Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü)
Abstract
:1. Introduction
2. Results
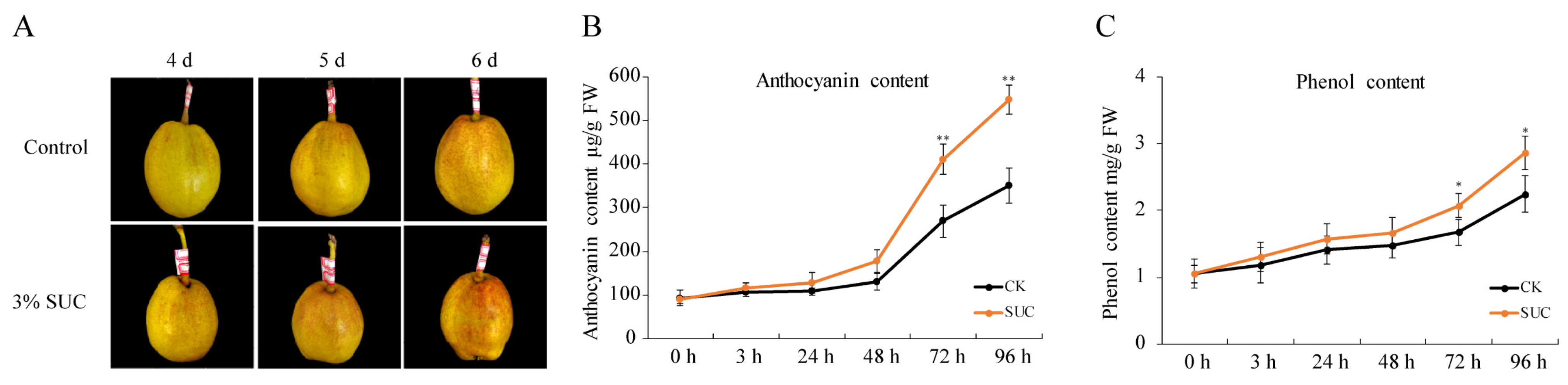
2.1. Sucrose Application Enhanced Anthocyanin Accumulation in ‘Kuerle Xiangli’
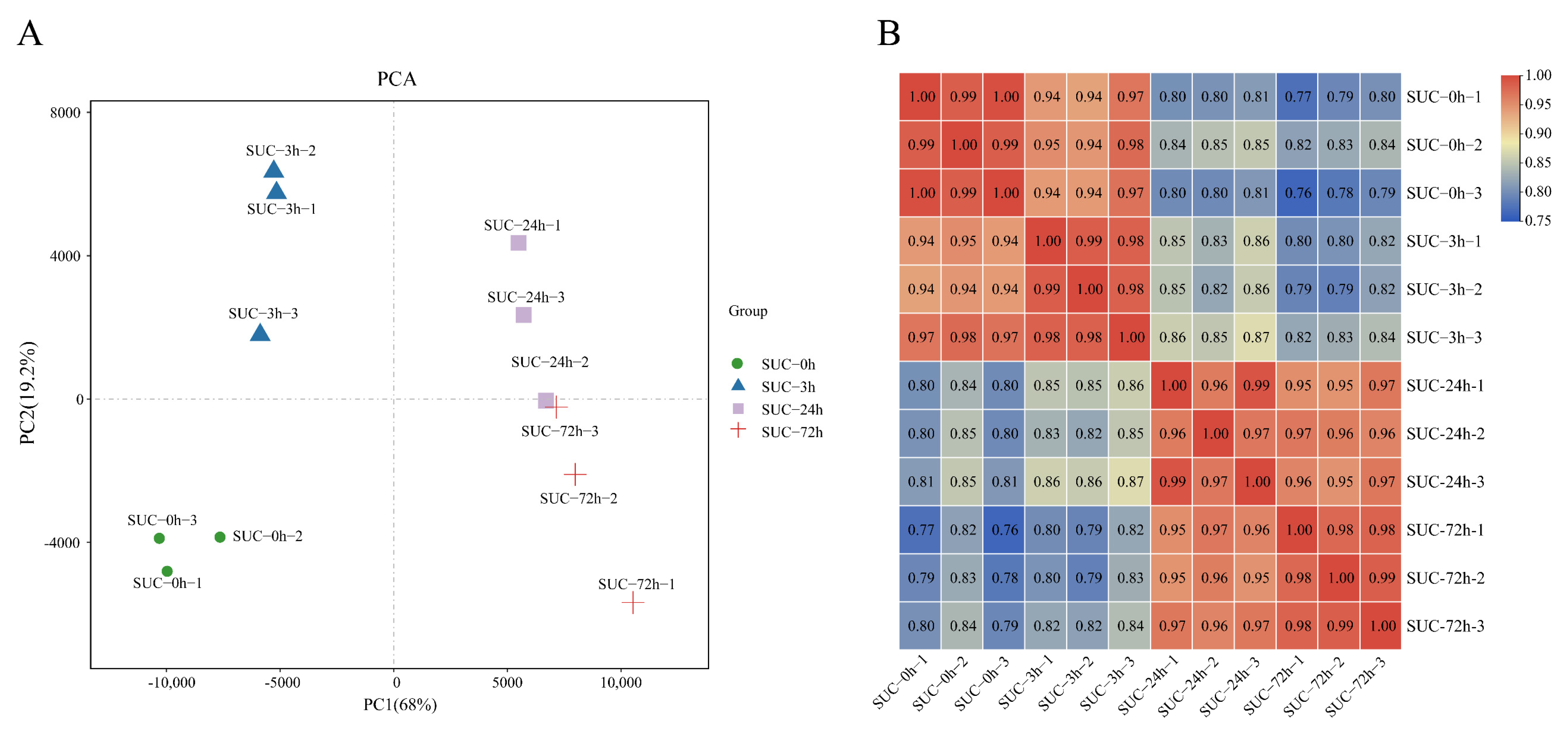
2.2. Analysis of Pear Transcriptome following Sucrose Treatment
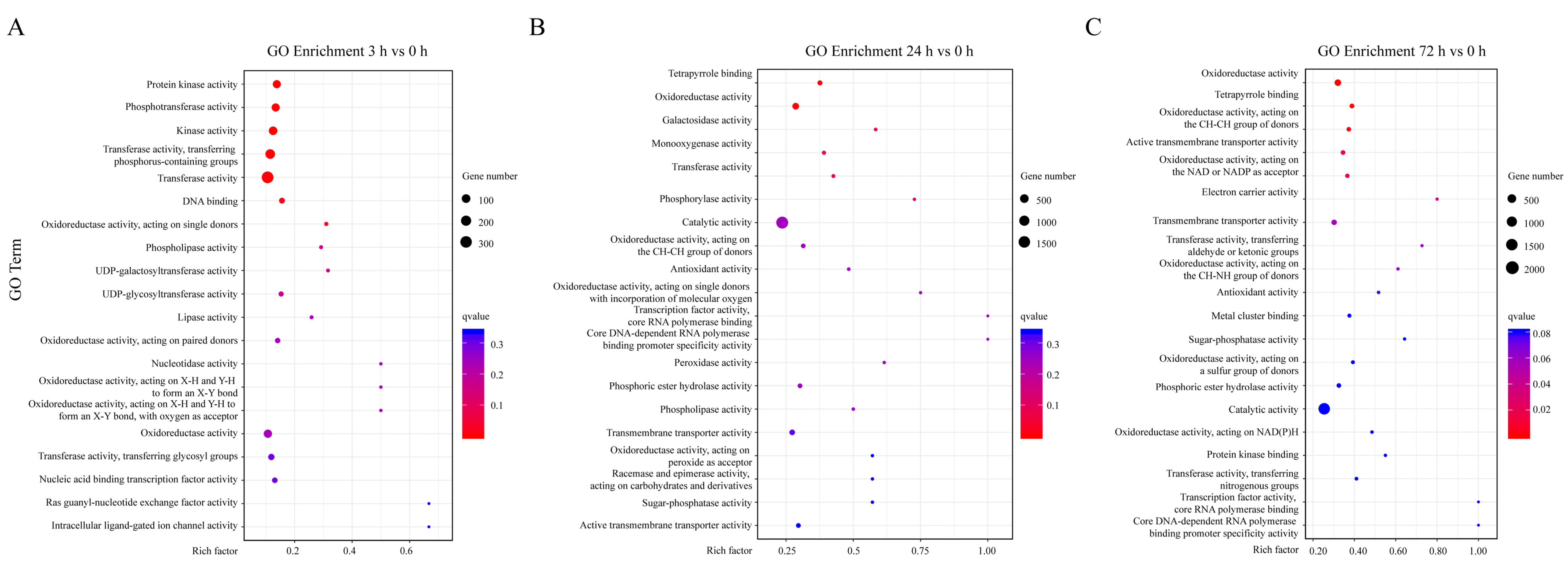
2.3. Gene Ontology (GO) Enrichment Analysis of DEGs under Sucrose Treatment
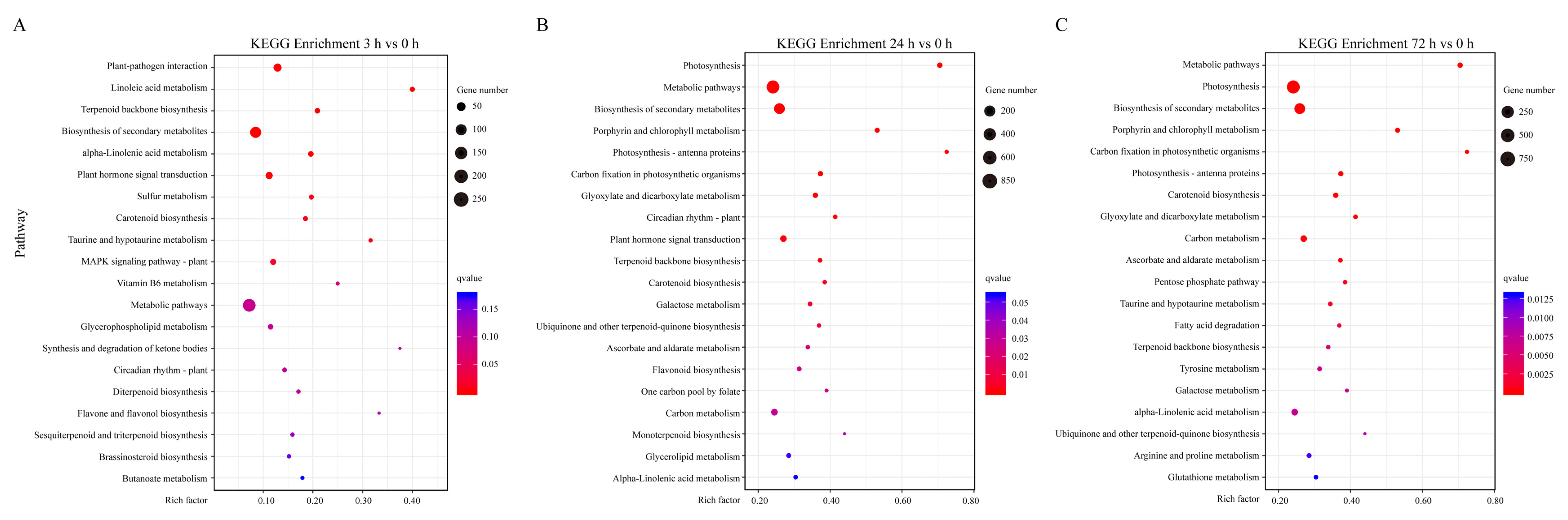
2.4. KEGG Enrichment Analysis of DEGs under Sucrose Treatment
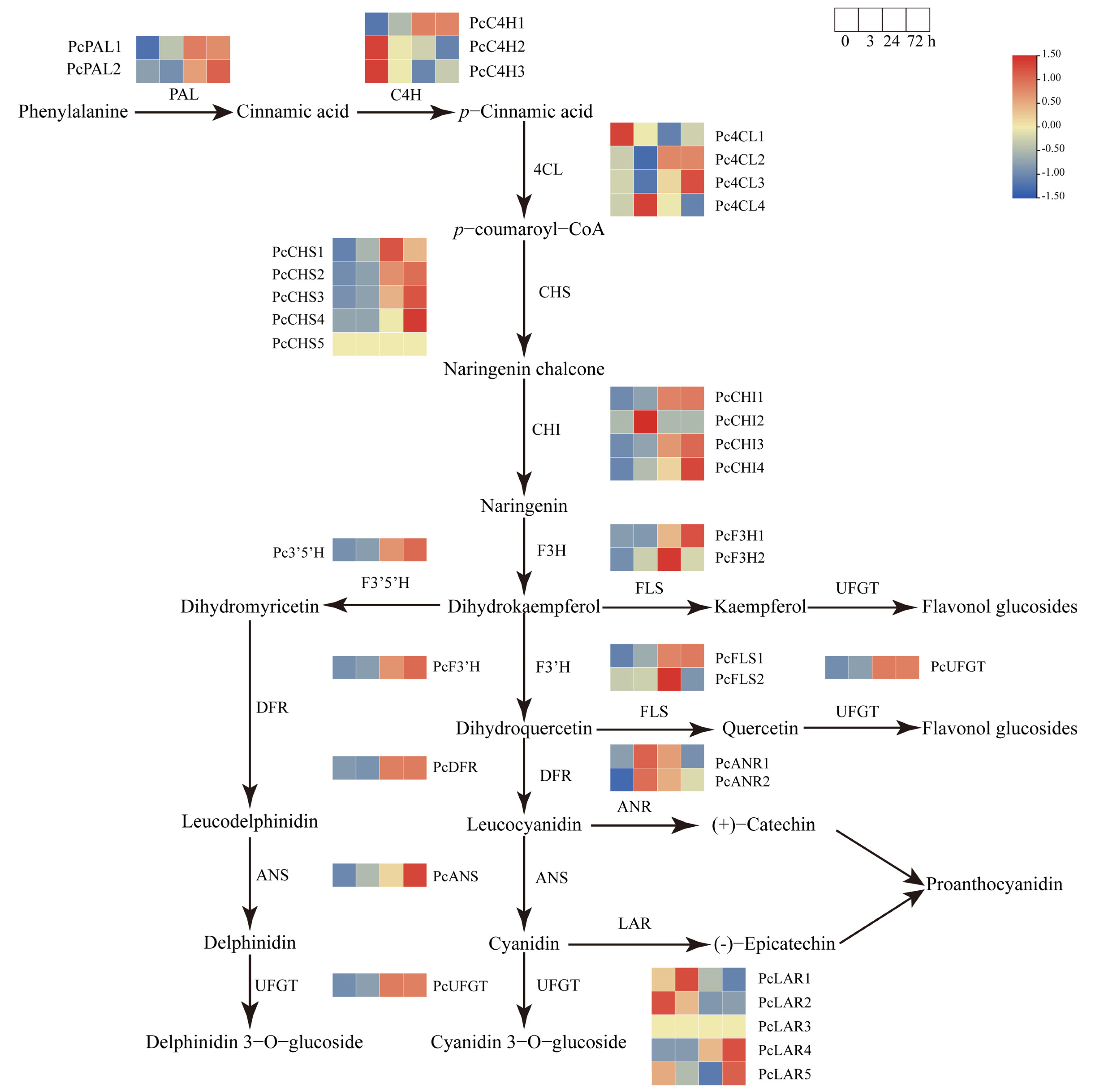
2.5. Analysis of Anthocyanin Biosynthetic Structural Genes under Sucrose Treatment
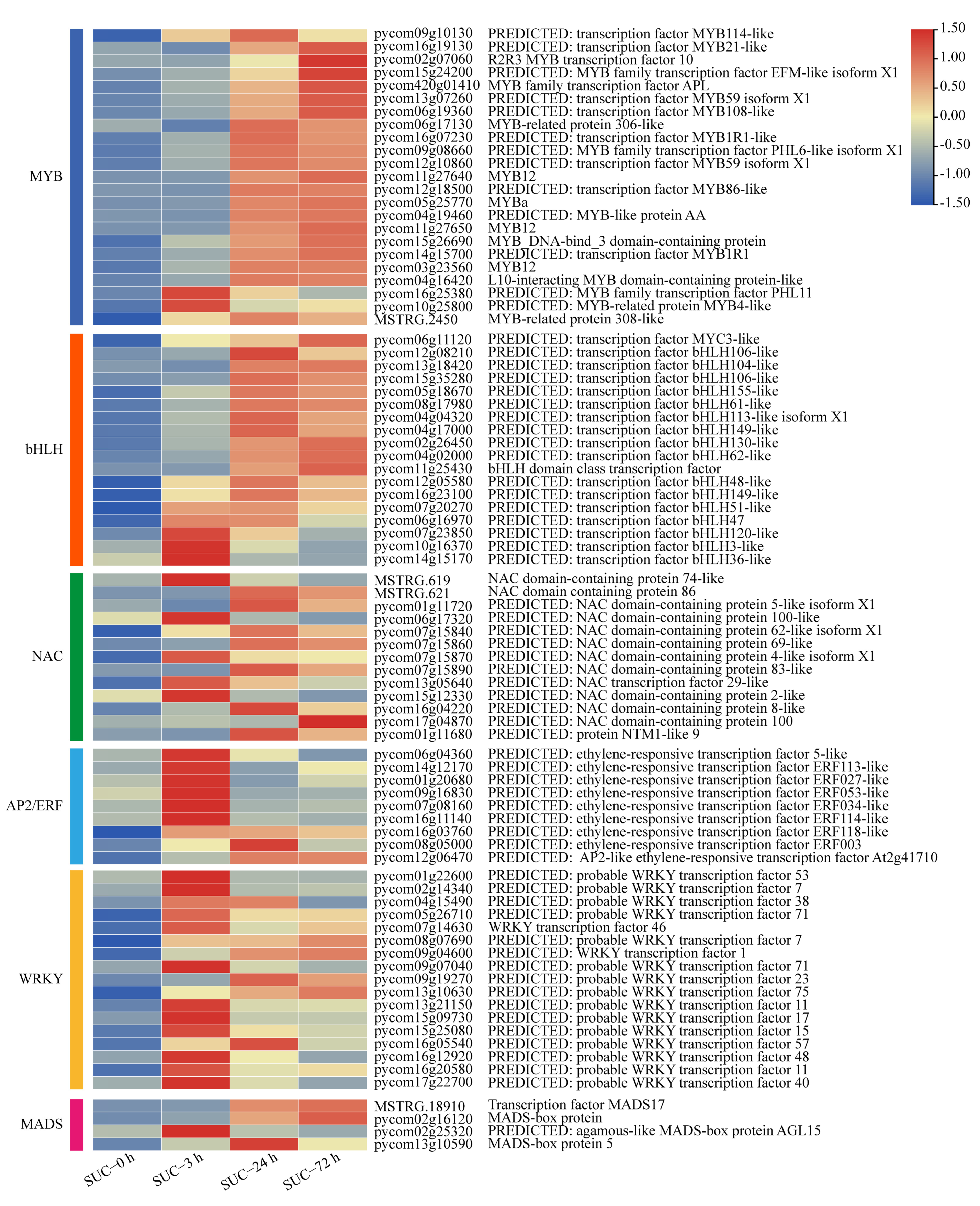
2.6. Analysis of Anthocyanin Biosynthetic Regulatory Genes under Sucrose Treatment
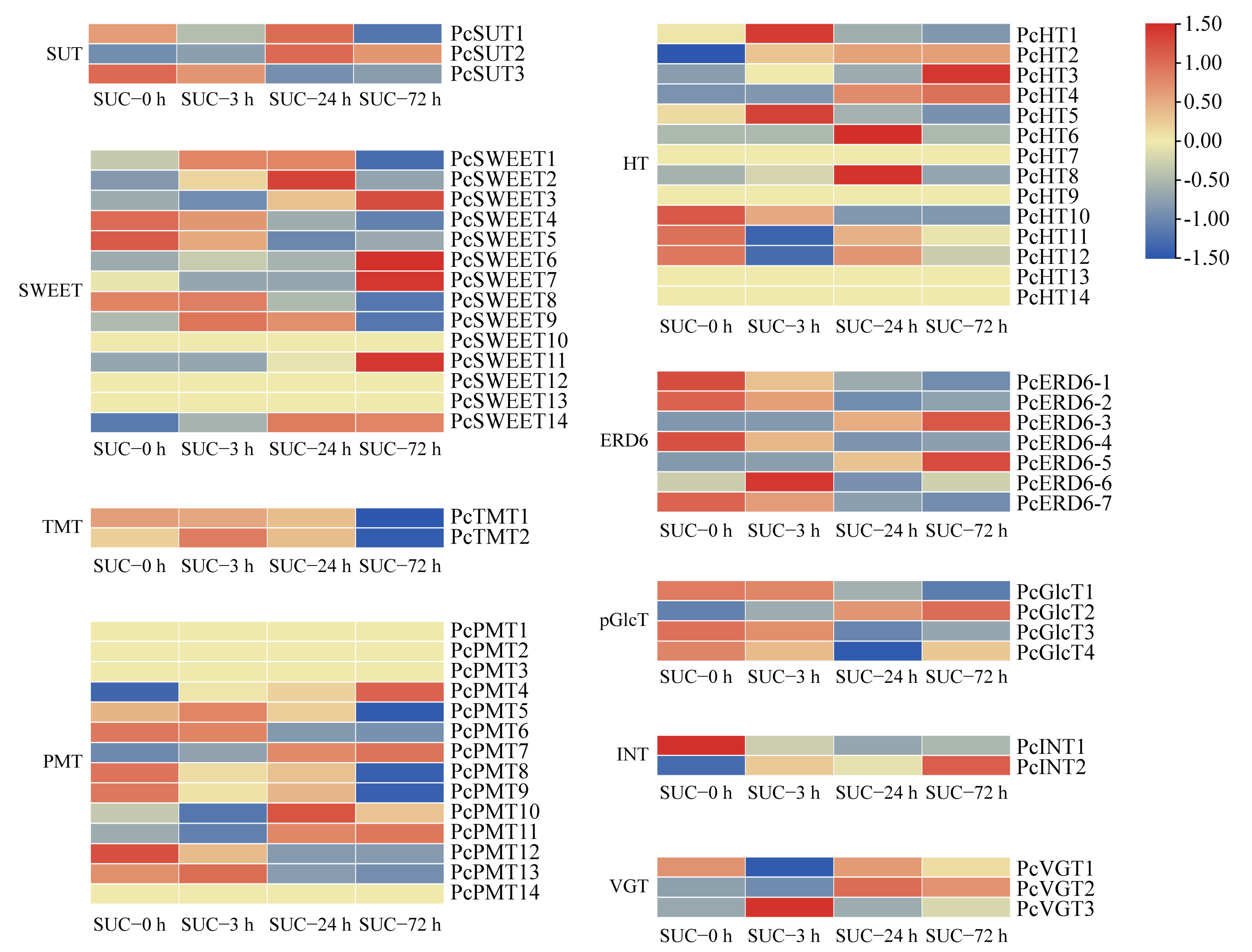
2.7. Analysis of Sugar Transporter Genes under Sucrose Treatment
2.8. WGCNA of Critical Genes Involved in Sucrose-Mediated Coloration
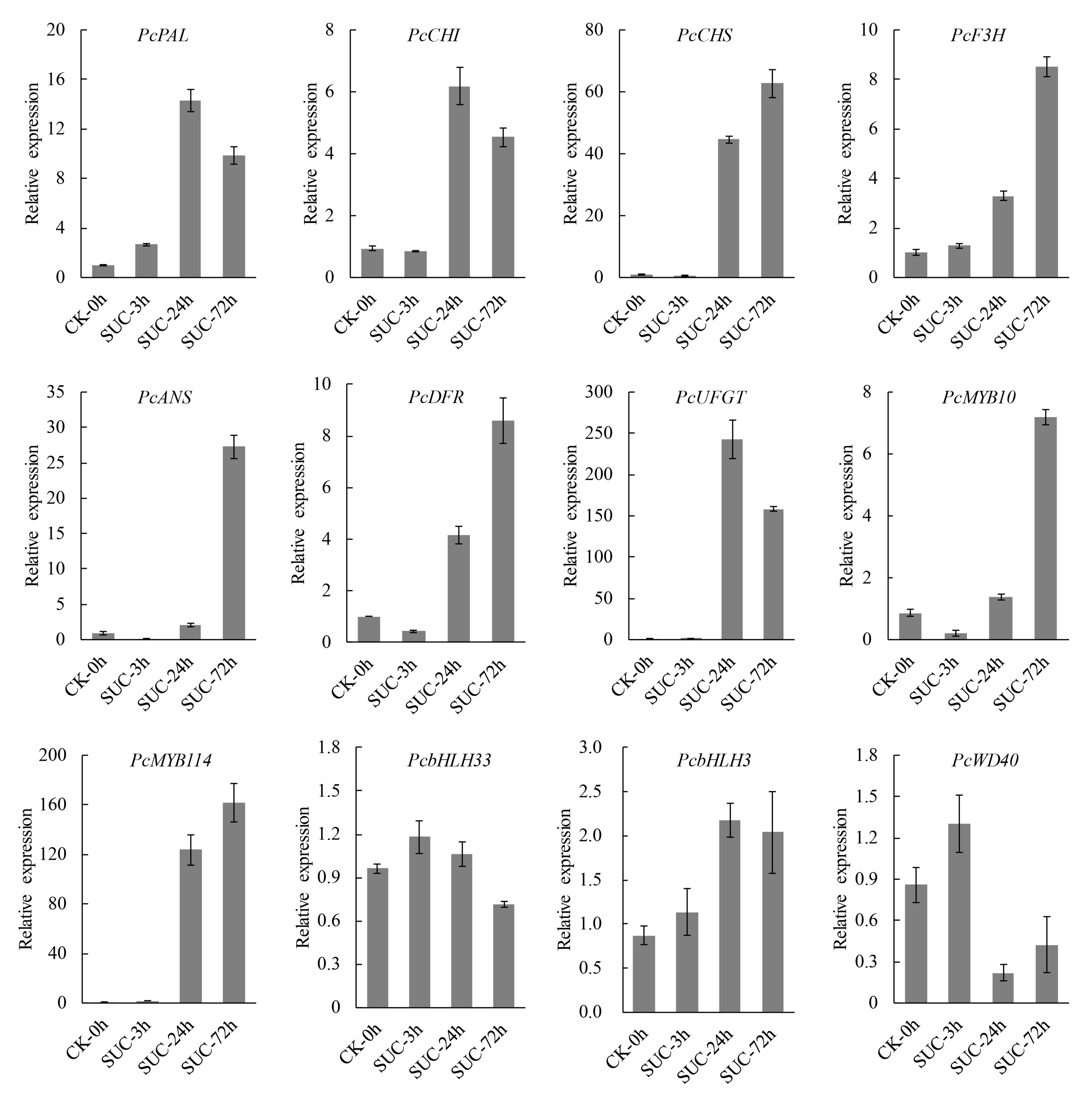
2.9. RT-qPCR Analysis of Transcript Level of Several Anthocyanin Biosynthetic Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Treatments
4.2. Measurement of Total Phenol and Anthocyanin Content
4.3. Library Preparation and RNA Sequencing
4.4. RNA Isolation and RT-qPCR Analysis
4.5. DEGs, GO, and KEGG analysis
4.6. WGCNA and Gene Network Construction
4.7. Principal Component Analysis
4.8. Statistical Analysis
4.9. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xue, H.; Zhang, P.; Shi, T.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Zhang, H.; Qiao, Y.; Li, X. Genome-wide characterization of simple sequence repeats in Pyrus bretschneideri and their application in an analysis of genetic diversity in pear. BMC Genom. 2018, 19, 473. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Shi, T.; Wang, F.; Zhou, H.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Zhang, Z.; Qiao, Y. Interval mapping for red/green skin color in Asian pears using a modified QTL-seq method. Hortic. Res. 2017, 4, 17053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Wang, C.; Qi, T.; An, Z.; Wu, M.; Qu, L.; Li, J.; Wen, Y.; Shi, Q.; Zhai, R. Effect of natural light on the phenolic compounds contents and coloration in the peel of ‘Xiyanghong’(Pyrus bretschneideri × Pyrus communis). Sci. Hortic. 2020, 266, 109052. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Gould, K.S. Role of anthocyanins in plant defence. In Anthocyanins; Springer: Berlin, Germany, 2008; pp. 22–28. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.E.; Frederiksen, H.; Struntze Krogholm, K.; Poulsen, L. Dietary proanthocyanidins: Occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol. Nutr. Food Res. 2005, 49, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Lila, M.A.; Burton-Freeman, B.; Grace, M.; Kalt, W. Unraveling anthocyanin bioavailability for human health. Annu. Rev. Food Sci. Technol. 2016, 7, 375–393. [Google Scholar] [CrossRef]
- Pojer, E.; Mattivi, F.; Johnson, D.; Stockley, C.S. The case for anthocyanin consumption to promote human health: A review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 483–508. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Zhang, X.; Duan, R.; Han, C.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Wang, L.; Dong, Y. Genomic analysis of the glutathione S-transferase family in pear (Pyrus communis) and functional identification of PcGST57 in anthocyanin accumulation. Int. J. Mol. Sci. 2022, 23, 746. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, C.D.; Casati, P.; Walbot, V. A multidrug resistance–associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in Pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, J.; Hu, K.-D.; Wei, S.-W.; Sun, H.-Y.; Hu, L.-Y.; Han, Z.; Yao, G.-F.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef] [Green Version]
- Jaakola, L.; Poole, M.; Jones, M.O.; Kämäräinen-Karppinen, T.; Koskimäki, J.J.; Hohtola, A.; Häggman, H.; Fraser, P.D.; Manning, K.; King, G.J. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010, 153, 1619–1629. [Google Scholar] [CrossRef] [Green Version]
- Qian, M.; Ni, J.; Niu, Q.; Bai, S.; Bao, L.; Li, J.; Sun, Y.; Zhang, D.; Teng, Y. Response of miR156-SPL module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef] [Green Version]
- Ou, C.; Zhang, X.; Wang, F.; Zhang, L.; Zhang, Y.; Fang, M.; Wang, J.; Wang, J.; Jiang, S.; Zhang, Z. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of “Zaosu Red” pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Van den Ende, W.; Rolland, F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol. Plant. 2014, 7, 570–572. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Cho, L.-H.; Tun, W.; Jeon, J.-S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef]
- Hara, M.; Oki, K.; Hoshino, K.; Kuboi, T. Enhancement of anthocyanin biosynthesis by sugar in radish (Raphanus sativus) hypocotyl. Plant Sci. 2003, 164, 259–265. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, R.; Wang, Z.; Yang, C.; Lin-Wang, K.; Espley, R.; Liu, J.; Li, X.; Wu, Z.; Li, P.; Guan, Q. PbGA2ox8 induces vascular-related anthocyanin accumulation and contributes to red stripe formation on pear fruit. Hortic. Res. 2019, 6, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Wang, Y.; Sun, M.; Li, B.; Han, Y.; Zhao, Y.; Li, X.; Ding, N.; Li, C.; Ji, W. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013, 198, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-M.; Lo, S.-F.; Ho, T.-H.D. Source–sink communication: Regulated by hormone, nutrient, and stress cross-signaling. Trends Plant Sci. 2015, 20, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.M.; Wang, L.; Ruan, Y.-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signaling to enhance crop yield and food security. J. Exp. Bot. 2014, 65, 1713–1735. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Jiu, S.; Zhang, C.; Wang, C.; Tariq, P.; Liu, Z.; Wang, B.; Cui, L.; Fang, J. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol. J. 2016, 14, 2045–2065. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [Green Version]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Horacio, P.; Martinez-Noel, G. Sucrose signaling in plants: A world yet to be explored. Plant Signal Behav. 2013, 8, e23316. [Google Scholar] [CrossRef] [Green Version]
- Okumura, M.; Inoue, S.-I.; Kuwata, K.; Kinoshita, T. Photosynthesis activates plasma membrane H+-ATPase via sugar accumulation. Plant Physiol. 2016, 171, 580–589. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Ma, H.; Li, Y.; Zhang, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J. 2021, 106, 1728–1745. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-J.; Yeh, K.-W.; Tsai, C.-Y. Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 2001, 161, 635–644. [Google Scholar] [CrossRef]
- Huang, H.; Xie, S.; Xiao, Q.; Wei, B.; Zheng, L.; Wang, Y.; Cao, Y.; Zhang, X.; Long, T.; Li, Y. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci Rep. 2016, 6, 27590. [Google Scholar] [CrossRef] [PubMed]
- Ciereszko, I.; Johansson, H.; Kleczkowski, L.A. Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem J. 2001, 354, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; Povero, G.; Novi, G.; Solfanelli, C.; Alpi, A.; Perata, P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008, 179, 1004–1016. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.Y.; Ye, Y.Q.; Fan, S.K.; Jin, C.W.; Zheng, S.J. Increased sucrose accumulation regulates iron-deficiency responses by promoting auxin signaling in Arabidopsis plants. Plant Physiol. 2016, 170, 907–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, S.; Vallarino, J.G.; Szecowka, M.; Ufaz, S.; Tzin, V.; Angelovici, R.; Galili, G.; Fernie, A.R. Alteration of the interconversion of pyruvate and malate in the plastid or cytosol of ripening tomato fruit invokes diverse consequences on sugar but similar effects on cellular organic acid, metabolism, and transitory starch accumulation. Plant Physiol. 2013, 161, 628–643. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, K.; Khadilkar, A.S.; Sulpice, R.; Pant, B.; Scheible, W.-R.; Fisahn, J.; Stitt, M.; Ayre, B.G. Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiol. 2014, 165, 715–731. [Google Scholar] [CrossRef] [Green Version]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant. 2011, 4, 377–394. [Google Scholar] [CrossRef] [Green Version]
- Daloso, D.M.; Dos Anjos, L.; Fernie, A.R. Roles of sucrose in guard cell regulation. New Phytol. 2016, 211, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.A.; Espley, R.V.; Allan, A.C. Genomic analysis uncovers functional variation in the C-terminus of anthocyanin-activating MYB transcription factors. Hortic. Res. 2021, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Qu, Y.; Sha, G.; Zhang, S.; Ma, Y.; Chen, M.; Zhai, R.; Yang, C.; Xu, L.; Wang, Z. PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol Plant. 2021, 173, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Zuo, W.; Wang, Y.; Liu, W.; Zhang, Z.; Wang, N.; Chen, X. The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct. Plant Biol. 2022. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef]
- Ning, Z.; Hu, K.; Zhou, Z.; Zhao, D.; Tang, J.; Wang, H.; Li, L.; Ding, C.; Chen, X.; Yao, G.; et al. IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato (Ipomoea batatas L.). Plant Cell Rep. 2021, 40, 157–169. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Feng, X.; An, Y.; Zheng, J.; Sun, M.; Wang, L. Proteomics and SSH analyses of ALA-promoted fruit coloration and evidence for the involvement of a MADS-box gene, MdMADS1. Front. Plant Sci. 2016, 7, 1615. [Google Scholar] [CrossRef] [Green Version]
- Mazza, G.; Fukumoto, L.; Delaquis, P.; Girard, B.; Ewert, B. Anthocyanins, phenolics, and color of cabernet franc, merlot, and pinot noir wines from British Columbia. J. Agric. Food Chem. 1999, 47, 4009–4017. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Sample | Raw Data | Clean Data (%) | Q20 (%) | Q30 (%) | GC (%) | Total Mapped (%) |
|---|---|---|---|---|---|---|
| SUC-0 h-1 | 82,667,734 | 82,450,850 (99.74%) | 98.14% | 94.09% | 45.72% | 71,106,922 (86.29%) |
| SUC-0 h-2 | 47,461,574 | 47,337,338 (99.74%) | 98.00% | 93.70% | 45.57% | 40,788,425 (86.21%) |
| SUC-0 h-3 | 59,384,286 | 59,245,816 (99.77%) | 98.19% | 94.17% | 45.73% | 51,538,794 (87.05%) |
| SUC-3 h-1 | 50,820,492 | 50,699,094 (99.76%) | 98.22% | 94.29% | 45.76% | 44,155,124 (87.15%) |
| SUC-3 h-2 | 59,876,342 | 59,730,970 (99.76%) | 98.21% | 94.21% | 45.77% | 51,955,390 (87.05%) |
| SUC-3 h-3 | 49,816,612 | 49,697,046 (99.76%) | 98.27% | 94.41% | 45.82% | 42,609,191 (85.85%) |
| SUC-24 h-1 | 53,803,782 | 53,669,030 (99.75%) | 98.10% | 94.07% | 46.13% | 46,304,024 (86.34%) |
| SUC-24 h-2 | 66,094,172 | 65,934,960 (99.76%) | 98.14% | 94.12% | 46.15% | 56,921,111 (86.44%) |
| SUC-24 h-3 | 51,003,520 | 50,893,952 (99.79%) | 98.16% | 94.15% | 46.18% | 44,200,763 (86.97%) |
| SUC-72 h-1 | 50,473,534 | 50,349,396 (99.75%) | 98.14% | 94.17% | 46.52% | 43,343,719 (86.31%) |
| SUC-72 h-2 | 57,464,526 | 57,323,698 (99.75%) | 98.18% | 94.27% | 46.20% | 49,017,640 (85.89%) |
| SUC-72 h-3 | 61,896,448 | 61,749,868 (99.76%) | 98.11% | 94.11% | 46.21% | 53,437,535 (86.70%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Li, B.; Duan, R.; Han, C.; Wang, L.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Xue, H. Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü). Genes 2022, 13, 1064. https://doi.org/10.3390/genes13061064
Zhang X, Li B, Duan R, Han C, Wang L, Yang J, Wang L, Wang S, Su Y, Xue H. Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü). Genes. 2022; 13(6):1064. https://doi.org/10.3390/genes13061064
Chicago/Turabian StyleZhang, Xiangzhan, Bo Li, Ruiwei Duan, Chunhong Han, Lei Wang, Jian Yang, Long Wang, Suke Wang, Yanli Su, and Huabai Xue. 2022. "Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü)" Genes 13, no. 6: 1064. https://doi.org/10.3390/genes13061064
APA StyleZhang, X., Li, B., Duan, R., Han, C., Wang, L., Yang, J., Wang, L., Wang, S., Su, Y., & Xue, H. (2022). Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü). Genes, 13(6), 1064. https://doi.org/10.3390/genes13061064






