Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.)
Abstract
1. Introduction
2. Material and Methods
2.1. Databases and Whole-Genome Identification of Filamin Gene Family Members in Plants
2.2. Phylogenetic Tree Construction and Sequence Analysis of Filamin Gene Family Members
2.3. Chromosomal Distribution, Gene Duplication and Synteny Analysis of the Filamin Gene Family Members for Four Cotton Species
2.4. Calculation of Ka/Ks of Filamin Gene Family Members in Three Cotton Species
2.5. Gene Structure, Conserved Motif and Gene Ontology Analysis of Filamin Gene Family
2.6. 3D Structure of Filamin Protein in G. hirsutum
2.7. Promoter Region Cis-Element Analysis and Expression Profile Analysis of Different Filamin Gene Family Members
2.8. RNA-Seq and Quantitative RT-PCR (qRT-PCR) for Filamin Gene Family Members in G. hirsutum
2.9. Protein Interaction Network Analysis of Filamin Proteins
3. Results and Discussion
3.1. Identification, Physicochemical Property Analysis and Subcellular Localization Prediction of Filamin Proteins
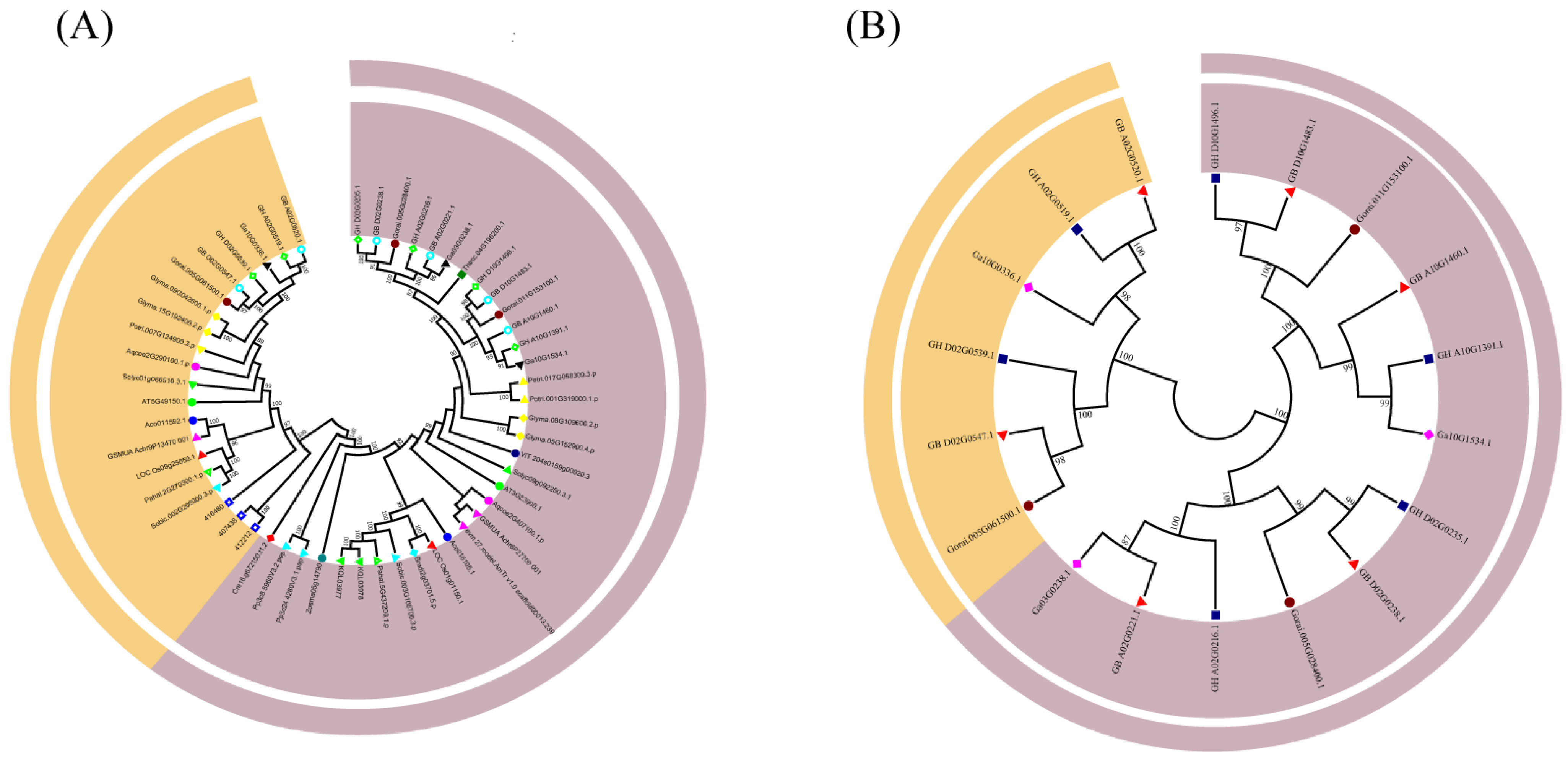
3.2. Sequence and Evolutionary Analysis of Filamin Gene Family Members in 23 Plant Species
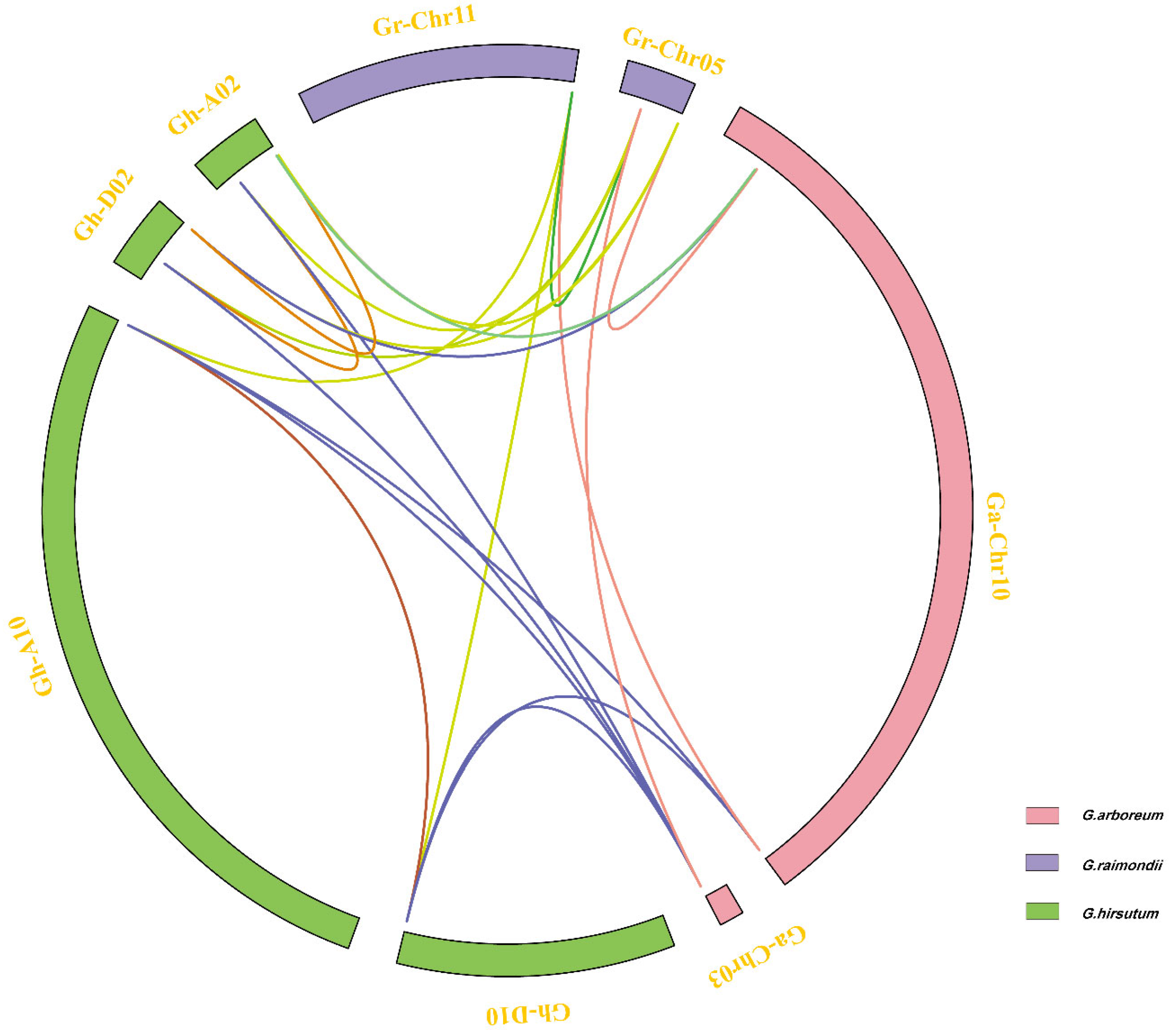
3.3. Chromosome Distribution of Filamin Genes for Four Cotton Species
3.4. Collinearity Analysis and Ka/Ks Analysis of Filamin Gene Family Members
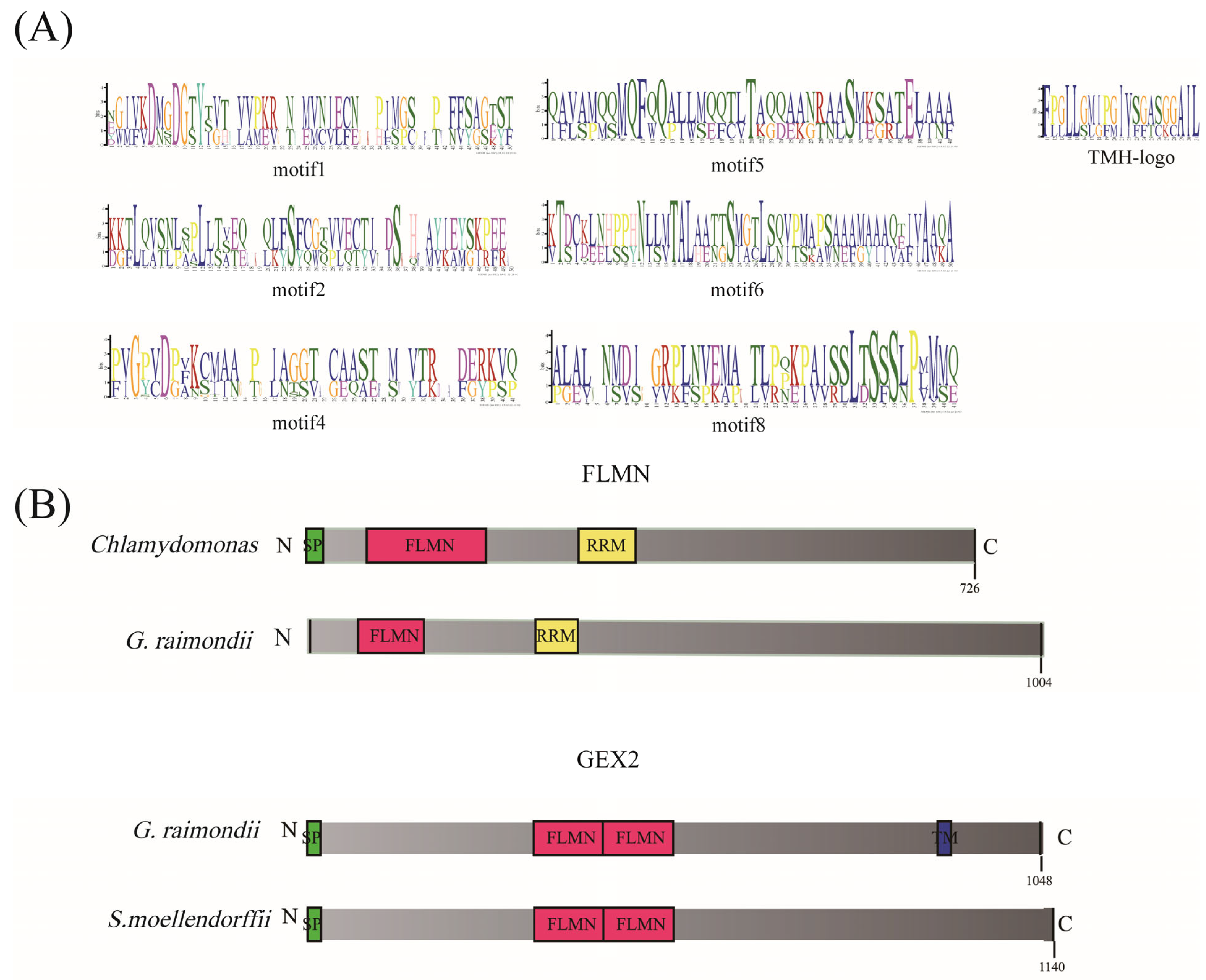
3.5. Gene Structure and Conserved Domain Analysis of Filamin Gene Family Members in Cotton
3.6. Protein 3D Structure Prediction of Filamin Proteins
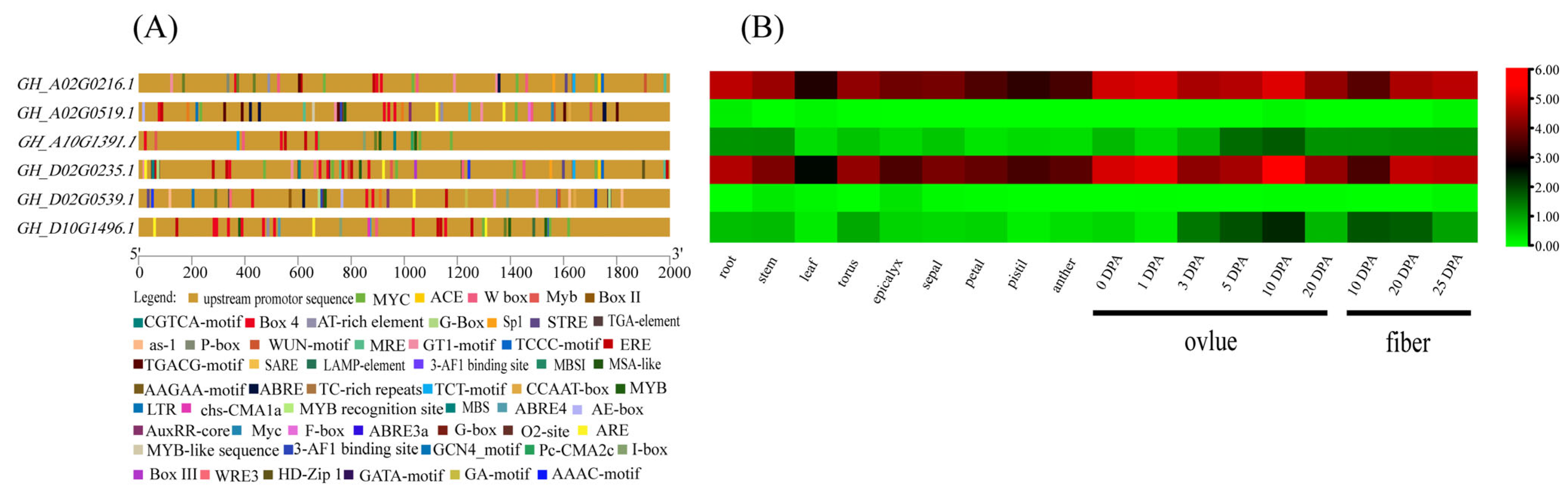
3.7. Analysis of Promoter Cis-Elements of Filamin Gene Family Members in Upland Cotton
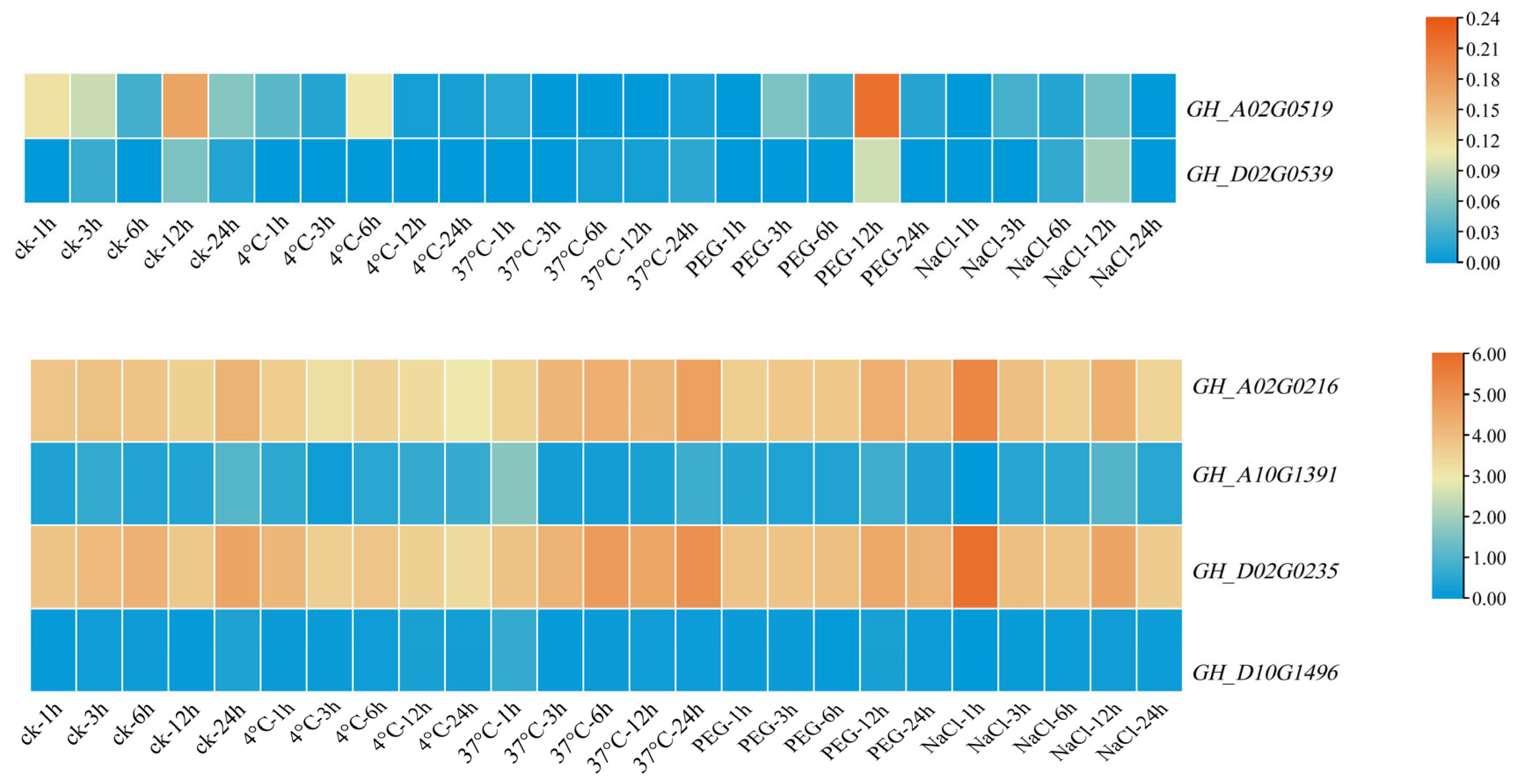
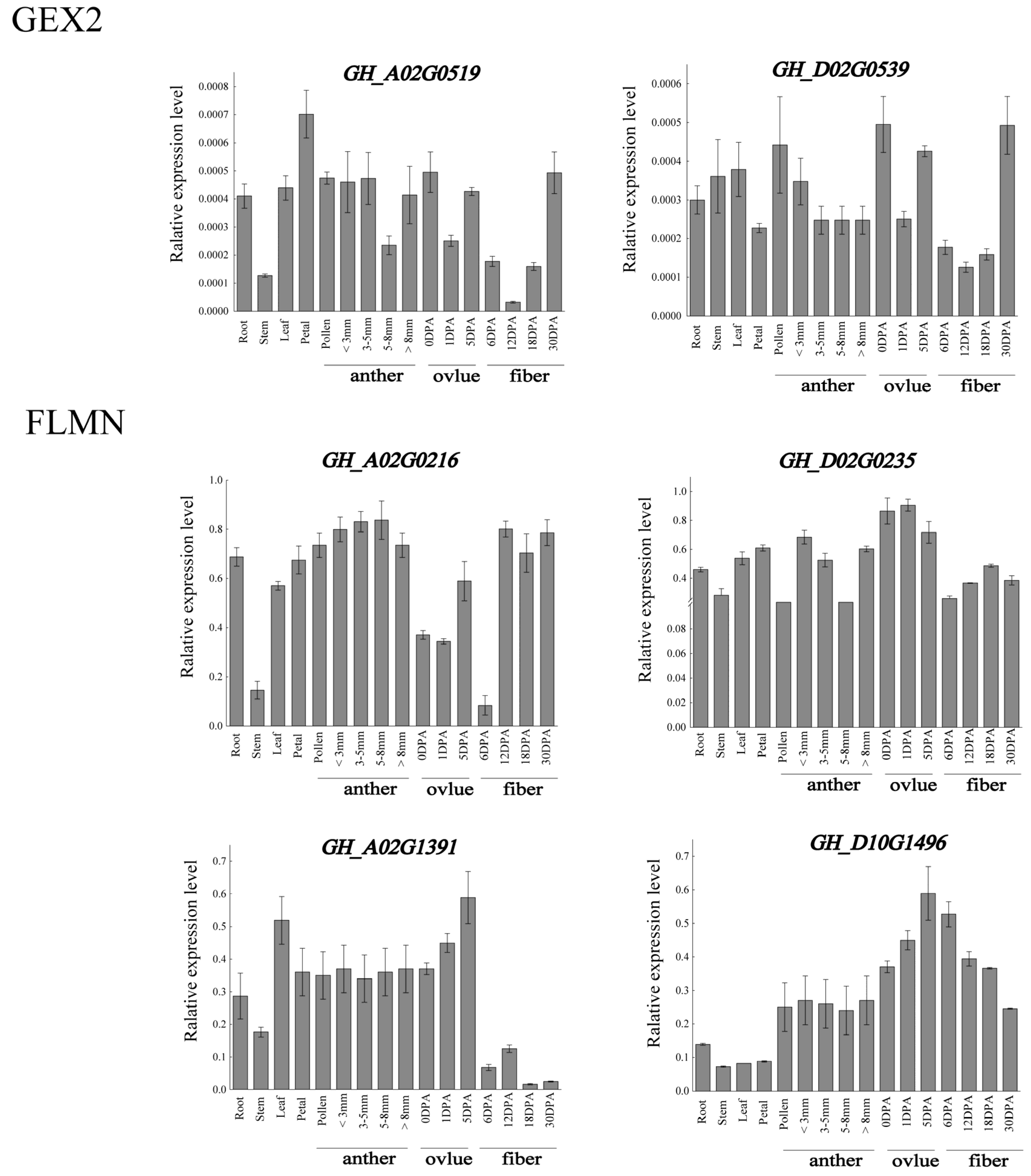
3.8. Expression Pattern Analysis of Filamin Gene Family Members in Upland Cotton under Different Tissues and Abiotic Stresses
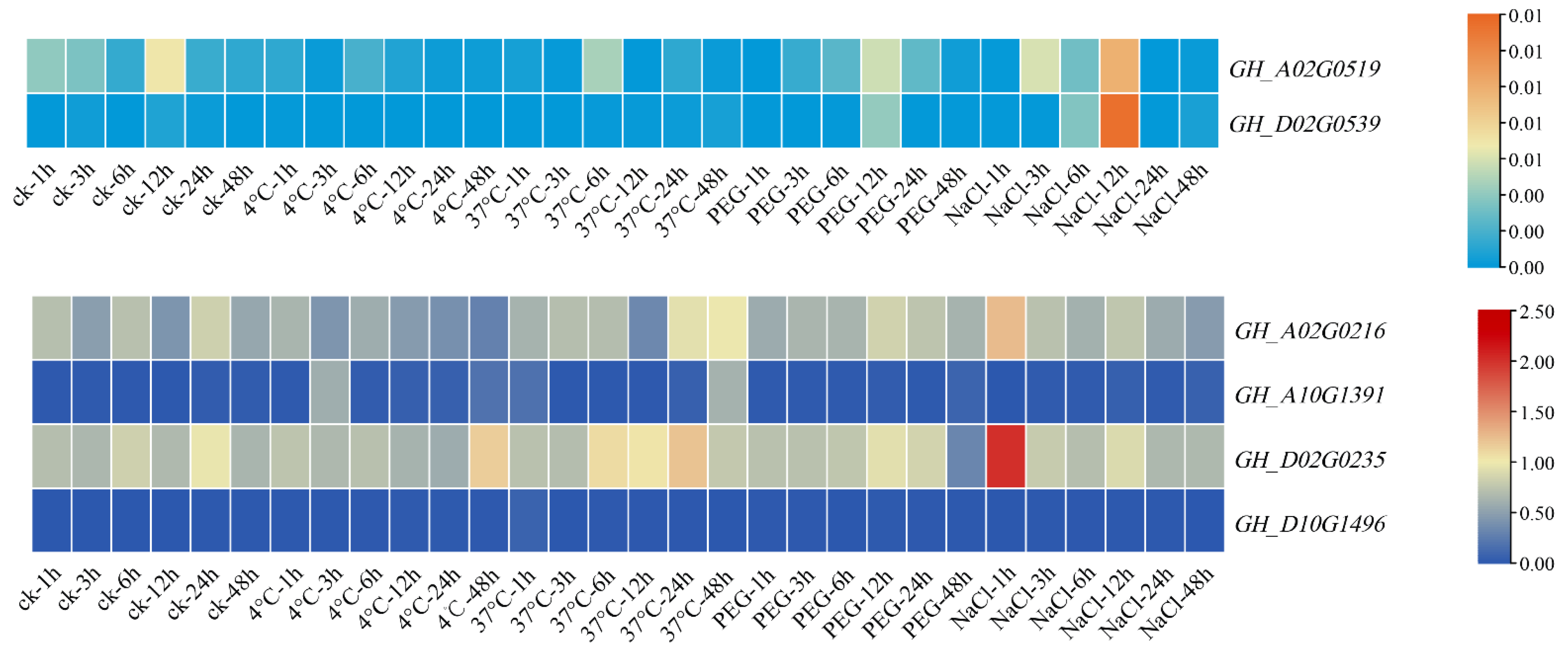
3.9. qRT-PCR Analysis of Filamin Gene Family Members in Upland Cotton under Different Tissues and Abiotic Stresses
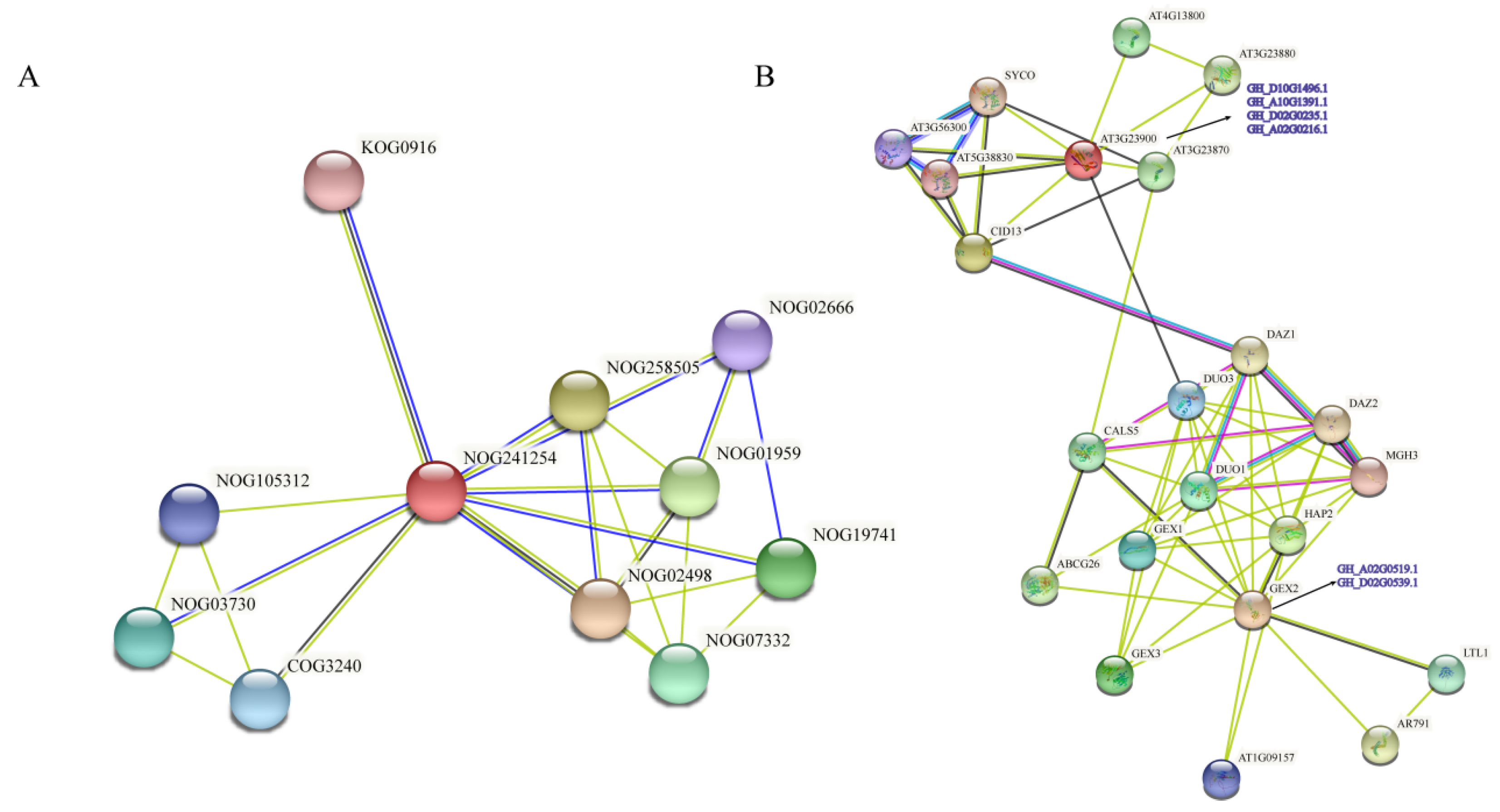
3.10. Interaction Network of Filamin Proteins in Upland Cotton
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, J.; Dong, X.; Pinello, J.; Zhang, J.; Lu, C.; Iacob, R.E.; Engen, J.R.; Snell, W.J.; Springer, T.A. Fusion surface structure, function, and dynamics of gamete fusogen HAP2. eLife 2018, 7, e39772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pinello, J.F.; Fernández, I.; Baquero, E.; Fedry, J.; Rey, F.A.; Snell, W.J. Species-specific gamete recognition initiates fusion-driving trimer formation by conserved fusogen HAP2. Nat. Commun. 2021, 12, 4380. [Google Scholar] [CrossRef] [PubMed]
- Valansi, C.; Moi, D.; Leikina, E.; Matveev, E.; Graña, M.; Chernomordik, L.V.; Romero, H.; Aguilar, P.S.; Podbilewicz, B. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 2017, 216, 571–581. [Google Scholar] [CrossRef]
- Von Besser, K.; Frank, A.C.; Johnson, M.A.; Preuss, D. Arabidopsis HAP2(GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 2006, 133, 4761–4769. [Google Scholar] [CrossRef]
- Brukman, N.G.; Li, X.; Podbilewicz, B. Fusexins, HAP2/GCS1 and Evolution of Gamete Fusion. Front. Cell Dev. Biol. 2022, 9, 824024. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, S.-I.; Yamaguchi, Y.; Suzuki, C.; Yabe, A.; Sato, Y.; Kurihara, D.; Sato, Y.; Susaki, D.; Higashiyama, T.; Maruyama, D. Arabidopsis GEX1 Is a Nuclear Membrane Protein of Gametes Required for Nuclear Fusion During Reproduction. Front. Plant Sci. 2020, 11, 548032. [Google Scholar] [CrossRef]
- Yabe, A.; Nishikawa, S.-I. Expression of GEX1 Orthologs of Brassica rapa and Oryza sativa Rescued the Nuclear Fusion Defect of the Arabidopsis GEX1 Mutant. Plants 2022, 11, 1808. [Google Scholar] [CrossRef]
- Shah, S.; Chen, Y.; Bhattacharya, D.; Chan, C.X. Sex in Symbiodiniaceae dinoflagellates: Genomic evidence for independent loss of the canonical synaptonemal complex. Sci. Rep. 2020, 10, 979. [Google Scholar] [CrossRef]
- Berdieva, M.A.; Pozdnyakov, I.A.; Kalinina, V.O.; Skarlato, S.O. Putative Meiotic Toolkit in the Dinoflagellate Prorocentrum cordatum: Additional Evidence for Sexual Process from Transcriptome. J. Eukaryot. Microbiol. 2021, 68, e12845. [Google Scholar] [CrossRef]
- Misamore, M.J.; Gupta, S.; Snell, W.J. The Chlamydomonas Fus1 Protein Is Present on the Mating Type plus Fusion Organelle and Required for a Critical Membrane Adhesion Event during Fusion with minus Gametes. Mol. Biol. Cell 2003, 14, 2530–2542. [Google Scholar] [CrossRef]
- Lamsoul, I.; Dupré, L.; Lutz, P.G. Molecular Tuning of Filamin A Activities in the Context of Adhesion and Migration. Front. Cell Dev. Biol. 2020, 8, 591323. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, K.; Hamanaka, K.; Riku, Y.; Hattori, Y.; Hara, N.; Iguchi, Y.; Ishigaki, S.; Hashizume, A.; Miyatake, S.; Mitsuhashi, S.; et al. Actin-binding protein filamin-A drives tau aggregation and contributes to progressive supranuclear palsy pathology. Sci. Adv. 2022, 8, eabm5029. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Nakamura, F. Structure and Function of Filamin C in the Muscle Z-Disc. Int. J. Mol. Sci. 2020, 21, 2696. [Google Scholar] [CrossRef] [PubMed]
- Fucini, P.; Renner, C.; Herberhold, C.; Noegel, A.A.; Holak, T.A. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat. Struct. Biol. 1997, 4, 223–230. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Fucini, P.; Noegel, A.A.; Stewart, M. Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod. Nat. Struct. Biol. 1999, 6, 836–841. [Google Scholar] [CrossRef]
- Nakamura, F.; Stossel, T.P.; Hartwig, J.H. The filamins: Organizers of cell structure and function. Cell Adhes. Migr. 2011, 5, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Savoy, R.M.; Ghosh, P.M. The dual role of filamin A in cancer: Can’t live with (too much of) it, can’t live without it. Endocr.-Relat. Cancer 2013, 20, R341–R356. [Google Scholar] [CrossRef]
- Dash, L.; McEwan, R.E.; Montes, C.; Mejia, L.; Walley, J.W.; Dilkes, B.P.; Kelley, D.R. Slim shady is a novel allele of PHYTOCHROME B present in the T-DNA line SALK_015201. Plant Direct 2021, 5, e00326. [Google Scholar] [CrossRef] [PubMed]
- Dressano, K.; Weckwerth, P.R.; Poretsky, E.; Takahashi, Y.; Villarreal, C.; Shen, Z.; Schroeder, J.I.; Briggs, S.P.; Huffaker, A. Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing. Nat. Plants 2020, 6, 1008–1019. [Google Scholar] [CrossRef]
- Engel, M.L.; Holmes-Davis, R.; McCormick, S. Green Sperm. Identification of Male Gamete Promoters in Arabidopsis. Plant Physiol. 2005, 138, 2124–2133. [Google Scholar] [CrossRef]
- Mori, T.; Igawa, T.; Tamiya, G.; Miyagishima, S.-Y.; Berger, F. Gamete Attachment Requires GEX2 for Successful Fertilization in Arabidopsis. Curr. Biol. 2014, 24, 170–175. [Google Scholar] [CrossRef]
- Mori, T.; Igawa, T. Gamete attachment process revealed in flowering plant fertilization. Plant Signal. Behav. 2014, 9, e977715. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Wang, W.; Sun, M.-X. Endosperm development is an autonomously programmed process independent of embryogenesis. Plant Cell 2021, 33, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Warman, C.; Panda, K.; Vejlupkova, Z.; Hokin, S.; Unger-Wallace, E.; Cole, R.A.; Chettoor, A.M.; Jiang, D.; Vollbrecht, E.; Evans, M.M.S.; et al. High expression in maize pollen correlates with genetic contributions to pollen fitness as well as with coordinated transcription from neighboring transposable elements. PLoS Genet. 2020, 16, e1008462. [Google Scholar] [CrossRef] [PubMed]
- Volokhina, I.V.; Moiseeva, Y.M.; Gusev, Y.S.; Gutorova, O.V.; Chumakov, M.I. Analysis of the Gamete-Fusion Genes in the Haploid-inducing ZMS-P Maize Line. Ontogenez 2017, 48, 134–139. [Google Scholar] [PubMed]
- Engel, M.L.; Chaboud, A.; Dumas, C.; McCormick, S. Sperm cells of Zeamays have a complex complement of mRNAs. Plant J. 2003, 34, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Snell, W.J. Uncovering an ancestral green ménage à trois: Contributions of Chlamydomonas to the discovery of a broadly conserved triad of plant fertilization proteins. Curr. Opin. Plant Biol. 2022, 69, 102275. [Google Scholar] [CrossRef]
- Pinello, J.F.; Liu, Y.; Snell, W.J. MAR1 links membrane adhesion to membrane merger during cell-cell fusion in Chlamydomonas. Dev. Cell 2021, 56, 3380–3392.e9. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41, D348–D352. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, M.; Liu, H.; Yang, Y. Prediction of Protein Subcellular Localization Based on Microscopic Images via Multi-Task Multi-Instance Learning. Chin. J. Electron. 2022, 31, 888–896. [Google Scholar] [CrossRef]
- Yu, C.-S.; Cheng, C.-W.; Su, W.-C.; Chang, S.-C.; Huang, S.-W.; Hwang, J.-K.; Lu, C.-H. CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating γ-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, in press. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.-Y.; Zhu, Q.-H.; Chen, X.; Luo, J.-C. GSDS: A gene structure display server. Yi Chuan = Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Boekhorst, J.; Francke, C.; Siezen, R.J. LocateP: Genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinform. 2008, 9, 173. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117. [Google Scholar] [CrossRef]
- Dai, F.; Chen, J.; Zhang, Z.; Liu, F.; Li, J.; Zhao, T.; Hu, Y.; Zhang, T.; Fang, L. COTTONOMICS: A comprehensive cotton multi-omics database. Database 2022, 2022, baac080. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, X.; Chen, W.; Yao, J.; Li, Y.; Fang, S.; Lv, Y.; Li, X.; Pan, J.; Liu, C.; et al. Cotton DMP gene family: Characterization, evolution, and expression profiles during development and stress. Int. J. Biol. Macromol. 2021, 183, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 291, 2199–2213. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, X.; Malik, W.A.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Chen, C.; Guo, L.; Ye, W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 2020, 146, 569–578. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Fasken, M.B.; Corbett, A.H.; Stewart, M. Structure-function relationships in the Nab2 polyadenosine-RNA binding Zn finger protein family. Protein Sci. 2019, 28, 513–523. [Google Scholar] [CrossRef]
- Ruff, K.M.; Pappu, R.V. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Shi, X.; Han, X.; Lu, T.-G. Callose synthesis during reproductive development in monocotyledonous and dicotyledonous plants. Plant Signal. Behav. 2016, 11, e1062196. [Google Scholar] [CrossRef]
- Alandete-Saez, M.; Ron, M.; Leiboff, S.; McCormick, S. Arabidopsis thaliana GEX1 has dual functions in gametophyte development and early embryogenesis. Plant J. 2011, 68, 620–632. [Google Scholar] [CrossRef]
- Borg, M.; Brownfield, L.; Khatab, H.; Sidorova, A.; Lingaya, M.; Twell, D. The R2R3 MYB Transcription Factor DUO1 Activates a Male Germline-Specific Regulon Essential for Sperm Cell Differentiation in Arabidopsis. Plant Cell 2011, 23, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Kawashima, T.; Borg, M.; Zhao, M.; López-Vidriero, I.; Sakayama, H.; Montgomery, S.A.; Sekimoto, H.; Hackenberg, D.; Shimamura, M.; et al. Transcription factor DUO1 generated by neo-functionalization is associated with evolution of sperm differentiation in plants. Nat. Commun. 2018, 9, 5283. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Rutley, N.; Kagale, S.; Hamamura, Y.; Gherghinoiu, M.; Kumar, S.; Sari, U.; Esparza-Franco, M.A.; Sakamoto, W.; Rozwadowski, K.; et al. An EAR-Dependent Regulatory Module Promotes Male Germ Cell Division and Sperm Fertility in Arabidopsis. Plant Cell 2014, 26, 2098–2113. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J.; Hu, J.-Y.; Zhou, Y.; He, F.; Dong, X.; Liu, L.-Y.; Coupland, G.; Turck, F.; De Meaux, J. Uncovering Male Germline Development in Arabidopsis: The Gametophyte Revealed. Plant Cell 2014, 26, 1837. [Google Scholar] [CrossRef]
- Borges, F.; Gardner, R.; Lopes, T.; Calarco, J.P.; Boavida, L.C.; Slotkin, R.K.; Martienssen, R.A.; Becker, J.D. FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 2012, 8, 44. [Google Scholar] [CrossRef]
- Shen, H.; Luong, P.; Huq, E. The F-Box Protein MAX2 Functions as a Positive Regulator of Photomorphogenesis in Arabidopsis. Plant Physiol. 2007, 145, 1471–1483. [Google Scholar] [CrossRef]
- Kuroda, H.; Takahashi, N.; Shimada, H.; Seki, M.; Shinozaki, K.; Matsui, M. Classification and Expression Analysis of Arabidopsis F-Box-Containing Protein Genes. Plant Cell Physiol. 2002, 43, 1073–1085. [Google Scholar] [CrossRef]
- Kägi, C.; Baumann, N.; Nielsen, N.; Stierhof, Y.-D.; Groß-Hardt, R. The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc. Natl. Acad. Sci. USA 2010, 107, 22350–22355. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Friedberg, F. Single and multiple CH (calponin homology) domain containing multidomain proteins in Arabidopsis and Saccharomyces: An inventory. Mol. Biol. Rep. 2011, 38, 213–218. [Google Scholar] [CrossRef]
- Ma, H.; Liu, M. The microtubule cytoskeleton acts as a sensor for stress response signaling in plants. Mol. Biol. Rep. 2019, 46, 5603–5608. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wu, L.; Zhu, S.; Chen, W.; Yao, J.; Li, Y.; Li, T.; Shang, H.; Zhang, Y. Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.). Genes 2022, 13, 2313. https://doi.org/10.3390/genes13122313
Wang M, Wu L, Zhu S, Chen W, Yao J, Li Y, Li T, Shang H, Zhang Y. Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.). Genes. 2022; 13(12):2313. https://doi.org/10.3390/genes13122313
Chicago/Turabian StyleWang, Mingyang, Lanxin Wu, Shouhong Zhu, Wei Chen, Jinbo Yao, Yan Li, Tengyu Li, Haihong Shang, and Yongshan Zhang. 2022. "Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.)" Genes 13, no. 12: 2313. https://doi.org/10.3390/genes13122313
APA StyleWang, M., Wu, L., Zhu, S., Chen, W., Yao, J., Li, Y., Li, T., Shang, H., & Zhang, Y. (2022). Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.). Genes, 13(12), 2313. https://doi.org/10.3390/genes13122313





