Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of m6A Regulatory Genes in Litchi
2.2. Physicochemical Properties Analysis
2.3. Phylogenetic Analysis
2.4. Gene Structure, Conserved Domain, and Conserved Motif Analysis
2.5. Prediction of MiRNA Target Site
2.6. Chromosomal Arrangement and Gene Ontology (GO) Enrichment Analysis
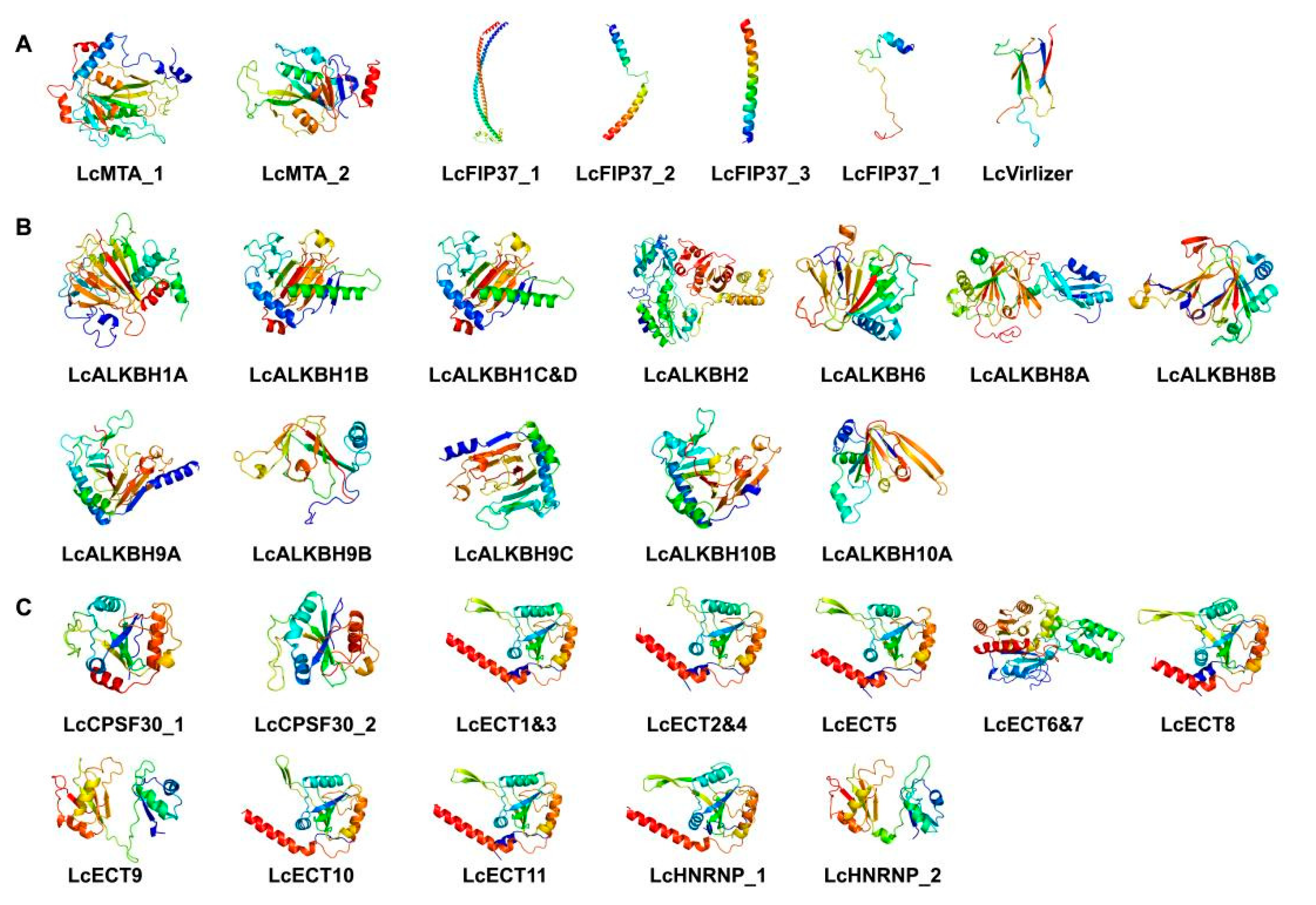
2.7. The 3D Protein Structure Analysis
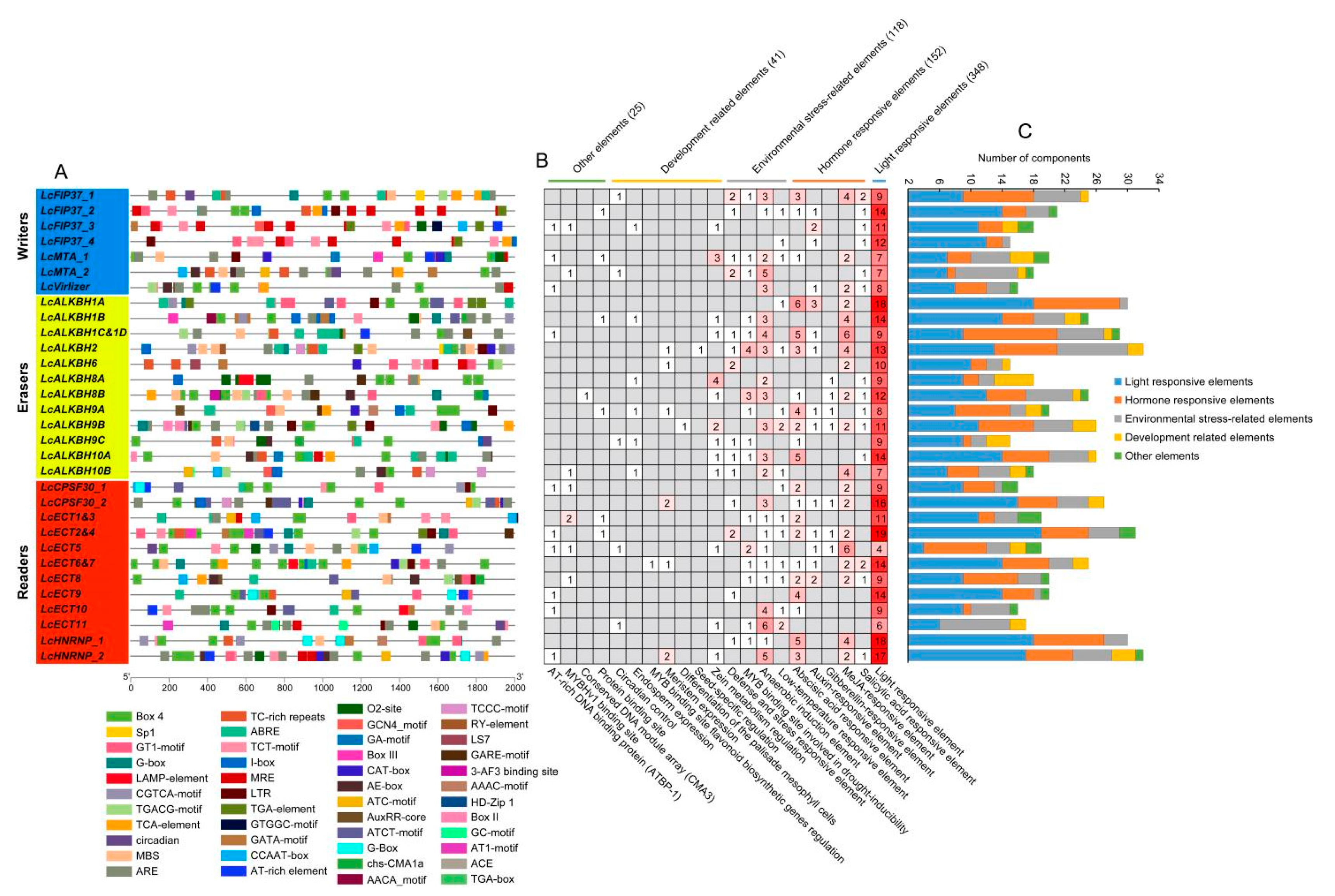
2.8. Cis-Regulatory Elements Analysis
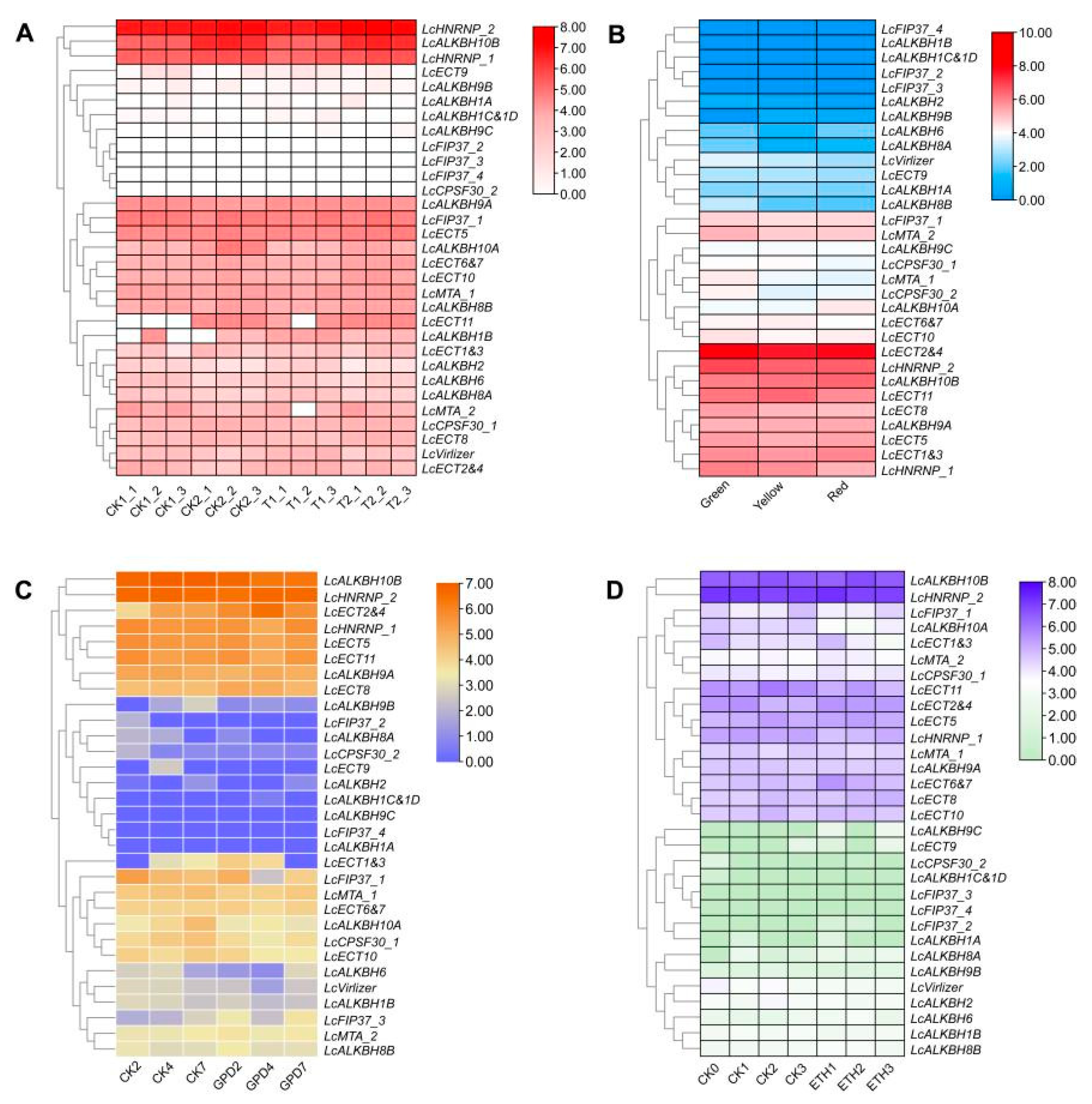
2.9. Expression Analysis of Litchi m6A Regulatory Genes RNA-Seq Data
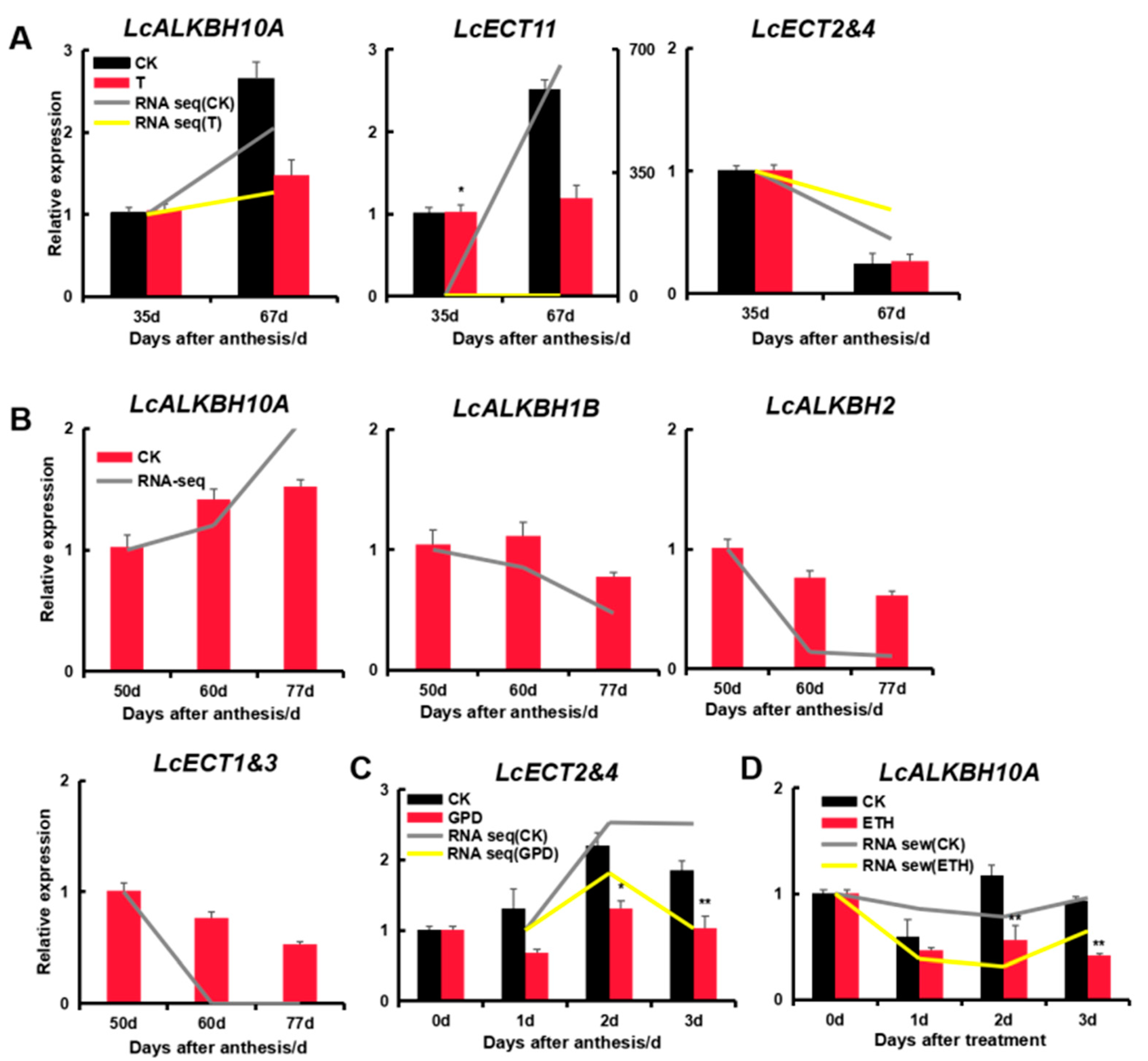
2.10. Expression Analysis of Litchi m6A Regulatory Genes Identified by Quantitative qRT-PCR
3. Results
3.1. Identification of Litchi m6A Regulatory Genes
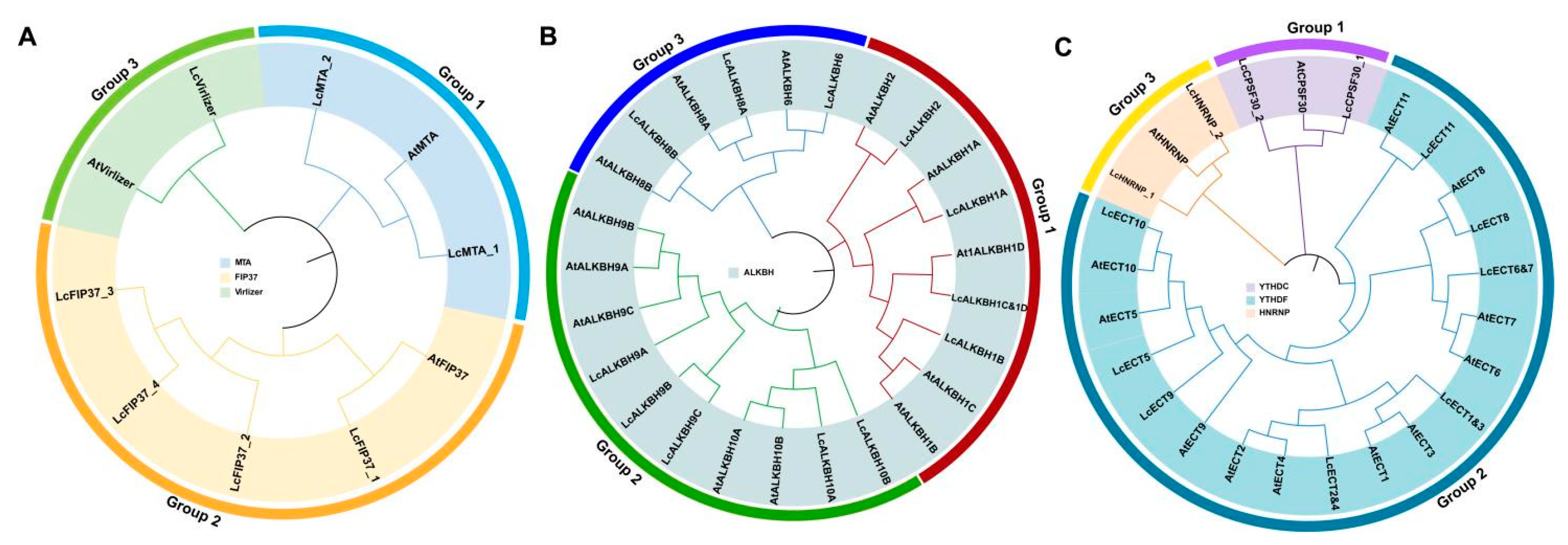
3.2. Phylogenetic Relationship Analysis
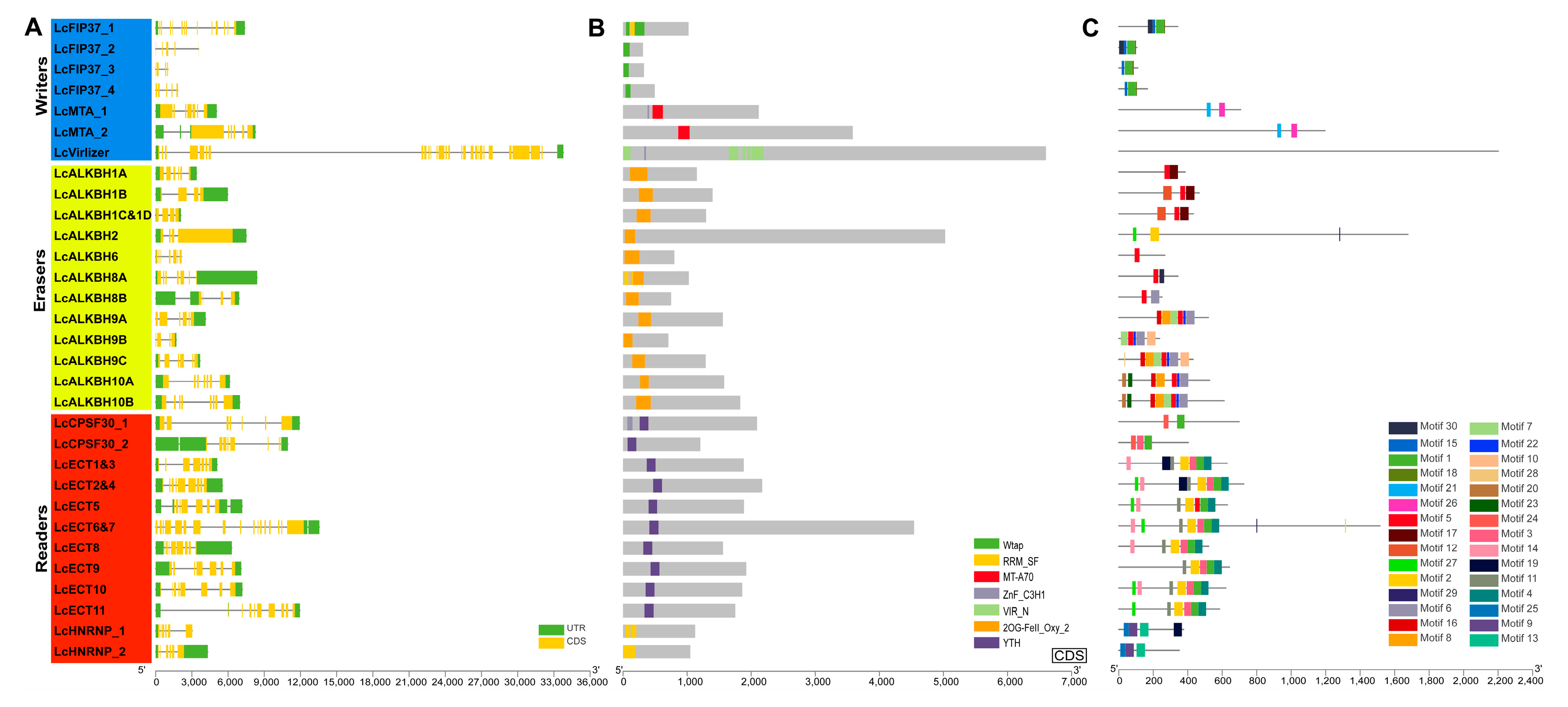
3.3. Structural Features Analysis
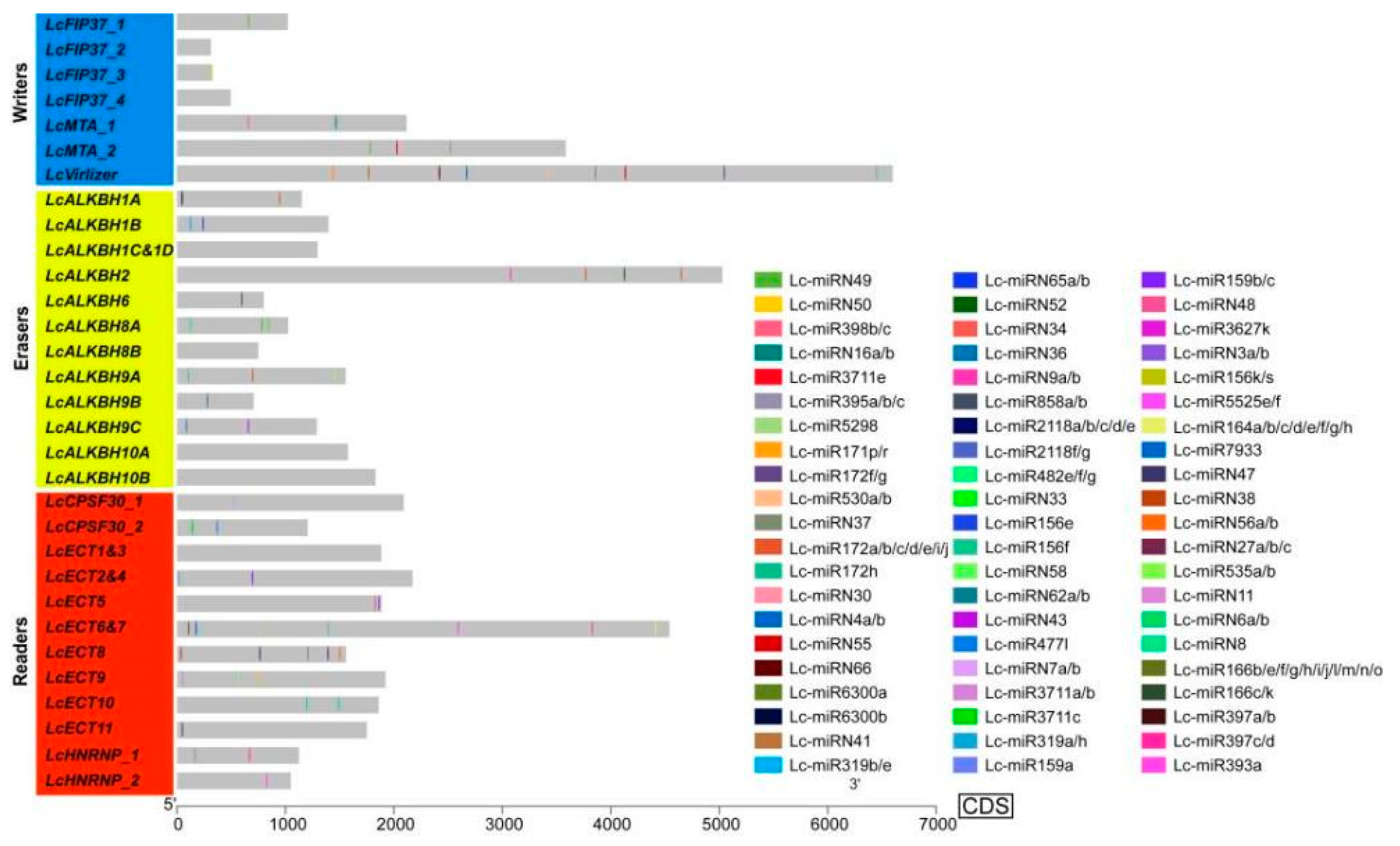
3.4. The miRNA Target Site Prediction
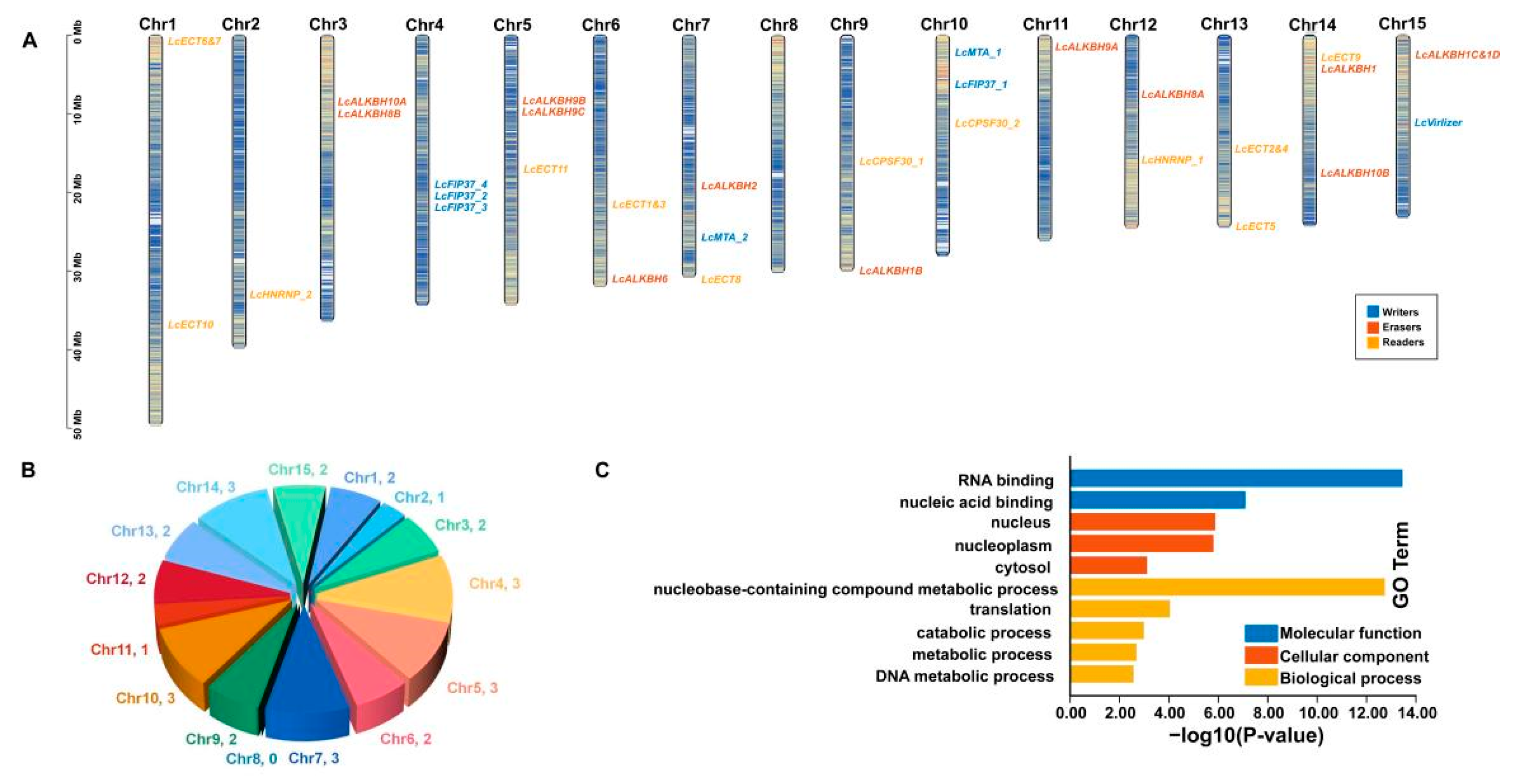
3.5. Chromosomal Arrangement and GO Enrichment Analysis
3.6. Cis-Regulatory Elements Analysis
3.7. The 3D Protein Structure Analysis
3.8. Expression Analysis of Litchi m6A Regulatory Genes by RNA-Seq Data
3.9. Expression Analysis of Litchi m6A Regulatory Genes by Quantitative qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Kierzek, E. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003, 31, 4472–4480. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, F.; Li, Y.; Wang, J.; Zhao, X.; Li, Z.; Xu, J.; Wang, W.; Fu, B. Global N6-Methyladenosine Profiling Revealed the Tissue-Specific Epitranscriptomic Regulation of Rice Responses to Salt Stress. Int. J. Mol. Sci. 2022, 23, 2091. [Google Scholar] [CrossRef]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Duan, H.; Wei, L.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2018, 29, 2995–3011. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Bio. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Liu, J.; Harada, B.T.; He, C. Regulation of Gene Expression by N-methyladenosine in Cancer. Trends Cell Biol. 2019, 29, 487–499. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Wang, J.; Liu, X.; Lin, F.; Lu, J. N6-methyladenosine RNA methylation is involved in virulence of the rice blast fungus Pyricularia oryzae (syn. Magnaporthe oryzae). FEMS Microbiol. Lett. 2018, 366, fny286. [Google Scholar] [CrossRef]
- Huang, T.; He, W.; Li, C.; Zhang, J.; Liao, Y.; Song, B.; Yang, P. Transcriptome-wide analyses of RNA m6A methylation in hexaploid wheat reveal its roles in mRNA translation regulation. Front. Plant Sci. 2022, 13, 917335. [Google Scholar] [CrossRef]
- Reichel, M.; Köster, T.; Staiger, D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 2019, 11, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cai, J.; Umme, A.; Chen, Y.; Xu, T.; Kang, H. Unique features of mRNA m6A methylomes during expansion of tomato (Solanum lycopersicum) fruits. Plant Physiol. 2022, 188, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, L.; Chen, H.; Huang, D.; Song, B. m6A-Maize: Weakly supervised prediction of m6A-carrying transcripts and m6A-affecting mutations in maize (Zea mays). Methods 2022, 203, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 2017, 215, 157–172. [Google Scholar] [CrossRef]
- Cheng, P.; Bao, S.; Li, C.; Tong, J.; Shen, L.; Yu, H. RNA N6-methyladenosine modification promotes auxin biosynthesis required for male meiosis in rice. Dev. Cell 2022, 57, 246–259. [Google Scholar] [CrossRef]
- Chen, D.; Fu, L.; Su, T.; Xiong, J.; Chen, Y.; Shen, Q.; Kuang, L.; Wu, D. N6-methyladenosine methylation analysis reveals transcriptome-wide expression response to salt stress in rice roots. Environ. Exp. Bot. 2022, 201, 104945. [Google Scholar] [CrossRef]
- Ok, S.H.; Jeong, H.J.; Bae, J.M.; Shin, J.; Luan, S.; Kim, K. Novel CIPK1-Associated Proteins in Arabidopsis Contain an Evolutionarily Conserved C-Terminal Region That Mediates Nuclear Localization. Plant Physiol. 2005, 139, 138–150. [Google Scholar] [CrossRef]
- Yue, X.; Zhang, Y.; Yang, C.; Li, J.; Rui, X.; Ding, F.; Hu, F.; Wang, X.; Ma, W.; Zhou, K. Genome-wide identification and expression analysis of carotenoid cleavage oxygenase genes in Litchi (Litchi chinensis Sonn.). BMC Plant Biol. 2022, 22, 398. [Google Scholar] [CrossRef]
- Zhao, M.; Li, J. Molecular Events Involved in Fruitlet Abscission in Litchi. Plants 2020, 9, 151. [Google Scholar] [CrossRef]
- Hu, G.; Feng, J.; Xiang, X.; Wang, J.; Salojärvi, J.; Liu, C.; Wu, Z.; Zhang, J.; Liang, X.; Jiang, Z.; et al. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 2022, 54, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Chen, C.; Liu, Y.; Zeng, M.; Meyers, B.C.; Li, J.; Xia, R. Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol 2018, 217, 1535–1550. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Hu, B.; Qin, Y.; Zhao, J.; Wang, H.; Hu, G. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genom. 2015, 16, 225. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Huang, X.; Li, J.; Wang, H.; Li, J. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 2015, 6, 439. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Ying, P.; Ma, W.; Li, J. Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front. Plant Sci. 2015, 6, 502. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Zhong, H.; Chen, J.; Li, C.; Chen, L.; Wu, J.; Chen, J.; Lu, W.; Li, J. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011, 30, 641–653. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Brodersen, P. Occurrence and Functions of m6A and Other Covalent Modifications in Plant mRNA. Plant Physiol. 2020, 182, 79–96. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, S.; Zhou, C.; Xie, S.; Chen, G.; Tian, C.; Xu, K.; Lin, Y.; Lai, Z.; Guo, Y. Genome-Wide Investigation of N6-Methyladenosine Regulatory Genes and Their Roles in Tea (Camellia sinensis) Leaves During Withering Process. Front. Plant Sci. 2021, 12, 1183. [Google Scholar] [CrossRef]
- Chen, X. microRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Somasundaram, K. mRNA Traffic Control Reviewed: N6-Methyladenosine (m6A) Takes the Driver’s Seat. Bioessays 2018, 40, 1700093. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Teng, C.; Xia, R.; Meyers, B.C. PhasiRNAs in Plants: Their Biogenesis, Genic Sources, and Roles in Stress Responses, Development, and Reproduction. Plant Cell 2020, 32, 3059–3080. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Wang, H.; Huang, X.; Huang, H. A study on the causative factors retarding pigmentation in the fruit of ‘Feizixiao’ litchi. Acta Hortic. Sinica 2002, 29, 408–412. [Google Scholar]
- Hu, B.; Li, J.; Wang, D.; Wang, H.; Qin, Y.; Hu, G.; Zhao, J. Transcriptome profiling of Litchi chinensis pericarp in response to exogenous cytokinins and abscisic acid. Plant Growth Regul. 2018, 84, 437–450. [Google Scholar] [CrossRef]
- Lai, B.; Li, X.J.; Hu, B.; Qin, Y.H.; Huang, X.M.; Wang, H.C.; Hu, G.B. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Liu, X.S.; Luo, Y.C.; Wang, S.W.; Wang, H.C.; Harpaz-Saad, S.; Huang, X.M. Residue Analysis and the Effect of Preharvest Forchlorfenuron (CPPU) Application on On-Tree Quality Maintenance of Ripe Fruit in “Feizixiao” Litchi (Litchi chinensis Sonn.). Front. Plant Sci. 2022, 13, 829635. [Google Scholar] [CrossRef]
- Singh, S.P.; Saini, M.K.; Singh, J.; Pongener, A.; Sidhu, G.S. Preharvest application of abscisic acid promotes anthocyanins accumulation in pericarp of litchi fruit without adversely affecting postharvest quality. Postharvest Biol. Technol. 2014, 96, 14–22. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, R.; Li, X.; Tian, S.; Li, B.; Qin, G. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021, 22, 168. [Google Scholar] [CrossRef]
| Plant Materials for RNA-Seq Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|
| Data Set | Variety | Sample ID | Samples | Biological Replicates | Platform | LibraryLayout | Accesion Number | Data Sources |
| 1 | ‘Feizixiao’ (27-year-old) | CK1_1, CK1_2, CK1_3, CK2_1, CK2_2,CK2_3, T1_1, T1_2, T1_3, T2_1, T2_1, T2_1. | Peel: CK1 and T1: Green stage (35 d after anthesis); CK2 and T2: (57 d after anthesis). | 3 | ILLUMINA | PAIRED | Home data | |
| 2 | ‘Nuomici’ (Adult tree) | Green (SRX700596), Yellow (SRX700598), Red (SRX700599). | Peel: Green: 52 d after anthesis; Yellow: 62 d after anthesis; Red: 72 d after anthesis. Grown in normal conditions. | 1 (mixed sample of 3 biological replicates) | ILLUMINA | SINGLE | SRP047115 | Lai et al., 2015 [24] |
| 3 | ‘Wuye’ (9-year-old) | CK2 (SRX847812), CK4 (SRX847822), CK7(SRX847823), GPD2 (SRX847824), GPD4 (SRX847825), GPD7(SRX847826). | Fruitlet: CK: Control group; GPD: Treatment group; 2, 4, and 7 d represented fruitlet abscission at 2, 4, and 7 days after being treated with GPD treatment at 35 days after anthesis. | 1 (Mix sample) | ILLUMINA | SINGLE | SRA234477 | Li et al., 2015 [25] |
| 4 | ‘Feizixiao’ (9-year-old) | CK0 (SRX5126892), CK1 (SRX5126893), CK2 (SRX5126894), CK3 (SRX5126895), ETH1 (SRX5126896), ETH2 (SRX5126897), ETH3 (SRX5126898).. | Abscission zone: CK: Control group; GPD: Treatment group; 0, 1, 2, and 3 d represented fruitlet abscission at 0, 1, 2, and 3 days after being treated with 250 mg/L ethephon solution at 25 days after anthesis. | 1 | ILLUMINA | SINGLE | SRP173341 | Li et al., 2015 [26] |
| Plant materials for qRT-PCR analysis | ||||||||
| 1 | ‘Feizixiao’ (27-year-old) | 35 d and 67 d. | Peel collected at 35 and 67 days treated with 4 mg/L CPPU after anthesis. | 3 | Home data | |||
| 2 | ‘Nuomici’ (27-year-old) | 50 d, 60 d, and 77 d. | Peel collected at 50, 60, and 77 days after anthesis, which grew in normal condition. | 3 | Home data | |||
| 3 | ‘Feizixiao’ (27-year-old) | 0 d, 1 d, 2 d, and 3 d. | Fruitlet collected at 0, 1, 2, and 3 days treated with GPD treatment at 35 days after anthesis. | 3 | Home data | |||
| 4 | ‘Feizixiao’ (27-year-old) | 0 d, 1 d, 2 d, and 3 d. | Abscission zone tissues collected at 0, 1, 2, and 3 days treated with 250 mg/Lexogenous ethephon (ETH) after anthesis. | 3 | Home data | |||
| Genes | Gene ID in Genome | Number of Amino Acids (aa) | PI | MW (kDa) | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization Prediction |
|---|---|---|---|---|---|---|---|---|
| Writers | ||||||||
| Lc_FIP37_1 | LITCHI022446.m2 | 341 | 5.13 | 38.74 | 56.79 | 79.88 | −0.83 | Nucleus |
| Lc_FIP37_2 | LITCHI031231.m1 | 104 | 7.86 | 11.82 | 50.26 | 93.94 | −0.591 | Nucleus |
| Lc_FIP37_3 | LITCHI031301.m1 | 109 | 4.9 | 12.39 | 43.18 | 95.78 | −0.299 | Nucleus |
| Lc_FIP37_4 | LITCHI031225.m1 | 165 | 5.83 | 18.67 | 47.28 | 99.33 | −0.358 | Nucleus |
| Lc_MTA_1 | LITCHI021922.m2 | 706 | 5.71 | 78.63 | 47.91 | 76.4 | −0.526 | Nucleus |
| Lc_MTA_2 | LITCHI009397.m2 | 1195 | 7.68 | 134.28 | 60.59 | 40.72 | −1.349 | Nucleus |
| Lc_Virlizer | LITCHI018982.m2 | 2200 | 5.41 | 240.57 | 52.3 | 96.91 | −0.079 | Nucleus |
| Erasers | ||||||||
| Lc_ALKBH1A | LITCHI004660.m2 | 384 | 8.05 | 43.52 | 61.01 | 79.3 | −0.287 | Cytoplasm |
| Lc_ALKBH1B | LITCHI029924.m2 | 466 | 7.72 | 51.25 | 45.99 | 66.31 | −0.72 | Nucleus |
| Lc_ALKBH1C&1D | LITCHI018059.m1 | 432 | 9.51 | 48.40 | 53.24 | 77.36 | −0.516 | Nucleus |
| Lc_ALKBH2 | LITCHI008899.m3 | 1676 | 8.86 | 190.29 | 42.36 | 82.82 | −0.362 | Plasma membrane |
| Lc_ALKBH6 | LITCHI004157.m1 | 267 | 6.25 | 29.92 | 47.8 | 89.36 | −0.281 | Nucleus |
| Lc_ALKBH8A | LITCHI020056.m1 | 342 | 6.51 | 38.28 | 51.41 | 80.91 | −0.256 | Chloroplast |
| Lc_ALKBH8B | LITCHI026423.m1 | 250 | 4.75 | 28.35 | 52.21 | 78 | −0.406 | Nucleus |
| Lc_ALKBH9A | LITCHI006480.m1 | 519 | 5.76 | 57.89 | 54.08 | 73.04 | −0.638 | Nucleus |
| Lc_ALKBH9B | LITCHI000266.m1 | 236 | 8.85 | 26.13 | 66.51 | 81.74 | −0.242 | Cytoplasm |
| Lc_ALKBH9C | LITCHI000270.m1 | 430 | 8.38 | 48.34 | 55.71 | 81.33 | −0.443 | Nucleus |
| Lc_ALKBH10A | LITCHI026300.m1 | 526 | 5.64 | 58.08 | 54.42 | 80.82 | −0.421 | Chloroplast |
| Lc_ALKBH10B | LITCHI005944.m1 | 610 | 8.25 | 66.15 | 58.06 | 70.97 | −0.541 | Nucleus |
| Readers | ||||||||
| Lc_ECT1&3 | LITCHI003339.m2 | 628 | 6.23 | 68.53 | 37.06 | 57.75 | −0.731 | Nucleus |
| Lc_ECT2&4 | LITCHI024309.m2 | 724 | 6.96 | 79.28 | 33.38 | 57.64 | −0.714 | Nucleus |
| Lc_ECT5 | LITCHI025245.m1 | 629 | 5.33 | 68.72 | 48.82 | 54.74 | −0.716 | Nucleus |
| Lc_ECT6&7 | LITCHI014289.m1 | 1514 | 5.63 | 168.82 | 4.02 | 75.09 | −0.44 | Chloroplast |
| Lc_ECT8 | LITCHI009804.m1 | 520 | 6.83 | 57.50 | 36.75 | 58.56 | −0.583 | Nucleus |
| Lc_ECT9 | LITCHI004655.m12 | 641 | 6.83 | 72.08 | 47.45 | 70.23 | −0.5 | Extracellular space |
| Lc_ECT10 | LITCHI016506.m1 | 620 | 5.28 | 68.19 | 44.72 | 63.39 | −0.694 | Nucleus |
| Lc_ECT11 | LITCHI000687.m1 | 584 | 6.62 | 65.00 | 28.17 | 52.43 | −0.864 | Nucleus |
| Lc_HNRNP_1 | LITCHI020595.m1 | 375 | 6.27 | 38.96 | 25.54 | 39.81 | −0.74 | Nucleus |
| Lc_HNRNP_2 | LITCHI013653.m1 | 350 | 9.07 | 36.25 | 30.65 | 44.57 | −0.678 | Nucleus |
| Lc_CPSF30_1 | LITCHI028685.m1 | 697 | 6.12 | 75.97 | 54.57 | 42.31 | −0.859 | Nucleus |
| Lc_CPSF30_2 | LITCHI022917.m5 | 402 | 5.85 | 45.82 | 57.48 | 64.78 | −0.807 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Xue, J.; Ren, X.; Zhang, Y.; Du, L.; Ding, F.; Zhou, K.; Ma, W. Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.). Genes 2022, 13, 2284. https://doi.org/10.3390/genes13122284
Tang L, Xue J, Ren X, Zhang Y, Du L, Ding F, Zhou K, Ma W. Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.). Genes. 2022; 13(12):2284. https://doi.org/10.3390/genes13122284
Chicago/Turabian StyleTang, Liwen, Jiali Xue, Xingyu Ren, Yue Zhang, Liqing Du, Feng Ding, Kaibing Zhou, and Wuqiang Ma. 2022. "Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.)" Genes 13, no. 12: 2284. https://doi.org/10.3390/genes13122284
APA StyleTang, L., Xue, J., Ren, X., Zhang, Y., Du, L., Ding, F., Zhou, K., & Ma, W. (2022). Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.). Genes, 13(12), 2284. https://doi.org/10.3390/genes13122284






