An Investigation of the Etiologies of Non-Immune Hydrops Fetalis in the Era of Next-Generation Sequence—A Single Center Experience
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
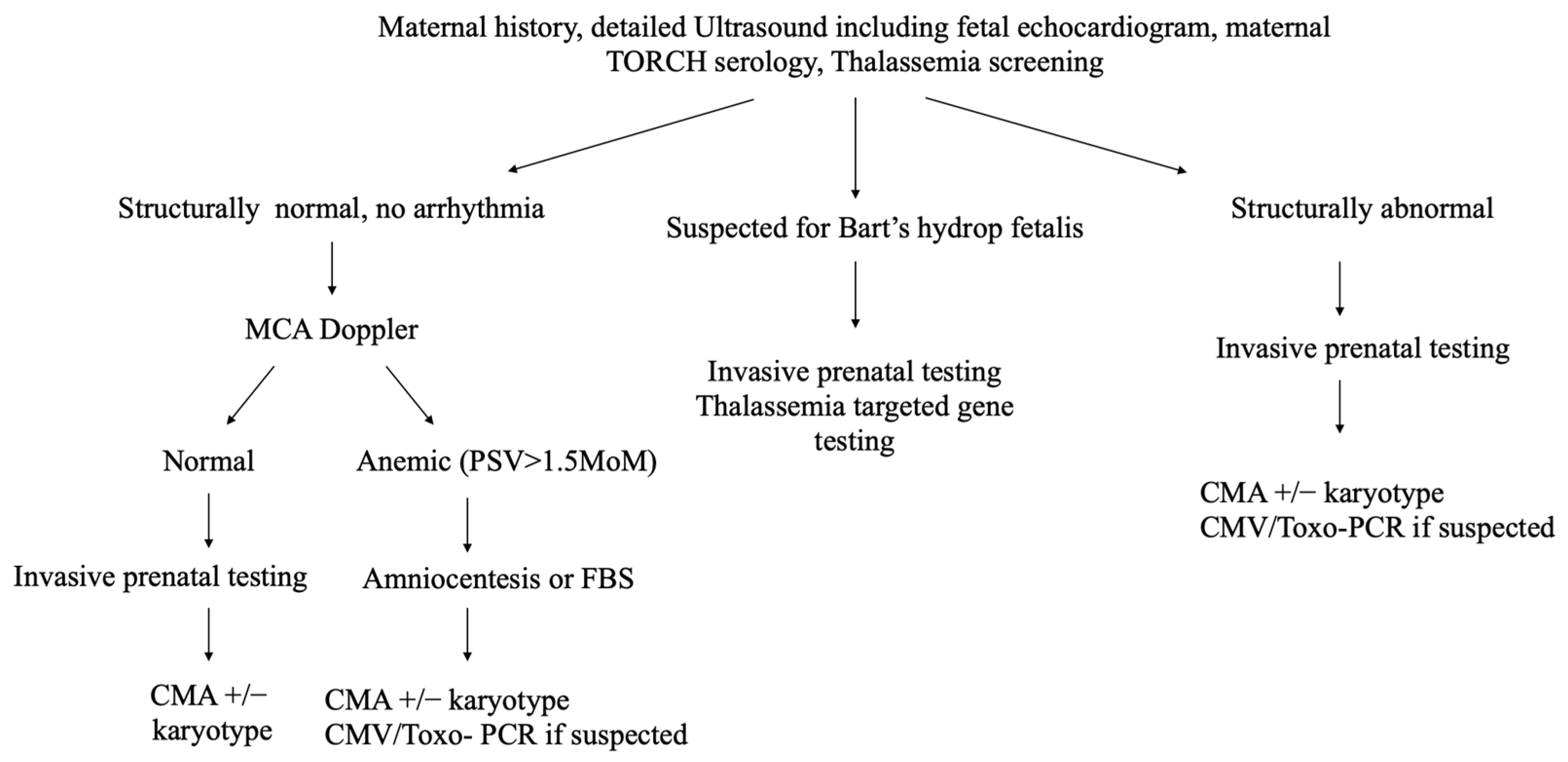
2.2. Procedure
2.3. Outcomes
2.4. Statistical Analysis
3. Results
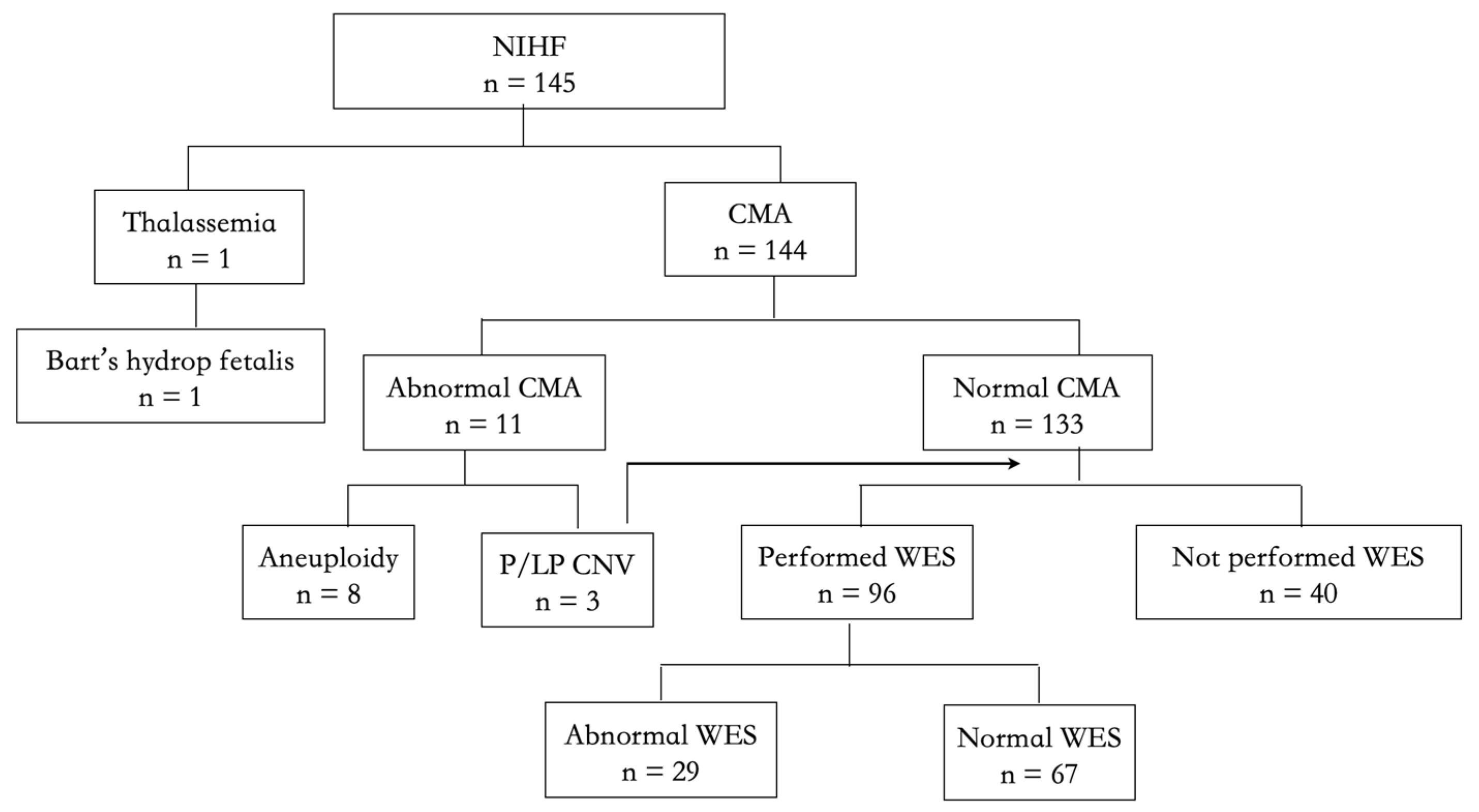
3.1. Study Participants
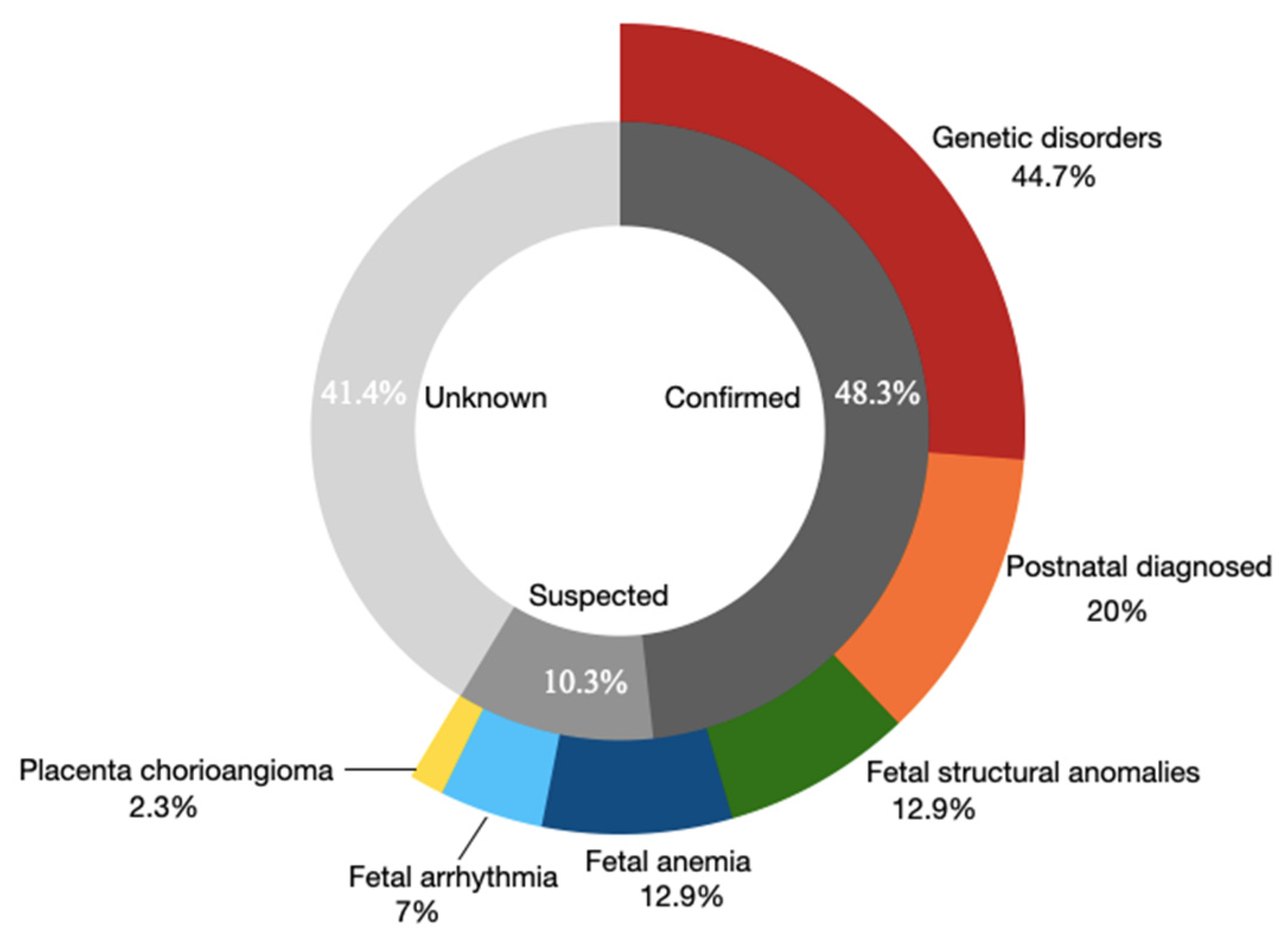
3.2. The Overall Etiologies in the NIHF Cohort
3.3. The Detection Yield of CMA
3.4. The Detection Yield of ES
3.5. The Etiologies of Cases That Did Not Perform ES
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santolaya, J.; Alley, D.; Jaffe, R.; Warsof, S.L. Antenatal classification of hydrops fetalis. Obstet. Gynecol. 1992, 79, 256–259. [Google Scholar] [PubMed]
- Ismail, K.M.; Martin, W.L.; Ghosh, S.; Whittle, M.J.; Kilby, M.D. Etiology and outcome of hydrops fetalis. J. Matern. Fetal Med. 2001, 10, 175–181. [Google Scholar] [CrossRef]
- Steurer, M.A.; Peyvandi, S.; Baer, R.J.; MacKenzie, T.; Li, B.C.; Norton, M.E.; Jelliffe-Pawlowski, L.L.; Moon-Grady, A.J. Epidemiology of Live Born Infants with Nonimmune Hydrops Fetalis-Insights from a Population-Based Dataset. J. Pediatr. 2017, 187, 182–188.e3. [Google Scholar] [CrossRef]
- Nassr, A.A.; Ness, A.; Hosseinzadeh, P.; Salmanian, B.; Espinoza, J.; Berger, V.; Werner, E.; Erfani, H.; Welty, S.; Bateni, Z.H.; et al. Outcome and Treatment of Antenatally Diagnosed Nonimmune Hydrops Fetalis. Fetal Diagn. Ther. 2018, 43, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Whybra, C.; Källén, K.; Hansson, S.R.; Gunnarsson, R. Non-immune hydrops fetalis was rare in Sweden during 1997–2015, but cases were associated with complications and poor prognosis. Acta Paediatr. 2020, 109, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Society for Maternal-Fetal Medicine (SMFM); Norton, M.E.; Chauhan, S.P.; Dashe, J.S. Society for maternal-fetal medicine (SMFM) clinical guideline #7: Nonimmune hydrops fetalis. Am. J. Obstet. Gynecol. 2015, 212, 127–139. [Google Scholar]
- Sparks, T.N.; Thao, K.; Lianoglou, B.R.; Boe, N.M.; Bruce, K.G.; Datkhaeva, I.; Field, N.T.; Fratto, V.M.; Jolley, J.; Laurent, L.C.; et al. University of California Fetal–Maternal Consortium (UCfC). Nonimmune hydrops fetalis: Identifying the underlying genetic etiology. Genet. Med. 2019, 21, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Mardy, A.H.; Rangwala, N.; Hernandez-Cruz, Y.; Gosnell, K.A.; Gonzalez, J.M.; Norton, M.E.; Sparks, T.N. Utility of chromosomal microarray for diagnosis in cases of nonimmune hydrops fetalis. Prenat. Diagn. 2020, 40, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Hui, A.S.; Chau, M.H.K.; Chan, Y.M.; Cao, Y.; Kwan, A.H.; Zhu, X.; Kwok, Y.K.; Chen, Z.; Lao, T.T.; Choy, K.W.; et al. The role of chromosomal microarray analysis among fetuses with normal karyotype and single system anomaly or nonspecific sonographic findings. Acta Obstet. Et Gynecol. Scand. 2021, 100, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Fu, F.; Yu, Q.; Li, R.; Li, F.; Wang, D.; Lei, T.; Yang, X.; Liao, C. Nonimmune hydrops fetalis: Genetic analysis and clinical outcome. Prenat Diagn. 2020, 40, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Sparks, T.N.; Lianoglou, B.R.; Adami, R.R.; Pluym, I.D.; Holliman, K.; Duffy, J.; Downum, S.L.; Patel, S.; Faubel, A.; Boe, N.M.; et al. University of California Fetal–Maternal Consortium; University of California, San Francisco Center for Maternal–Fetal Precision Medicine. Exome Sequencing for Prenatal Diagnosis in Nonimmune Hydrops Fetalis. N. Engl. J. Med. 2020, 383, 1746–1756. [Google Scholar] [CrossRef] [PubMed]
- Al-Kouatly, H.B.; Makhamreh, M.M.; Rice, S.M.; Smith, K.; Harman, C.; Quinn, A.; Valcarcel, B.N.; Firman, B.; Liu, R.; Hegde, M.; et al. High diagnosis rate for nonimmune hydrops fetalis with prenatal clinical exome from the Hydrops-Yielding Diagnostic Results of Prenatal Sequencing (HYDROPS) Study. Genet. Med. 2021, 23, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhou, J.; Wei, X.; Yao, R.; Yang, Y.; Deng, L.; Zou, G.; Wang, X.; Yang, Y.; Duan, T.; et al. Value of Exome Sequencing in Diagnosis and Management of Recurrent Non-immune Hydrops Fetalis: A Retrospective Analysis. Front. Genet. 2021, 12, 616392. [Google Scholar] [CrossRef] [PubMed]
- Norton, M.E.; Ziffle, J.V.; Lianoglou, B.R.; Hodoglugil, U.; Devine, W.P.; Sparks, T.N. Exome sequencing vs targeted gene panels for the evaluation of nonimmune hydrops fetalis. Am. J. Obstet. Gynecol. 2022, 226, 128.e1–128.e11. [Google Scholar] [CrossRef] [PubMed]
- Fetal Medicine Subgroup, Chinese Society of Perinatal Medicine, Chinese Medical Association; Obstetrics Subgroup, Chinese Society of Obstetrics and Gynecology, Chinese Medical Association. Guideline for nonimmune hydrops fetalis. Zhonghua Fu Chan Ke Za Zhi 2017, 52, 721–727. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Beke, A.; Joó, J.G.; Csaba, A.; Lázár, L.; Bán, Z.; Papp, C.; Tóth-Pál, E.; Papp, Z. Incidence of chromosomal abnormalities in the presence of fetal subcutaneous oedema, such as nuchal oedema, cystic hygroma and non-immune hydrops. Fetal Diagn. Ther. 2009, 25, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Donarini, G.; Paladini, D.; Calevo, M.G.; Bellini, T.; Ramenghi, L.A.; Hennekam, R.C. Etiology of non-immune hydrops fetalis: An update. Am. J. Med. Genet. Part A 2015, 167, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Sileo, F.G.; Kulkarni, A.; Branescu, I.; Homfray, T.; Dempsey, E.; Mansour, S.; Thilaganathan, B.; Bhide, A.; Khalil, A. Non-immune fetal hydrops: Etiology and outcome according to gestational age at diagnosis. Ultrasound Obstet. Gynecol. 2020, 56, 416–421. [Google Scholar] [CrossRef]
- Turan, O.; Hirfanglu, I.M.; Beken, S.; Biri, A.; Efeturk, T.; Atalay, Y. Prenatally detected congenital cystic adenomatoid malformation and postnatally diagnosed trisomy 13: Case report and review of the literature. Turk J. Pediatr. 2011, 53, 337–341. [Google Scholar]
- Sirotkina, M.; Douroudis, K.; Westgren, M.; Papadogiannakis, N. Genetic Analysis of Copy Number Variation in Large Chorangiomas. Pediatr. Dev. Pathol. 2019, 22, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.M. Fetal Arrhythmias: Genetic Background and Clinical Implications. Pediatr. Cardiol. 2019, 40, 247–256. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Values |
|---|---|
| Maternal age, years, mean ± SD | 29.6 ± 4.6 |
| Spontaneous conception, n (%) | 132 (91.0) |
| Recurrent NIHF §, n (%) | 31 (21.4) |
| GA at diagnosis, wks, median (range) | 24.3 (13.0, 35.0) |
| <16 weeks, n (%) | 15 (10.3) |
| Accompanied with abnormal structure ‡, n (%) | 57 (39.3%) |
| Termination of pregnancy, n (%) | 102 (70.3%) |
| Continued pregnancy, n (%) | 43 (29.7%) |
| Intrauterine fetal death, n (%) | 5 (11.6%) |
| Neonatal death, n (%) | 14 (36.8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Yang, Y.; Zhou, J.; Zhou, X.; Xiong, S.; Chen, J.; Zhou, F.; Zou, G.; Sun, L. An Investigation of the Etiologies of Non-Immune Hydrops Fetalis in the Era of Next-Generation Sequence—A Single Center Experience. Genes 2022, 13, 2231. https://doi.org/10.3390/genes13122231
Wei X, Yang Y, Zhou J, Zhou X, Xiong S, Chen J, Zhou F, Zou G, Sun L. An Investigation of the Etiologies of Non-Immune Hydrops Fetalis in the Era of Next-Generation Sequence—A Single Center Experience. Genes. 2022; 13(12):2231. https://doi.org/10.3390/genes13122231
Chicago/Turabian StyleWei, Xing, Yingjun Yang, Jia Zhou, Xinyao Zhou, Shiyi Xiong, Jianping Chen, Fenhe Zhou, Gang Zou, and Luming Sun. 2022. "An Investigation of the Etiologies of Non-Immune Hydrops Fetalis in the Era of Next-Generation Sequence—A Single Center Experience" Genes 13, no. 12: 2231. https://doi.org/10.3390/genes13122231
APA StyleWei, X., Yang, Y., Zhou, J., Zhou, X., Xiong, S., Chen, J., Zhou, F., Zou, G., & Sun, L. (2022). An Investigation of the Etiologies of Non-Immune Hydrops Fetalis in the Era of Next-Generation Sequence—A Single Center Experience. Genes, 13(12), 2231. https://doi.org/10.3390/genes13122231




