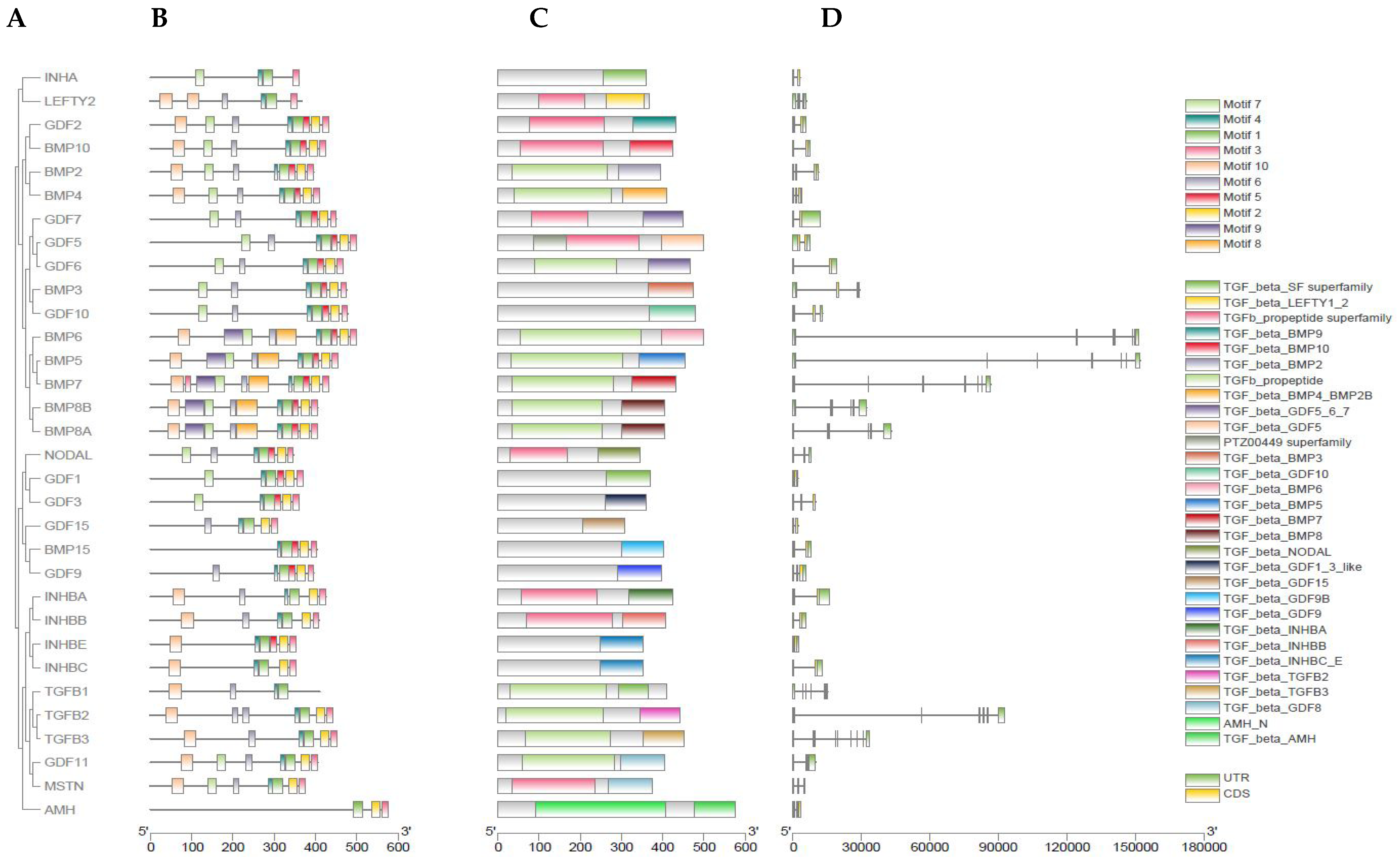
3.3. Gene Structure Analysis of Buffalo TGF-β Genes
3.4. Characterization of Physicochemical Properties of Buffalo TGF-β Genes
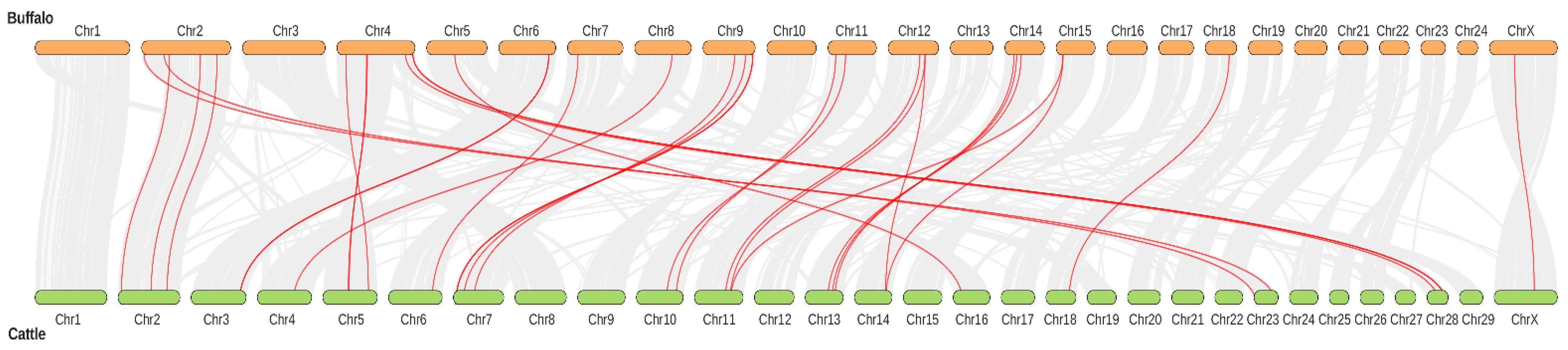
3.5. Buffalo and Cattle TGF-β Genes Collinearity Analysis and Chromosomal Distribution
3.6. Comparative Amino Acid and Functional Mutation Effect Analysis of Buffalo TGF-β Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moses, H.L.; Roberts, A.B.; Derynck, R. The Discovery and Early Days of TGF-β: A Historical Perspective. Cold Spring Harb. Perspect. Biol. 2016, 8, a021865. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.; Gong, W.; Gressner, A.M. Expression of Different Isoforms of TGF-β and the Latent TGF-β Binding Protein (LTBP) by Rat Kupffer Cells. J. Hepatol. 1998, 29, 915–922. [Google Scholar] [CrossRef]
- Capdevila, J.; Belmonte, J.C.I. Patterning Mechanisms Controlling Vertebrate Limb Development. Annu. Rev. Cell Dev. Biol. 2001, 17, 87–132. [Google Scholar] [CrossRef] [PubMed]
- Rissi, M.; Wittbrodt, J.; Délot, E.; Naegeli, M.; Rosa, F.M. Zebrafish Radar: A New Member of the TGF-β Superfamily Defines Dorsal Regions of the Neural Plate and the Embryonic Retina. Mech. Dev. 1995, 49, 223–234. [Google Scholar] [CrossRef]
- Grimaud, E.; Heymann, D.; Rédini, F. Recent Advances in TGF-β Effects on Chondrocyte Metabolism: Potential Therapeutic Roles of TGF-β in Cartilage Disorders. Cytokine Growth Factor Rev. 2002, 13, 241–257. [Google Scholar] [CrossRef]
- Wu, M.Y.; Hill, C.S. TGF-β Superfamily Signaling in Embryonic Development and Homeostasis. Dev. Cell 2009, 16, 329–343. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef]
- Kang, J.S.; Liu, C.; Derynck, R. New Regulatory Mechanisms of TGF-β Receptor Function. Trends Cell Biol. 2009, 19, 385–394. [Google Scholar] [CrossRef]
- Jiang, D.; Liang, J. A Long Noncoding RNA Links TGF-β Signaling in Lung Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 200, 123–125. [Google Scholar] [CrossRef]
- Lei, X.; Shi, D.; Zhang, H.; Jiang, J.; Su, J.; Cui, K.; Liu, Q.; Li, Z. BMP-1 Participates in the Selection and Dominance Of Buffalo Follicles by Regulating the Proliferation And Apoptosis of Granulosa Cells. Theriogenology 2016, 85, 999–1012. [Google Scholar] [CrossRef]
- Li, Y.; Jing, J.; Dang, W.; Jia, K.; Guo, X.; Kebreab, E.; Lyu, L.; Zhao, J. Cross-Talk between NOTCH2 and BMP4/SMAD Signaling Pathways in Bovine Follicular Granulosa Cells. Theriogenology 2022, 187, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.-Q.; Li, X.-L.; Wang, W.-J.; Hao, H.-S.; Zou, H.-Y.; Pang, Y.-W.; Zhao, X.-M.; Zhu, H.-B.; Du, W.-H. Knockdown of Bone Morphogenetic Protein 4 Gene Induces Apoptosis and Inhibits Proliferation of Bovine Cumulus Cells. Theriogenology 2022, 188, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chauhan, M.S. Relative Expression of the Developmentally Important Candidate Genes in Immature Oocytes and in Vitro-Produced Embryos of Buffalo (Bubalus Bubalis). Zygote 2022, 1–7, Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Taqi, M.O.; Sosa, A.S.A.; El-Naby, A.-S.A.-H.; Mahmoud, K.G.M.; Darwish, H.R.H.; Abd El Hameed, A.R.; Nawito, M.F. Spatiotemporal Expression Pattern of MiR-205, MiR-26a-5p, MiR-17-5p, Let-7b-5p, and Their Target Genes during Different Stages of Corpus Luteum in Egyptian Buffaloes. J. Genet. Eng. Biotechnol. 2022, 20, 37. [Google Scholar] [CrossRef] [PubMed]
- Babera, S.C.; Uy, M.R.D.; Tubalinal, G.A.S.P.; Mamuad, F.V.; Mingala, C.N. Molecular Characterization and Polymorphism of Inhibin (INHβA) Gene in Water Buffalo (Bubalus Bubalis) Bulls. Philipp. J. Sci. 2022, 151, 275–280. [Google Scholar]
- Zhu, P.; Li, H.; Huang, G.; Cui, J.; Zhang, R.; Cui, K.; Yang, S.; Shi, D. Molecular Cloning, Identification, and Expression Patterns of Myostatin Gene in Water Buffalo (Bubalus Bubalis). Anim. Biotechnol. 2018, 29, 26–33. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Available online: http://www.Fao.Org/Faostat/En/#home (accessed on 10 April 2020).
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER Web Server: 2015 Update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. Proteom. Protoc. Handb. 2005, 112, 571–607. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Shafique, L.; Yousuf, M.R.; Liu, Q.; Ahmed, J.Z.; Riaz, H. Spectrophotometric Calibration and Comparison of Different Semen Evaluation Methods in Nili-Ravi Buffalo Bulls. Pak. Vet. J. 2019, 39, 568–572. [Google Scholar] [CrossRef]
- Rehman, S.U.; Nadeem, A.; Javed, M.; Hassan, F.; Luo, X.; Khalid, R.B.; Liu, Q. Genomic Identification, Evolution and Sequence Analysis of the Heat-Shock Protein Gene Family in Buffalo. Genes 2020, 11, 1388. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Hassan, F.; Luo, X.; Li, Z.; Liu, Q. Whole-Genome Sequencing and Characterization of Buffalo Genetic Resources: Recent Advances and Future Challenges. Animals 2021, 11, 904. [Google Scholar] [CrossRef]
- Wu, S.; Hassan, F.; Luo, Y.; Fatima, I.; Ahmed, I.; Ihsan, A.; Safdar, W.; Liu, Q.; Rehman, S.U. Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors. Biology 2021, 10, 1207. [Google Scholar] [CrossRef]
- Hassan, F.; Nadeem, A.; Li, Z.; Javed, M.; Liu, Q.; Azhar, J.; Rehman, M.S.; Cui, K.; Rehman, S.U. Role of Peroxisome Proliferator-Activated Receptors (PPARs) in Energy Homeostasis of Dairy Animals: Exploiting Their Modulation through Nutrigenomic Interventions. Int. J. Mol. Sci. 2021, 22, 12463. [Google Scholar] [CrossRef]
- Rehman, S.U.; Feng, T.; Wu, S.; Luo, X.; Lei, A.; Luobu, B.; Hassan, F.; Liu, Q. Comparative Genomics, Evolutionary and Gene Regulatory Regions Analysis of Casein Gene Family in Bubalus Bubalis. Front. Genet. 2021, 12, 662609. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, D. The TGF- Superfamily: New Members, New Receptors, and New Genetic Tests of Function in Different Organisms. Genes Dev. 1994, 8, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.L. Bone Morphogenetic Proteins: Multifunctional Regulators of Vertebrate Development. Genes Dev. 1996, 10, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Long, J.; Liu, Z.; Tao, W.; Wang, D. Identification and Evolution of TGF-β Signaling Pathway Members in Twenty-Four Animal Species and Expression in Tilapia. Int. J. Mol. Sci. 2018, 19, 1154. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Wotton, D. Transcriptional Control by the TGF-β/Smad Signaling System. EMBO J. 2000, 19, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Massagué, J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Massague, J. TGF-β Signal Transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef]
- Tanaka, C.; Sakuma, R.; Nakamura, T.; Hamada, H.; Saijoh, Y. Long-Range Action of Nodal Requires Interaction with GDF1. Genes Dev. 2007, 21, 3272–3282. [Google Scholar] [CrossRef]
- Pelliccia, J.L.; Jindal, G.A.; Burdine, R.D. Gdf3 Is Required for Robust Nodal Signaling during Germ Layer Formation and Left-Right Patterning. Elife 2017, 6, e28635. [Google Scholar] [CrossRef]
- Jamin, S.P.; Arango, N.A.; Mishina, Y.; Hanks, M.C.; Behringer, R.R. Requirement of Bmpr1a for Müllerian Duct Regression during Male Sexual Development. Nat. Genet. 2002, 32, 408–410. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of Skeletal Muscle Mass in Mice by a New TGF-p Superfamily Member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; McPherron, A.C. Regulation of Myostatin Activity and Muscle Growth. Proc. Natl. Acad. Sci. USA 2001, 98, 9306–9311. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.P.; Yeo, C.-Y.; Lee, Y.; Schrewe, H.; Whitman, M.; Li, E. Activin Type IIA and IIB Receptors Mediate Gdf11 Signaling in Axial Vertebral Patterning. Genes Dev. 2002, 16, 2749–2754. [Google Scholar] [CrossRef] [PubMed]
- Egerman, M.A.; Cadena, S.M.; Gilbert, J.A.; Meyer, A.; Nelson, H.N.; Swalley, S.E.; Mallozzi, C.; Jacobi, C.; Jennings, L.L.; Clay, I.; et al. GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell Metab. 2015, 22, 164–174. [Google Scholar] [CrossRef]
- Sartori, R.; Schirwis, E.; Blaauw, B.; Bortolanza, S.; Zhao, J.; Enzo, E.; Stantzou, A.; Mouisel, E.; Toniolo, L.; Ferry, A.; et al. BMP Signaling Controls Muscle Mass. Nat. Genet. 2013, 45, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Mallet, C.; Mazerbourg, S.; Feige, J.-J.; Bailly, S. Identification of BMP9 and BMP10 as Functional Activators of the Orphan Activin Receptor-like Kinase 1 (ALK1) in Endothelial Cells. Blood 2007, 109, 1953–1961. [Google Scholar] [CrossRef] [PubMed]
- Kleinberg, D.L.; Wood, T.L.; Furth, P.A.; Lee, A.V. Growth Hormone and Insulin-like Growth Factor-I in the Transition from Normal Mammary Development to Preneoplastic Mammary Lesions. Endocr. Rev. 2009, 30, 51–74. [Google Scholar] [CrossRef]
- Pardali, E.; Goumans, M.-J.; ten Dijke, P. Signaling by Members of the TGF-β Family in Vascular Morphogenesis and Disease. Trends Cell Biol. 2010, 20, 556–567. [Google Scholar] [CrossRef]
- Andriopoulos, B., Jr.; Corradini, E.; Xia, Y.; Faasse, S.A.; Chen, S.; Grgurevic, L.; Knutson, M.D.; Pietrangelo, A.; Vukicevic, S.; Lin, H.Y.; et al. BMP6 Is a Key Endogenous Regulator of Hepcidin Expression and Iron Metabolism. Nat. Genet. 2009, 41, 482–487. [Google Scholar] [CrossRef]
- Meynard, D.; Kautz, L.; Darnaud, V.; Canonne-Hergaux, F.; Coppin, H.; Roth, M.-P. Lack of the Bone Morphogenetic Protein BMP6 Induces Massive Iron Overload. Nat. Genet. 2009, 41, 478–481. [Google Scholar] [CrossRef]
- Mottershead, D.G.; Ritter, L.J.; Gilchrist, R.B. Signalling Pathways Mediating Specific Synergistic Interactions between GDF9 and BMP15. Mol. Hum. Reprod. 2012, 18, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Gamer, L.W.; Cox, K.; Carlo, J.M.; Rosen, V. Overexpression of BMP3 in the Developing Skeleton Alters Endochondral Bone Formation Resulting in Spontaneous Rib Fractures. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009, 238, 2374–2381. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-J.; Lin, T.-Y.; Sung, L.-Y.; Chang, W.-F.; Wu, P.-C.; Luo, C.-W. BMP8A Sustains Spermatogenesis by Activating Both SMAD1/5/8 and SMAD2/3 in Spermatogonia. Sci. Signal. 2017, 10, eaal1910. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.-Q.; Deng, K.; Labosky, P.A.; Liaw, L.; Hogan, B.L. The Gene Encoding Bone Morphogenetic Protein 8B Is Required for the Initiation and Maintenance of Spermatogenesis in the Mouse. Genes Dev. 1996, 10, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Settle, S.H., Jr.; Rountree, R.B.; Sinha, A.; Thacker, A.; Higgins, K.; Kingsley, D.M. Multiple Joint and Skeletal Patterning Defects Caused by Single and Double Mutations in the Mouse Gdf6 and Gdf5 Genes. Dev. Biol. 2003, 254, 116–130. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene Duplication as a Major Force in Evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Lipinski, K.J.; Farslow, J.C.; Fitzpatrick, K.A.; Lynch, M.; Katju, V.; Bergthorsson, U. High Spontaneous Rate of Gene Duplication in Caenorhabditis Elegans. Curr. Biol. 2011, 21, 306–310. [Google Scholar] [CrossRef]
- Lynch, V.J. Inventing an Arsenal: Adaptive Evolution and Neofunctionalization of Snake Venom Phospholipase A2 Genes. BMC Evol. Biol. 2007, 7, 2. [Google Scholar] [CrossRef][Green Version]
- Conant, G.C.; Wolfe, K.H. Turning a Hobby into a Job: How Duplicated Genes Find New Functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. TIG 2002, 18, 486. [Google Scholar] [CrossRef]
- Martin, A.P. Increasing Genomic Complexity by Gene Duplication and the Origin of Vertebrates. Am. Nat. 1999, 154, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.Y.; Beppu, H.; Kawai, N.; Li, E.; Bloch, K.D. Bone Morphogenetic Protein (BMP) Type II Receptor Deletion Reveals BMP Ligand-Specific Gain of Signaling in Pulmonary Artery Smooth Muscle Cells. J. Biol. Chem. 2005, 280, 24443–24450. [Google Scholar]
- Antsiferova, M.; Werner, S. The Bright and the Dark Sides of Activin in Wound Healing and Cancer. J. Cell Sci. 2012, 125, 3929–3937. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Q.; Wigglesworth, K.; Rangarajan, A.; Kattamuri, C.; Peterson, R.T.; Eppig, J.J.; Thompson, T.B.; Matzuk, M.M. Growth Differentiation Factor 9: Bone Morphogenetic Protein 15 Heterodimers Are Potent Regulators of Ovarian Functions. Proc. Natl. Acad. Sci. USA 2013, 110, E776–E785. [Google Scholar] [CrossRef]
- Knight, P.G.; Glister, C. TGF-β Superfamily Members and Ovarian Follicle Development. Reproduction 2006, 132, 191–206. [Google Scholar] [CrossRef]
- Shi, J.; Yoshino, O.; Osuga, Y.; Akiyama, I.; Harada, M.; Koga, K.; Fujimoto, A.; Yano, T.; Taketani, Y. Growth Differentiation Factor 3 Is Induced by Bone Morphogenetic Protein 6 (BMP-6) and BMP-7 and Increases Luteinizing Hormone Receptor Messenger RNA Expression in Human Granulosa Cells. Fertil. Steril. 2012, 97, 979–983. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Moore, S.S.; Schenkel, F.S.; Stothard, P. Genome-Wide Scan for Positional and Functional Candidate Genes Affecting Milk Production Traits in Canadian Holstein Cattle. In Proceedings of the 9th World Congress on Genetic Applied to Livestock Production (WCGALP), Leipzig, Germany, 1–6 August 2010. [Google Scholar]
- Ye, M.; Berry-Wynne, K.M.; Asai-Coakwell, M.; Sundaresan, P.; Footz, T.; French, C.R.; Abitbol, M.; Fleisch, V.C.; Corbett, N.; Allison, W.T.; et al. Mutation of the Bone Morphogenetic Protein GDF3 Causes Ocular and Skeletal Anomalies. Hum. Mol. Genet. 2010, 19, 287–298. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Biyanwila, J.; Kovach, P.; Bardakjian, T.; Traboulsi, E.I.; Ragge, N.K.; Schneider, A.; Malicki, J. Genetic Defects of GDF6 in the Zebrafish out of Sight Mutant and in Human Eye Developmental Anomalies. BMC Genet. 2010, 11, 102. [Google Scholar] [CrossRef]
- Hreinsson, J.G.; Scott, J.E.; Rasmussen, C.; Swahn, M.L.; Hsueh, A.J.W.; Hovatta, O. Growth Differentiation Factor-9 Promotes the Growth, Development, and Survival of Human Ovarian Follicles in Organ Culture. J. Clin. Endocrinol. Metab. 2002, 87, 316–321. [Google Scholar] [CrossRef]
- Su, Y.-Q.; Wu, X.; O’Brien, M.J.; Pendola, F.L.; Denegre, J.N.; Matzuk, M.M.; Eppig, J.J. Synergistic Roles of BMP15 and GDF9 in the Development and Function of the Oocyte–Cumulus Cell Complex in Mice: Genetic Evidence for an Oocyte–Granulosa Cell Regulatory Loop. Dev. Biol. 2004, 276, 64–73. [Google Scholar] [CrossRef]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, G.H.; Powell, R.; Galloway, S.M. Mutations in the Genes for Oocyte-Derived Growth Factors GDF9 and BMP15 Are Associated with Both Increased Ovulation Rate and Sterility in Cambridge and Belclare Sheep (Ovis Aries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.-X.; Sang, L.-H.; Wang, J.-Y.; Fang, L.; Ye, S.-C. Study on BMP15 and GDF9 as Candidate Genes for Prolificacy of Small Tail Han Sheep. Yi Chuan Xue Bao/Acta Genet. Sin. 2005, 32, 38–45. [Google Scholar] [PubMed]
- Escobar-Chaparro, R.A.; Guillén, G.; Espejo-Galicia, L.U.; Meza-Villalvazo, V.M.; Peña-Castro, J.M.; Abad-Zavaleta, J. qPCR and HRM-Based Diagnosis of SNPs on Growth Differentiation Factor 9 (GDF9), a Gene Associated with Sheep (Ovis Aries) Prolificacy. 3 Biotech 2017, 7, 204. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Y.; Wu, Z.; Shafique, L.; Chen, S.; Xuan, Z.; Feng, T.; Luo, C.; Zhu, P.; Liu, Q. Molecular Cloning and Expression Patterns Study of Growth Differentiation Factor 9 Gene (GDF9) in Water Buffalo (Bubalus Bubalis). Int. J. Agric. Biol. 2019, 22, 1010–1016. [Google Scholar]
- Gendelman, M.; Roth, Z. Seasonal Effect on Germinal Vesicle-Stage Bovine Oocytes Is Further Expressed by Alterations in Transcript Levels in the Developing Embryos Associated with Reduced Developmental Competence. Biol. Reprod. 2012, 86, 1–9. [Google Scholar] [CrossRef]
- Liu, G.E.; Ventura, M.; Cellamare, A.; Chen, L.; Cheng, Z.; Zhu, B.; Li, C.; Song, J.; Eichler, E.E. Analysis of Recent Segmental Duplications in the Bovine Genome. BMC Genom. 2009, 10, 571. [Google Scholar] [CrossRef]
| MEME Motif | Amino Acid Sequence | Length | Pfam Domain |
|---|---|---|---|
| 1 | WIIAPKGYEANYCEGECPFPLASH | 24 | - |
| 2 | VPKPCCVPTKLSPJSILYFDD | 21 | - |
| 3 | LKKYPBMVVEECGCR | 15 | - |
| 4 | LYVDFQDLGW | 10 | - |
| 5 | LNPTNHAIIQTLVHL | 15 | - |
| 6 | SGWLVFDVTAAVRRW | 15 | - |
| 7 | FBLSSIPDGEAVTAAELRJYK | 21 | - |
| 8 | SPQKBLGLQLYVETDDGRSIBPGLAGLVGRQGPRSKQPFMVAFFKASEV | 49 | TGFb_propeptide |
| 9 | AGDPPLASGQDERFLGDADMVMSFVNLVERDKEFGHQEPHHKEFR | 45 | TGFb_propeptide |
| 10 | LEAIKRZILSKLGLPSRPRPSRPPPKPPL | 29 | - |
| Gene | Chr. | Exon count | A.A. | MW (Da) | pI | II | Al | GRAVY |
|---|---|---|---|---|---|---|---|---|
| TGFB1 | 18 | 8 | 410 | 46,776.39 | 7.98 | 53.22 | 80.88 | −0.463 |
| TGFB2 | 5 | 8 | 442 | 50,550.99 | 8.74 | 53.80 | 80.07 | −0.406 |
| TGFB3 | 11 | 8 | 452 | 51,418.09 | 8.28 | 50.18 | 86.26 | −0.392 |
| INHA | 2 | 2 | 360 | 38,828.70 | 6.91 | 65.11 | 85.42 | −0.093 |
| INHBA | 8 | 2 | 425 | 47,521.45 | 8.10 | 63.72 | 78.47 | −0.497 |
| INHBB | 2 | 2 | 408 | 45,056.70 | 8.72 | 56.03 | 80.27 | −0.262 |
| INHBC | 4 | 2 | 352 | 38,480.15 | 6.59 | 46.01 | 88.89 | −0.045 |
| INHBE | 4 | 2 | 352 | 38,731.98 | 9.95 | 57.93 | 91.45 | −0.171 |
| NODAL | 4 | 3 | 346 | 39,932.99 | 8.07 | 60.21 | 84.83 | −0.360 |
| MSTN | 2 | 3 | 375 | 42,495.89 | 6.05 | 41.03 | 85.79 | −0.332 |
| BMP3 | 7 | 3 | 475 | 53,458.10 | 9.04 | 66.66 | 78.44 | −0.556 |
| GDF10 | 4 | 3 | 478 | 52,683.43 | 9.55 | 55.40 | 76.63 | −0.450 |
| GDF11 | 4 | 3 | 355 | 37,982.55 | 8.28 | 59.40 | 78.30 | −0.428 |
| AMH | 9 | 5 | 575 | 60,812.20 | 8.63 | 54.46 | 96.50 | −0.044 |
| LEFTY2 | 5 | 4 | 367 | 41,460.84 | 8.91 | 52.23 | 92.18 | −0.251 |
| Gene | Chr. | Exon count | A.A. | MW (Da) | pI | II | Al | GRAVY |
|---|---|---|---|---|---|---|---|---|
| BMP2 | 14 | 4 | 395 | 44,569.82 | 8.96 | 55.51 | 79.75 | −0.422 |
| BMP4 | 11 | 6 | 409 | 46,639.95 | 8.57 | 59.49 | 80.54 | −0.543 |
| BMP5 | 2 | 7 | 454 | 51,323.27 | 9.10 | 52.12 | 79.49 | −0.414 |
| BMP6 | 2 | 7 | 499 | 54,621.73 | 7.96 | 55.48 | 71.24 | −0.413 |
| BMP7 | 14 | 7 | 431 | 49,288.88 | 7.36 | 53.76 | 76.96 | −0.410 |
| BMP8A | 6 | 7 | 404 | 44,786.52 | 9.12 | 66.86 | 86.71 | −0.288 |
| BMP8B | 6 | 7 | 405 | 44,683.35 | 8.91 | 65.49 | 85.78 | −0.285 |
| BMP10 | 12 | 2 | 424 | 48,424.90 | 4.97 | 49.13 | 84.60 | −0.428 |
| BMP15 | X | 2 | 403 | 46,096.52 | 9.67 | 48.27 | 93.08 | −0.292 |
| GDF1 | 9 | 2 | 369 | 39,252.61 | 9.51 | 71.00 | 93.44 | 0.002 |
| GDF2 | 4 | 2 | 431 | 48,303.95 | 6.21 | 54.20 | 77.77 | −0.424 |
| GDF3 | 4 | 3 | 360 | 40,354.43 | 7.58 | 62.16 | 88.83 | −0.214 |
| GDF5 | 14 | 2 | 499 | 55,133.38 | 9.80 | 45.22 | 72.99 | −0.586 |
| GDF6 | 15 | 2 | 466 | 51,852.09 | 9.48 | 66.01 | 69.55 | −0.604 |
| GDF7 | 12 | 2 | 450 | 46,750.93 | 9.62 | 53.66 | 81.09 | −0.133 |
| GDF9 | 9 | 3 | 396 | 45,364.01 | 9.27 | 56.71 | 73.13 | −0.480 |
| GDF15 | 9 | 2 | 308 | 34,638.86 | 10.42 | 64.60 | 84.94 | −0.472 |
| Gene Pairs | Chromosome | Duplication | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|
| TGFB1/TGFB3 | 18/10 | SD | 0.6711 | 0.6071 | 1.1 |
| TGFB2/TGFB3 | 16/10 | SD | 0.5789 | 0.3323 | 1.7 |
| BMP4/BMP2 | 10/13 | SD | 0.4284 | 0.8102 | 0.5 |
| BMP3/GDF10 | 6/28 | SD | 0.7044 | 0.6483 | 1.08 |
| BMP7/BMP5 | 13/23 | SD | 0.4044 | 0.6352 | 0.64 |
| BMP6/BMP5 | 23/23 | TD | 0.5073 | 0.5751 | 0.88 |
| BMP8A/BMP7 | 3/13 | SD | 0.5001 | 0.4318 | 1.15 |
| GDF9/BMP15 | 7/X | SD | 0.7914 | 1.0686 | 0.74 |
| GDF10/BMP15 | 28/X | SD | 0.8583 | 1.0661 | 0.8 |
| Mutations | Polyphen2 | Mupro | Provean | I-Mutant | Phd-Snp | SNAP2 | Predict SNP | Meta SNP | SIFT | Overall Effect | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TGFB1 | |||||||||||
| 1 | D154 > E | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | V155 > L | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| INHA | |||||||||||
| 1 | G15 > R | BE | DE | NE | IN | NE | EFF | NE | NE | NT | SY |
| 2 | L21 > P | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | L23 > V | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 4 | H58 > P | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 5 | T136 > I | BE | IN | NE | IN | NE | NE | NE | NE | NT | SY |
| 6 | M157 > T | BE | DE | NE | IN | NE | NE | NE | NE | NT | SY |
| 7 | P293 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 8 | P300 > S | BE | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 9 | V309 > I | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| INHBB | |||||||||||
| 1 | S21 > W | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 2 | S255 > G | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| INHBC | |||||||||||
| 1 | H77 > Q | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | E103 > Q | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | T175 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | E203 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | R214 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | V221 > M | PD | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 7 | T310 > A | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| INHBE | |||||||||||
| 1 | L4 > P | UNK | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| 2 | T33 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | Q130 > H | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | P195 > L | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | T203 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | A210 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | R222 > Q | BE | DE | NE | DE | DIS | EFF | NE | NE | NT | SY |
| NODAL | |||||||||||
| 1 | Q4 > H | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | C5 > R | PD | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| 3 | T172 > M | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | S173 > P | PD | IN | NE | IN | DIS | NE | NE | DIS | TOL | SY |
| 5 | S174 > T | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | R182 > Q | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | S185 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| MSTN | |||||||||||
| 1 | E116 > D | PD | DE | NE | DE | NE | EFF | DEL | NE | TOL | NSY |
| 2 | T117 > A | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | K141 > Q | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | H275 > R | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| BMP2 | |||||||||||
| 1 | V16 > I | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| BMP3 | |||||||||||
| 1 | E82 > D | PD | IN | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 2 | P86 > Q | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | P92 > L | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | K233 > T | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | Q278 > H | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | S281 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | R287 > W | UNK | DE | DEL | DE | NE | EFF | DIS | NE | NT | NSY |
| 8 | E316 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| BPM4 | |||||||||||
| 1 | D173 > E | BE | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| BMP5 | |||||||||||
| 1 | T25 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | M338 > V | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| BMP6 | |||||||||||
| 1 | G43 > S | BE | DE | NE | DE | NE | NE | NE | DIS | TOL | SY |
| 2 | D151 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | S160 > P | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 4 | Y419 > C | PD | DE | DEL | DE | DIS | EFF | DEL | DIS | NT | NSY |
| BMP8A | |||||||||||
| 1 | I23 > V | UNK | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | G57 > R | PD | IN | DEL | DE | NE | NE | NE | NE | TOL | SY |
| 3 | D87 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | D116 > N | BE | IN | DEL | DE | NE | NE | NE | NE | NT | SY |
| 5 | A145 > V | PD | IN | DEL | IN | DIS | EFF | DEL | DIS | NT | NSY |
| 6 | G258 > R | UNK | IN | NE | DE | DIS | NE | NE | NE | TOL | SY |
| 7 | P284 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 8 | N294 > D | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 9 | R305 > G | PD | DE | DEL | DE | DIS | EFF | DEL | DIS | NT | NSY |
| 10 | V375 > L | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| BMP8B | |||||||||||
| 1 | D87 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | D116 > N | BE | IN | DEL | DE | DIS | NE | NE | NE | NT | SY |
| 3 | G258 > A | UNK | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | R305 > G | PD | DE | DEL | DE | DIS | EFF | DEL | DIS | NT | NSY |
| BMP10 | |||||||||||
| 1 | E229 > K | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| BMP15 | |||||||||||
| 1 | V16 > A | UNK | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 2 | Q56 > L | PD | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | I62 > V | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | Q75 > H | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | I114 > V | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | S172 > T | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | L177 > S | PD | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 8 | G272 > R | PD | IN | DEL | DE | NE | EFF | NE | NE | TOL | NSY |
| 9 | L292 > Q | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 10 | E384 > Q | PD | IN | NE | DE | NE | EFF | NE | DIS | NT | NSY |
| GDF1 | |||||||||||
| 1 | S9 > G | UNK | DE | NE | DE | NE | NE | DEL | NE | NT | NSY |
| 2 | P254 > L | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| GDF2 | |||||||||||
| 1 | R3 > C | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | C14 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | G39 > R | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | I215 > V | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | G277 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | S308 > N | BE | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| 7 | T310 > M | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 8 | T323 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 9 | G324 > A | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| GDF3 | |||||||||||
| 1 | E60 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | A101 > T | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | I156 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | L211 > S | BE | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| GDF5 | |||||||||||
| 1 | P87 > S | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 2 | A214 > T | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| GDF6 | |||||||||||
| 1 | E253 > K | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | P257 > L | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 3 | G319 > R | PD | DE | NE | IN | NE | EFF | NE | NE | NT | SY |
| GDF7 | |||||||||||
| 1 | T98 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | V108 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | Q137 > E | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 4 | S190 > R | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 5 | S235 > R | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | R304 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| GDF9 | |||||||||||
| 1 | K6 > N | PD | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| 2 | L49 > F | PD | DE | NE | DE | NE | NE | DEL | DIS | NT | NSY |
| 3 | N67 > K | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | P77 > S | PD | DE | DEL | DE | NE | NE | DEL | DIS e | TOL | NSY |
| 5 | R84 > K | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | E184 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | L260 > V | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 8 | D291 > G | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 9 | M402 > Q | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| GDF10 | |||||||||||
| 1 | P42 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | T130 > M | BE | IN | NE | IN | NE | EFF | NE | DIS | NT | SY |
| 3 | P142 > H | PD | DE | NE | DE | NE | NE | NE | DIS | TOL | SY |
| 4 | P163 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 5 | T180 > N | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | S221 > A | PD | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | Q311 > H | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| GDF11 | |||||||||||
| 1 | A40 > G | PD | DE | NE | DE | DIS | NE | DEL | NE | TOL | NSY |
| 2 | G41 > A | PD | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| GDF15 | |||||||||||
| 1 | P80 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | S135 > R | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 3 | A175 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| AMH | |||||||||||
| 1 | F29 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 2 | L34 > S | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 3 | A50 > D | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 4 | S56 > P | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 5 | V89 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 6 | A115 > S | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 7 | N121 > D | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 8 | G122 > R | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 9 | P127 > A | BE | DE | NE | DE | NE | NE | NE | NE | NT | SY |
| 10 | V180 > L | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 11 | H216 > R | BE | IN | NE | DE | NE | NE | NE | NE | TOL | SY |
| 12 | S271 > P | BE | IN | NE | IN | NE | NE | NE | NE | TOL | SY |
| 13 | A273 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 14 | A317 > R | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 15 | A334 > T | PD | DE | NE | DE | NE | NE | DEL | NE | NT | NSY |
| 16 | S432 > G | UNK | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 17 | A468 > T | UNK | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 18 | T534 > A | UNK | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| LEFTY2 | |||||||||||
| 1 | Q2 > R | BE | DE | NE | IN | NE | NE | NE | NE | TOL | SY |
| 2 | V12 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 3 | T23 > M | PD | IN | NE | IN | NE | NE | DEL | NE | NT | SY |
| 4 | R26 > W | BE | DE | NE | DE | NE | EFF | NE | NE | NT | SY |
| 5 | D44 > N | BE | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 6 | A59 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 7 | G70 > A | BE | IN | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 8 | T91 > E | BE | IN | NE | DE | NE | NE | NE | NE | NT | SY |
| 9 | H97 > Y | PD | IN | NE | IN | NE | EFF | NE | NE | TOL | SY |
| 10 | T169 > S | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 11 | W230 > R | BE | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 12 | E260 > K | BE | IN | NE | DE | NE | EFF | NE | NE | NT | SY |
| 13 | A322 > T | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 14 | Q336 > R | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
| 15 | W356 > S | BE | DE | NE | DE | NE | EFF | NE | NE | TOL | SY |
| 16 | V359 > A | BE | DE | NE | DE | NE | NE | NE | NE | TOL | SY |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, M.S.-u.; Hassan, F.-u.; Rehman, Z.-u.; Ishtiaq, I.; Rehman, S.u.; Liu, Q. Molecular Characterization of TGF-Beta Gene Family in Buffalo to Identify Gene Duplication and Functional Mutations. Genes 2022, 13, 1302. https://doi.org/10.3390/genes13081302
Rehman MS-u, Hassan F-u, Rehman Z-u, Ishtiaq I, Rehman Su, Liu Q. Molecular Characterization of TGF-Beta Gene Family in Buffalo to Identify Gene Duplication and Functional Mutations. Genes. 2022; 13(8):1302. https://doi.org/10.3390/genes13081302
Chicago/Turabian StyleRehman, Muhammad Saif-ur, Faiz-ul Hassan, Zia-ur Rehman, Iqra Ishtiaq, Saif ur Rehman, and Qingyou Liu. 2022. "Molecular Characterization of TGF-Beta Gene Family in Buffalo to Identify Gene Duplication and Functional Mutations" Genes 13, no. 8: 1302. https://doi.org/10.3390/genes13081302
APA StyleRehman, M. S.-u., Hassan, F.-u., Rehman, Z.-u., Ishtiaq, I., Rehman, S. u., & Liu, Q. (2022). Molecular Characterization of TGF-Beta Gene Family in Buffalo to Identify Gene Duplication and Functional Mutations. Genes, 13(8), 1302. https://doi.org/10.3390/genes13081302









