Whole-Chromosome Karyotyping of Fetal Nucleated Red Blood Cells Using the Ion Proton Sequencing Platform
Abstract
1. Introduction
2. Methods
2.1. Sample Collection and Processing
2.2. Massively Parallel Sequencing
2.3. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, T. Combined first trimester screen or noninvasive prenatal testing or both. Singap. Med. J. 2015, 56, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Gadsbøll, K.; Petersen, O.B.; Gatinois, V.; Strange, H.; Jacobsson, B.; Wapner, R.; Vermeesch, J.R.; Vogel, I. Current use of noninvasive prenatal testing in Europe, Australia and the USA: A graphical presentation. Acta Obstet. Gynecol. Scand. 2020, 99, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Van der Meij, K.R.M.; Sistermans, E.A.; Macville, M.V.; Stevens, S.J.; Bax, C.J.; Bekker, M.N.; Bilardo, C.M.; Boon, E.M.; Boter, M.; Diderich, K.E.; et al. TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am. J. Hum. Genet. 2019, 105, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, K.H. Screening for fetal aneuploidies at 11 to 13 weeks. Prenat. Diagn. 2011, 31, 7–15. [Google Scholar] [CrossRef]
- Kagan, K.O.; Maier, V.; Sonek, J.; Abele, H.; Lüthgens, K.; Schmid, M.; Wagner, P.; Hoopmann, M. False-Positive Rate in First-Trimester Screening Based on Ultrasound and Cell-Free DNA versus First-Trimester Combined Screening with Additional Ultrasound Markers. Fetal Diagn. Ther. 2019, 45, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.W.K.; Chan, K.C.A.; Gao, Y.; Lau, V.Y.M.; Zheng, W.; Leung, T.Y.; Foo, C.H.F.; Xie, B.; Tsui, N.B.Y.; Lun, F.M.F.; et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 20458–20463. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef]
- Allyse, M.; Minear, M.; Rote, M.; Hung, A.; Chandrasekharan, S.; Berson, E.; Sridhar, S. Non-invasive prenatal testing: A review of international implementation and challenges. Int. J. Women’s Health 2015, 7, 113–126. [Google Scholar] [CrossRef]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gynecol. 2017, 50, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Phillips, S.; Freeman, K.; Geppert, J.; Agbebiyi, A.; Uthman, O.A.; Madan, J.; Clarke, A.; Quenby, S.; Clarke, A. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: A systematic review and meta-analysis. BMJ Open 2016, 6, e010002. [Google Scholar] [CrossRef] [PubMed]
- Advani, H.V.; Barrett, A.N.; Evans, M.I.; Choolani, M. Challenges in non-invasive prenatal screening for sub-chromosomal copy number variations using cell-free DNA. Prenat. Diagn. 2017, 37, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Cram, D.S.; Tan, H.; Linpeng, S.; Liu, Y.; Sun, H.; Zhang, Y.; Tian, F.; Zhu, H.; Xu, M.; et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet. Med. 2019, 21, 1998–2006. [Google Scholar] [CrossRef]
- Gregg, A.R.; Skotko, B.G.; Benkendorf, J.L.; Monaghan, K.G.; Bajaj, K.; Best, R.G.; Klugman, S.; Watson, M.S. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: A position statement of the American College of Medical Genetics and Genomics. Genet. Med. 2016, 18, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Bonanni, G.; Trevisan, V.; Zollino, M.; De Santis, M.; Romanzi, F.; Lanzone, A.; Bevilacqua, E. Case Report: Challenges of Non-Invasive Prenatal Testing (NIPT): A Case Report of Confined Placental Mosaicism and Clinical Considerations. Front. Genet. 2022, 13, 881284. [Google Scholar] [CrossRef] [PubMed]
- Tabor, A.; Alfirevic, Z. Update on Procedure-Related Risks for Prenatal Diagnosis Techniques. Fetal Diagn. Ther. 2010, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.M.; Molina, F.S.; Rodríguez-Fernández, M.; Delgado, J.L.; Carrillo, M.P.; Jani, J.; Plasencia, W.; Stratieva, V.; Maíz, N.; Carretero, P.; et al. New approach for estimating risk of miscarriage after chorionic villus sampling. Ultrasound Obstet. Gynecol. 2020, 56, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Akolekar, R.; Beta, J.; Picciarelli, G.; Ogilvie, C.; D’Antonio, F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: A systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Johnson, J.-A.; Langlois, S.; Lee, H.; Winsor, S.; Dineley, B.; Horniachek, M.; Lalatta, F.; Ronzoni, L.; Barrett, A.N.; et al. Preferences for prenatal tests for Down syndrome: An international comparison of the views of pregnant women and health professionals. Eur. J. Hum. Genet. 2016, 24, 968–975. [Google Scholar] [CrossRef]
- Choolani, M.; Mahyuddin, A.P.; Hahn, S. The promise of fetal cells in maternal blood. Best Pract. Res. Clin. Obstet. Gynaecol. 2012, 26, 655–667. [Google Scholar] [CrossRef]
- Choolani, M.; O’Donoghue, K.; Talbert, D.; Kumar, S.; Roberts, I.; Letsky, E.; Bennett, P.R.; Fisk, N. Characterization of first trimester fetal erythroblasts for non-invasive prenatal diagnosis. Mol. Hum. Reprod. 2003, 9, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Mavrou, A.; Kouvidi, E.; Antsaklis, A.; Souka, A.; Tzeli, S.K.; Kolialexi, A. Identification of nucleated red blood cells in maternal circulation: A second step in screening for fetal aneuploidies and pregnancy complications. Prenat. Diagn. 2007, 27, 150–153. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, K.; Guo, Y.; Wei, X.; Sun, Y.; Cai, B.; Shi, Y.; Du, Y.; Liu, Y.; Fan, C.; et al. The isolation and analysis of fetal nucleated red blood cells using multifunctional microbeads with a nanostructured coating toward early noninvasive prenatal diagnostics. J. Mater. Chem. B 2021, 9, 3047–3054. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Lewis, D.E.; Bischoff, F.Z.; Simpson, J.L. Isolation and genetic analysis of fetal nucleated red blood cells from maternal blood: The Baylor College of Medicine experience. Early Hum. Dev. 1996, 47, S85–S88. [Google Scholar] [CrossRef]
- Bianchi, D.W. Fetal cells in the maternal circulation: Feasibility for prenatal diagnosis. Br. J. Haematol. 1999, 105, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.; Barrett, A.N.; Tan, T.Z.; Huang, Z.; Mahyuddin, A.P.; Ponnusamy, S.; Sandhu, J.S.; Ho, S.S.Y.; Chan, J.K.Y.; Chong, S.; et al. Detection of aneuploidy from single fetal nucleated red blood cells using whole genome sequencing. Prenat. Diagn. 2015, 35, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Grati, F.R.; Malvestiti, F.; Ferreira, J.C.P.B.; Bajaj, K.; Gaetani, E.; Agrati, C.; Grimi, B.; Dulcetti, F.; Ruggeri, A.M.; De Toffol, S.; et al. Fetoplacental mosaicism: Potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet. Med. 2014, 16, 620–624. [Google Scholar] [CrossRef] [PubMed]
- FDA, U. Genetic Non-Invasive Prenatal Screening Tests May Have False Results: FDA Safety Communication. Available online: https://www.fda.gov/medical-devices/safety-communications/genetic-non-invasive-prenatal-screening-tests-may-have-false-results-fda-safety-communication (accessed on 1 November 2022).
- Breman, A.M.; Chow, J.C.; U’Ren, L.; Normand, E.A.; Qdaisat, S.; Zhao, L.; Henke, D.M.; Chen, R.; Shaw, C.A.; Jackson, L.; et al. Evidence for feasibility of fetal trophoblastic cell-based noninvasive prenatal testing. Prenat. Diagn. 2016, 36, 1009–1019. [Google Scholar] [CrossRef]
- Vossaert, L.; Wang, Q.; Salman, R.; McCombs, A.K.; Patel, V.; Qu, C.; Mancini, M.A.; Edwards, D.P.; Malovannaya, A.; Liu, P.; et al. Validation Studies for Single Circulating Trophoblast Genetic Testing as a Form of Noninvasive Prenatal Diagnosis. Am. J. Hum. Genet. 2019, 105, 1262–1273. [Google Scholar] [CrossRef]
- Crovetti, B.; Maktabi, M.A.; Erfani, H.; Panchalee, T.; Wang, Q.; Vossaert, L.; Van den Veyver, I. Circulating trophoblast numbers as a potential marker for pregnancy complications. Prenat. Diagn. 2022, 42, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Afshar, Y.; Dong, J.; Zhao, P.; Li, L.; Wang, S.; Zhang, R.Y.; Zhang, C.; Yin, O.; Han, C.S.; Einerson, B.D.; et al. Circulating trophoblast cell clusters for early detection of placenta accreta spectrum disorders. Nat. Commun. 2021, 12, 4408. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, I.; Benachi, A.; Saker, A.; Bonnefont, J.P.; Mouawia, H.; Broncy, L.; Frydman, R.; Brival, M.L.; Lacour, B.; Dachez, R.; et al. Cervical trophoblasts for non-invasive single-cell genotyping and prenatal diagnosis. Placenta 2016, 37, 56–60. [Google Scholar] [CrossRef]
- Bailey-Hytholt, C.M.; Sayeed, S.; Shukla, A.; Tripathi, A. Enrichment of Placental Trophoblast Cells from Clinical Cervical Samples Using Differences in Surface Adhesion on an Inclined Plane. Ann. Biomed. Eng. 2021, 49, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.V.; Kadam, L.; van Dijk, M.; Kohan-Ghadr, H.-R.; Kilburn, B.A.; Hartman, C.; Mazzorana, V.; Visser, A.; Hertz, M.; Bolnick, A.D.; et al. Fetal genome profiling at 5 weeks of gestation after noninvasive isolation of trophoblast cells from the endocervical canal. Sci. Transl. Med. 2016, 8, 3634. [Google Scholar] [CrossRef] [PubMed]
- Palis, J.; Yoder, M.C. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp. Hematol. 2001, 29, 927–936. [Google Scholar] [CrossRef]
- Van Handel, B.; Prashad, S.L.; Hassanzadeh-Kiabi, N.; Huang, A.; Magnusson, M.; Atanassova, B.; Chen, A.; Hamalainen, E.I.; Mikkola, H.K.A. The first trimester human placenta is a site for terminal maturation of primitive erythroid cells. Blood 2010, 116, 3321–3330. [Google Scholar] [CrossRef] [PubMed]
- Babochkina, T.; Mergenthaler, S.; De Napoli, G.; Hristoskova, S.; Tercanli, S.; Holzgreve, W.; Hahn, S. Numerous erythroblasts in maternal blood are impervious to fluorescent in situ hybridization analysis, a feature related to a dense compact nucleus with apoptotic character. Haematologica 2005, 90, 740–745. [Google Scholar]
- Bianchi, D.W.; Williams, J.M.; Sullivan, L.M.; Hanson, F.W.; Klinger, K.W.; Shuber, A.P. PCR Quantitation of Fetal Cells in Maternal Blood in Normal and Aneuploid Pregnancies. Am. J. Hum. Genet. 1997, 61, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.C.C.; Mahyuddin, A.P.; Liu, Y.; Wong, C.C.; Choolani, M. Isolation of fetal nucleated red blood cells using a microfilter chip for non-invasive prenatal diagnosis. Prenat. Diagn. 2014, 34, 22–86. [Google Scholar] [CrossRef]
- Kadam, P.; Ponnusamy, S.; Zhang, H.; Mahyuddin, A.P.; Ismail, N.S.; Shikkander, N.; Tan, S.L.; Elanvogan, A.; Choolani, M. A novel marker for isolation of fetal nucleated red blood cells for non-invasive prenatal diagnosis. Prenat. Diagn. 2012, 32, 1–128. [Google Scholar]
- Wang, Z.; Cheng, L.; Sun, Y.; Wei, X.; Cai, B.; Liao, L.; Zhang, Y.; Zhao, X.-Z. Enhanced Isolation of Fetal Nucleated Red Blood Cells by Enythrocyte-Leukocyte Hybrid Membrane-Coated Magnetic Nanoparticles for Noninvasive Pregnant Diagnostics. Anal. Chem. 2021, 93, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal Microarray versus Karyotyping for Prenatal Diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Yaron, Y.; Jani, J.; Schmid, M.; Oepkes, D. Current Status of Testing for Microdeletion Syndromes and Rare Autosomal Trisomies Using Cell-Free DNA Technology. Obstet. Gynecol. 2015, 126, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.K.; Karampetsou, E.; Boustred, C.; McKay, F.; Mason, S.; Hill, M.; Plagnol, V.; Chitty, L.S. Limited Clinical Utility of Non-invasive Prenatal Testing for Subchromosomal Abnormalities. Am. J. Hum. Genet. 2016, 98, 34–44. [Google Scholar] [CrossRef] [PubMed]
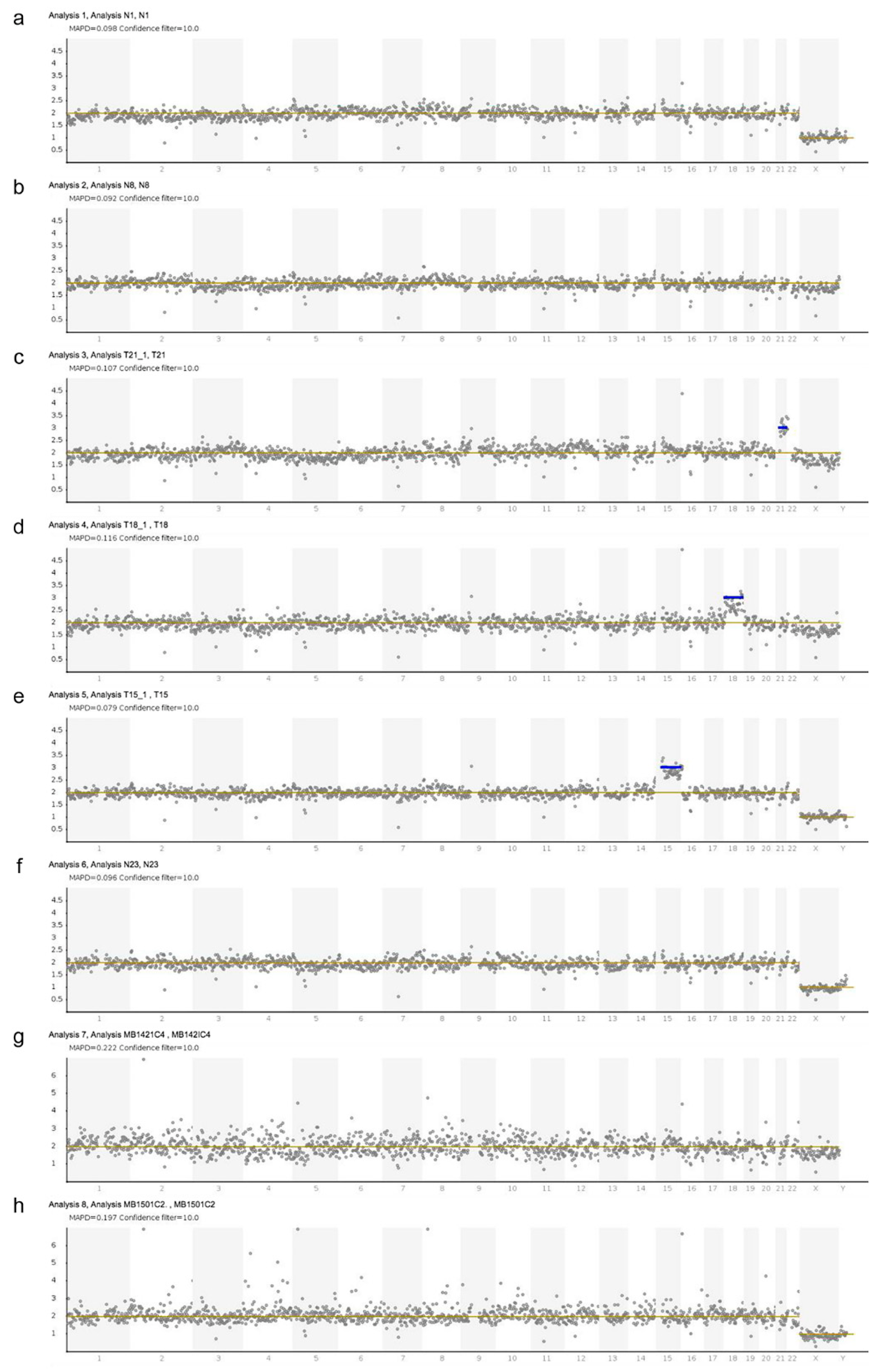
| Sample | Sample Type | Karyotype | Gestation (Weeks) | NIPD Genotype | NIPD Fetal Sex | MAPD |
|---|---|---|---|---|---|---|
| N1 | Post-TOP villi | 46,XY | 8 + 2 | Normal | M | 0.098 |
| N2 | Post-TOP villi | 46,XY | 8 + 2 | Normal | M | 0.092 |
| N3 | Post-TOP villi | 46,XX | 8 + 1 | Normal | F | 0.103 |
| N4 | Post-TOP villi | 46,XX | 8 + 1 | Normal | F | 0.088 |
| N5 | Post-TOP villi | 46,XX | 8 + 2 | Normal | F | 0.095 |
| N6 | Post-TOP villi | 46,XX | 8 + 2 | Normal | F | 0.096 |
| N7 | Post-TOP villi | 46,XX | 9 + 0 | Normal | F | 0.094 |
| N8 | Post-TOP villi | 46,XX | 9 + 0 | Normal | F | 0.092 |
| N9 | Post-TOP villi | 46,XY | 8 + 1 | Normal | M | 0.087 |
| N10 | Post-TOP villi | 46,XY | 8 + 1 | Normal | M | 0.098 |
| N11 | Post-TOP villi | 46,XX | 8 + 3 | Normal | F | 0.096 |
| N12 | Post-TOP villi | 46,XX | 8 + 3 | Normal | F | 0.106 |
| N13 | Post-TOP villi | 46,XY | 10 + 0 | Normal | M | 0.091 |
| N14 | Post-TOP villi | 46,XY | 10 + 0 | Normal | M | 0.102 |
| N15 | Post-TOP villi | 46,XX | 8 + 0 | Normal | F | 0.102 |
| N16 | Post-TOP villi | 46,XX | 8 + 0 | Normal | F | 0.110 |
| N17 | Post-TOP villi | 46,XX | 8 + 4 | Normal | F | 0.098 |
| N18 | Post-TOP villi | 46,XX | 8 + 4 | Normal | F | 0.100 |
| N19 | Post-TOP villi | 46,XX | 8 + 2 | Normal | F | 0.106 |
| N20 | Post-TOP villi | 46,XX | 9 + 0 | Normal | F | 0.100 |
| N21 | Post-TOP villi | 46,XX | 8 + 4 | Normal | F | 0.107 |
| N22 | Post-TOP villi | 46,XX | 8 + 4 | Normal | F | 0.100 |
| N23 | Post-TOP villi | 47,XY,t(15;18) (p10;q10),+18(4)/46,XY(16) | 8 + 6 | Normal | M | 0.096 |
| N24 | Post-TOP villi | 47,XY,t(15;18) (p10;q10),+18(4)/46,XY(16) | 8 + 6 | Normal | M | 0.098 |
| N25 | Post-TOP villi | 46,XY | 8 + 1 | Normal | M | 0.105 |
| N26 | Post-TOP villi | 46,XY | 8 + 4 | Normal | M | 0.099 |
| T21_1 | CVS villi | 47,XX,+21 | 12 + 0 | T21 | F | 0.107 |
| T21_2 | CVS villi | 47,XY,+21 | 13 + 0 | T21 | M | 0.099 |
| T18_1 | CVS villi | 47,XX,+18 | 12 + 4 | T18 | F | 0.116 |
| T18_2 | CVS villi | 47,XX,+18 | 12 + 4 | T18 | F | 0.133 |
| T15_1 | Post-TOP villi | 47,XY,+15 | 8 + 1 | T15 | M | 0.079 |
| T15_2 | Post-TOP villi | 47,XY,+15 | 8 + 1 | T15 | M | 0.090 |
| MB142_1c2 | Post-TOP MB | 46,XX | 8 + 2 | Normal | F | 0.224 |
| MB142_1c3 | Post-TOP MB | 46,XX | 8 + 2 | Normal | F | 0.201 |
| MB142_1c4 | Post-TOP MB | 46,XX | 8 + 2 | Normal | F | 0.222 |
| MB150_1c1 | Post-TOP MB | 46,XY | 9 + 0 | Normal | M | 0.171 |
| MB150_1c2 | Post-TOP MB | 46,XY | 9 + 0 | Normal | M | 0.197 |
| MB150_1c3 | Post-TOP MB | 46,XY | 9 + 0 | Normal | M | 1.251 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrett, A.N.; Huang, Z.; Aung, S.; Ho, S.S.Y.; Roslan, N.S.; Mahyuddin, A.P.; Biswas, A.; Choolani, M. Whole-Chromosome Karyotyping of Fetal Nucleated Red Blood Cells Using the Ion Proton Sequencing Platform. Genes 2022, 13, 2257. https://doi.org/10.3390/genes13122257
Barrett AN, Huang Z, Aung S, Ho SSY, Roslan NS, Mahyuddin AP, Biswas A, Choolani M. Whole-Chromosome Karyotyping of Fetal Nucleated Red Blood Cells Using the Ion Proton Sequencing Platform. Genes. 2022; 13(12):2257. https://doi.org/10.3390/genes13122257
Chicago/Turabian StyleBarrett, Angela N., Zhouwei Huang, Sarah Aung, Sherry S. Y. Ho, Nur Syazana Roslan, Aniza P. Mahyuddin, Arijit Biswas, and Mahesh Choolani. 2022. "Whole-Chromosome Karyotyping of Fetal Nucleated Red Blood Cells Using the Ion Proton Sequencing Platform" Genes 13, no. 12: 2257. https://doi.org/10.3390/genes13122257
APA StyleBarrett, A. N., Huang, Z., Aung, S., Ho, S. S. Y., Roslan, N. S., Mahyuddin, A. P., Biswas, A., & Choolani, M. (2022). Whole-Chromosome Karyotyping of Fetal Nucleated Red Blood Cells Using the Ion Proton Sequencing Platform. Genes, 13(12), 2257. https://doi.org/10.3390/genes13122257





