Plasmid Genomes Reveal the Distribution, Abundance, and Organization of Mercury-Related Genes and Their Co-Distribution with Antibiotic Resistant Genes in Gammaproteobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval of the Plasmid Genomic Sequences
2.2. Annotation of HRGs
2.3. Annotation of the ARGs
2.4. Co-Distribution Analysis of the ARGs and HRGs
2.5. Analysis of Preference of the HRGs for Specific ARG Classes
2.6. Statistical Analysis
3. Results and Discussion
3.1. Overview of the Plasmid Host Taxonomy
3.2. Distribution of the HRGs in Plasmid Genomes
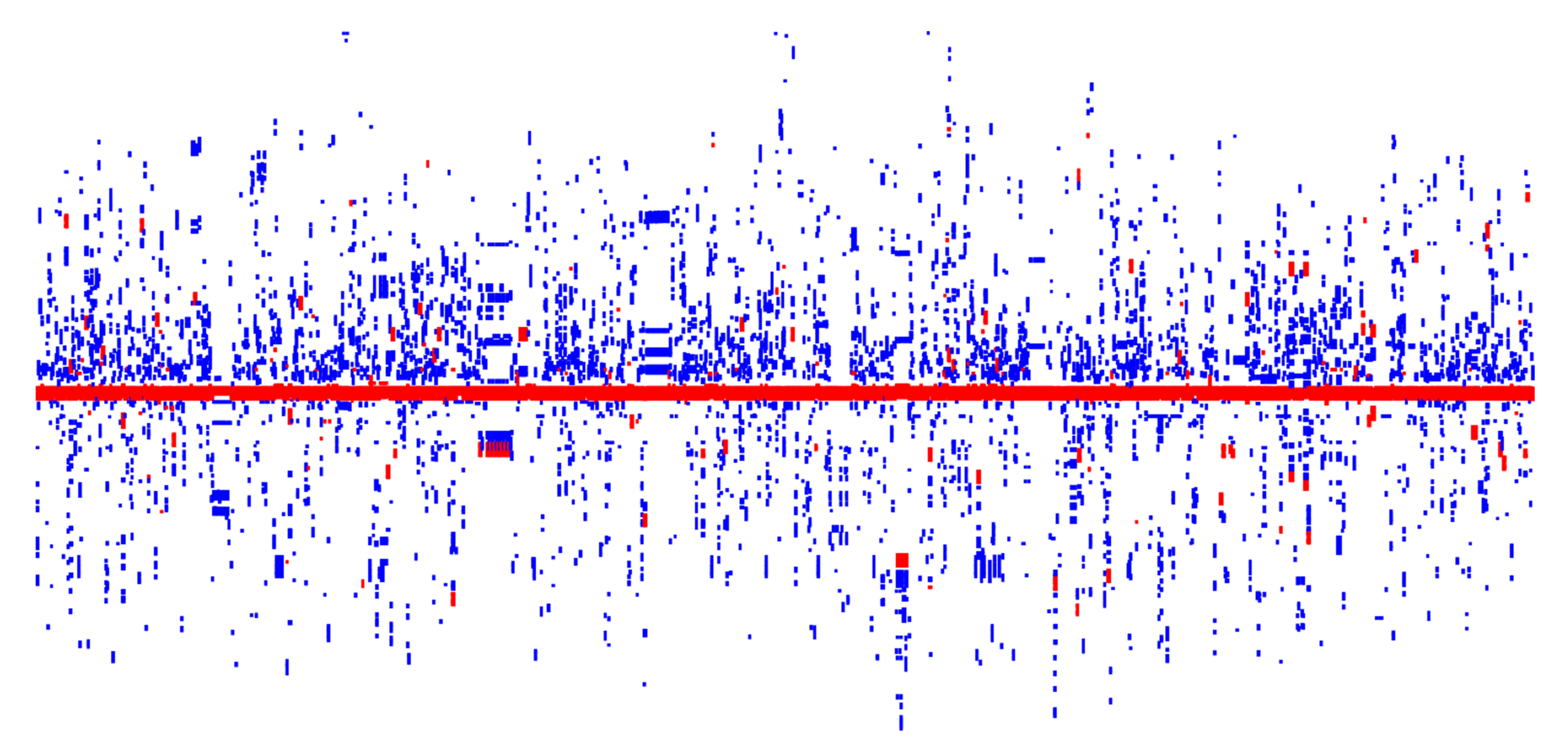
3.3. HRG Clusters Are Highly Abundant and Have an Extremely High Organizational Diversity
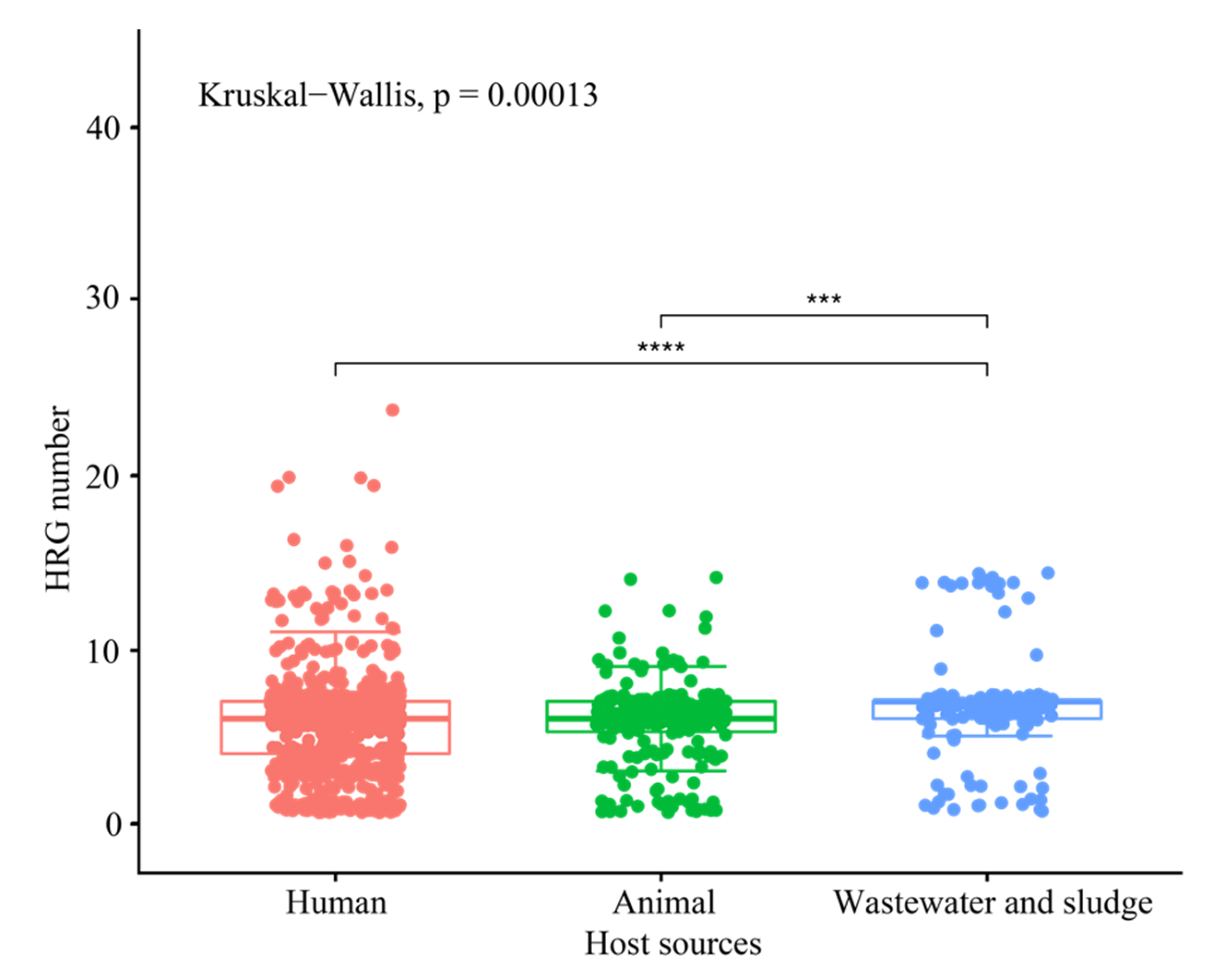
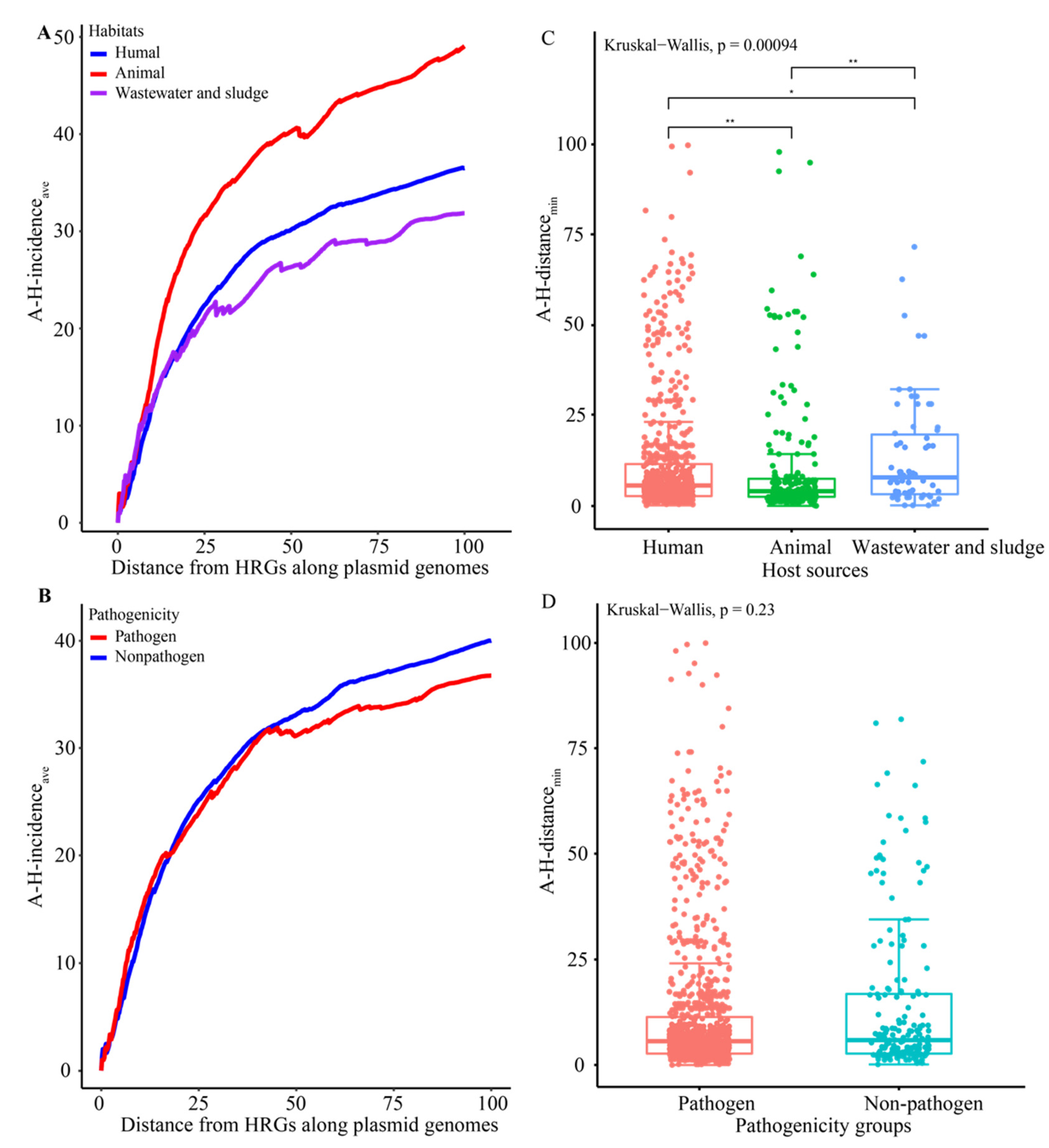
3.4. Co-Distribution of HRGs and ARGs in Plasmid Genomes
3.5. Preference of HRGs for Specific ARG Classes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Selin, N.E. Global Biogeochemical Cycling of Mercury: A Review. Annu. Rev. Environ. Resour. 2009, 34, 43–63. [Google Scholar] [CrossRef]
- Mergler, D.; Anderson, H.A.; Chan, L.H.; Mahaffey, K.R.; Murray, M.; Sakamoto, M.; Stern, A.H.; Panel on Health, R.; Toxicological Effects of, M. Methylmercury exposure and health effects in humans: A worldwide concern. Ambio 2007, 36, 3–11. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Johs, A.; Zhao, L.; Wang, T.; Yang, Z.; Lin, H.; Elias, D.A.; Pierce, E.M.; Liang, L.; et al. Anaerobic Mercury Methylation and Demethylation by Geobacter bemidjiensis Bem. Environ. Sci. Technol. 2016, 50, 4366–4373. [Google Scholar] [CrossRef] [PubMed]
- Barkay, T.; Miller, S.M.; Summers, A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. [Google Scholar] [CrossRef]
- Petrus, A.K.; Rutner, C.; Liu, S.; Wang, Y.; Wiatrowski, H.A. Mercury Reduction and Methyl Mercury Degradation by the Soil Bacterium Xanthobacter autotrophicus Py2. Appl. Environ. Microbiol. 2015, 81, 7833–7838. [Google Scholar] [CrossRef]
- Colombo, M.J.; Ha, J.; Reinfelder, J.R.; Barkay, T.; Yee, N. Oxidation of Hg(0) to Hg(II) by diverse anaerobic bacteria. Chem. Geol. 2014, 363, 334–340. [Google Scholar] [CrossRef]
- Natasha; Shahid, M.; Khalid, S.; Bibi, I.; Bundschuh, J.; Khan Niazi, N.; Dumat, C. A critical review of mercury speciation, bioavailability, toxicity and detoxification in soil-plant environment: Ecotoxicology and health risk assessment. Sci. Total Environ. 2020, 711, 134749. [Google Scholar] [CrossRef]
- Oremland, R.S.; Culbertson, C.W.; Winfrey, M.R. Methylmercury decomposition in sediments and bacterial cultures: Involvement of methanogens and sulfate reducers in oxidative demethylation. Appl. Environ. Microbiol. 1991, 57, 130–137. [Google Scholar] [CrossRef]
- Parks, J.M.; Johs, A.; Podar, M.; Bridou, R.; Hurt, R.A., Jr.; Smith, S.D.; Tomanicek, S.J.; Qian, Y.; Brown, S.D.; Brandt, C.C.; et al. The genetic basis for bacterial mercury methylation. Science 2013, 339, 1332–1335. [Google Scholar] [CrossRef]
- Boyd, E.S.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef]
- Barkay, T.; Kritee, K.; Boyd, E.; Geesey, G. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 2010, 12, 2904–2917. [Google Scholar] [CrossRef]
- Podar, M.; Gilmour, C.C.; Brandt, C.C.; Soren, A.; Brown, S.D.; Crable, B.R.; Palumbo, A.V.; Somenahally, A.C.; Elias, D.A. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv. 2015, 1, e1500675. [Google Scholar] [CrossRef]
- Gionfriddo, C.M.; Stott, M.B.; Power, J.F.; Ogorek, J.M.; Krabbenhoft, D.P.; Wick, R.; Holt, K.; Chen, L.X.; Thomas, B.C.; Banfield, J.F.; et al. Genome-Resolved Metagenomics and Detailed Geochemical Speciation Analyses Yield New Insights into Microbial Mercury Cycling in Geothermal Springs. Appl. Environ. Microbiol. 2020, 86, e00176-20. [Google Scholar] [CrossRef]
- Jones, D.S.; Johnson, N.W.; Mitchell, C.P.J.; Walker, G.M.; Bailey, J.V.; Pastor, J.; Swain, E.B. Diverse Communities of hgcAB(+) Microorganisms Methylate Mercury in Freshwater Sediments Subjected to Experimental Sulfate Loading. Environ. Sci. Technol. 2020, 54, 14265–14274. [Google Scholar] [CrossRef]
- Peterson, B.D.; McDaniel, E.A.; Schmidt, A.G.; Lepak, R.F.; Janssen, S.E.; Tran, P.Q.; Marick, R.A.; Ogorek, J.M.; DeWild, J.F.; Krabbenhoft, D.P.; et al. Mercury Methylation Genes Identified across Diverse Anaerobic Microbial Guilds in a Eutrophic Sulfate-Enriched Lake. Environ. Sci. Technol. 2020, 54, 15840–15851. [Google Scholar] [CrossRef]
- Barkay, T.; Gu, B. Demethylation─The Other Side of the Mercury Methylation Coin: A Critical Review. ACS Environ. Au 2021, 2, 77–97. [Google Scholar] [CrossRef]
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef]
- Lloyd, N.A.; Janssen, S.E.; Reinfelder, J.R.; Barkay, T. Co-selection of Mercury and Multiple Antibiotic Resistances in Bacteria Exposed to Mercury in the Fundulus heteroclitus Gut Microbiome. Curr. Microbiol. 2016, 73, 834–842. [Google Scholar] [CrossRef]
- Skurnik, D.; Ruimy, R.; Ready, D.; Ruppe, E.; Bernede-Bauduin, C.; Djossou, F.; Guillemot, D.; Pier, G.B.; Andremont, A. Is exposure to mercury a driving force for the carriage of antibiotic resistance genes? J. Med. Microbiol. 2010, 59, 804–807. [Google Scholar] [CrossRef]
- Yuan, L.; Li, Z.H.; Zhang, M.Q.; Shao, W.; Fan, Y.Y.; Sheng, G.P. Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci. Total Environ. 2019, 657, 1014–1022. [Google Scholar] [CrossRef]
- Gaeta, N.C.; de Carvalho, D.U.; Fontana, H.; Sano, E.; Moura, Q.; Fuga, B.; Munoz, P.M.; Gregory, L.; Lincopan, N. Genomic features of a multidrug-resistant and mercury-tolerant environmental Escherichia coli recovered after a mining dam disaster in South America. Sci. Total Environ. 2022, 823, 153590. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.; Wang, G. Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PLoS ONE 2014, 9, e92236. [Google Scholar] [CrossRef] [PubMed]
- Kothari, A.; Soneja, D.; Tang, A.; Carlson, H.K.; Deutschbauer, A.M.; Mukhopadhyay, A. Native Plasmid-Encoded Mercury Resistance Genes Are Functional and Demonstrate Natural Transformation in Environmental Bacterial Isolates. mSystems 2019, 4, e00588-19. [Google Scholar] [CrossRef] [PubMed]
- Perez-Palacios, P.; Delgado-Valverde, M.; Gual-de-Torrella, A.; Oteo-Iglesias, J.; Pascual, A.; Fernandez-Cuenca, F. Co-transfer of plasmid-encoded bla carbapenemases genes and mercury resistance operon in high-risk clones of Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2021, 105, 9231–9242. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Organism Groups. Available online: https://www.ncbi.nlm.nih.gov/pathogens/organisms/ (accessed on 8 October 2022).
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737-743. [Google Scholar] [CrossRef]
- Liu, Y.R.; Johs, A.; Bi, L.; Lu, X.; Hu, H.W.; Sun, D.; He, J.Z.; Gu, B. Unraveling Microbial Communities Associated with Methylmercury Production in Paddy Soils. Environ. Sci. Technol. 2018, 52, 13110–13118. [Google Scholar] [CrossRef]
- Zhang, L.; Philben, M.; Tas, N.; Johs, A.; Yang, Z.; Wullschleger, S.D.; Graham, D.E.; Pierce, E.M.; Gu, B. Unravelling biogeochemical drivers of methylmercury production in an Arctic fen soil and a bog soil. Environ. Pollut. 2022, 299, 118878. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Duan, D.; Peng, G.; Li, P.; Lei, P.; Zhong, H.; Tsui, M.T.; Pan, K. Effects and mechanisms of organic matter regulating the methylmercury dynamics in mangrove sediments. J. Hazard. Mater. 2022, 432, 128690. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Li, L.G.; Cai, L.; Zhang, X.X.; Zhang, T. Potentially novel copper resistance genes in copper-enriched activated sludge revealed by metagenomic analysis. Appl. Microbiol. Biotechnol. 2014, 98, 10255–10266. [Google Scholar] [CrossRef]
- Li, L.G.; Xia, Y.; Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Cooper, C.J.; Zheng, K.; Rush, K.W.; Johs, A.; Sanders, B.C.; Pavlopoulos, G.A.; Kyrpides, N.C.; Podar, M.; Ovchinnikov, S.; Ragsdale, S.W.; et al. Structure determination of the HgcAB complex using metagenome sequence data: Insights into microbial mercury methylation. Commun. Biol. 2020, 3, 320. [Google Scholar] [CrossRef]
- Yin, X.; Jiang, X.T.; Chai, B.; Li, L.; Yang, Y.; Cole, J.R.; Tiedje, J.M.; Zhang, T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Arango-Argoty, G.A.; Guron, G.K.P.; Garner, E.; Riquelme, M.V.; Heath, L.S.; Pruden, A.; Vikesland, P.J.; Zhang, L. ARGminer: A web platform for the crowdsourcing-based curation of antibiotic resistance genes. Bioinformatics 2020, 36, 2966–2973. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef]
- Moore, B. A new screen test and selective medium for the rapid detection of epidemic strains of Staphylococcus aureus. Lancet 1960, 276, 453–458. [Google Scholar] [CrossRef]
- Hintelmann, H. Organomercurials. Their formation and pathways in the environment. Met. Ions Life Sci. 2010, 7, 365–401. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshanee, M.; Chatterjee, S.; Rath, S.; Dash, H.R.; Das, S. Cellular and genetic mechanism of bacterial mercury resistance and their role in biogeochemistry and bioremediation. J. Hazard. Mater. 2022, 423, 126985. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Lingaswamy, B.; Koduru, T.N.; Nagu, P.P.; Jogadhenu, P.S.S. A putative merR family transcription factor Slr0701 regulates mercury inducible expression of MerA in the cyanobacterium Synechocystis sp. PCC6803. Microbiologyopen 2019, 8, e00838. [Google Scholar] [CrossRef] [PubMed]
- Cardona, G.I.; Escobar, M.C.; Acosta-Gonzalez, A.; Marin, P.; Marques, S. Highly mercury-resistant strains from different Colombian Amazon ecosystems affected by artisanal gold mining activities. Appl. Microbiol. Biotechnol. 2022, 106, 2775–2793. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Gomez-Alvarez, V.; Siponen, S.; Sarekoski, A.; Hokajarvi, A.M.; Kauppinen, A.; Torvinen, E.; Miettinen, I.T.; Pitkanen, T. Bacterial Genes Encoding Resistance Against Antibiotics and Metals in Well-Maintained Drinking Water Distribution Systems in Finland. Front. Microbiol. 2021, 12, 803094. [Google Scholar] [CrossRef]
- Li, X.; Chen, F.; Chen, Y. Gcluster: A simple-to-use tool for visualizing and comparing genome contexts for numerous genomes. Bioinformatics 2020, 36, 3871–3873. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Rodriguez-Beltran, J.; DelaFuente, J.; Leon-Sampedro, R.; MacLean, R.C.; San Millan, A. Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Mahbub, K.R.; King, W.L.; Siboni, N.; Nguyen, V.K.; Rahman, M.M.; Megharaj, M.; Seymour, J.R.; Franks, A.E.; Labbate, M. Long-lasting effect of mercury contamination on the soil microbiota and its co-selection of antibiotic resistance. Environ. Pollut. 2020, 265, 115057. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, H.W.; Su, J.Q.; Hao, X.; Guo, H.; Liu, Y.R.; Zhu, Y.G. Influence of Legacy Mercury on Antibiotic Resistomes: Evidence from Agricultural Soils with Different Cropping Systems. Environ. Sci. Technol. 2021, 55, 13913–13922. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef]

| Genus * | All Plasmids | Plasmids Harboring HRGs | Plasmids Harboring Both HRGs and ARGs | |||
|---|---|---|---|---|---|---|
| Plasmid Number | Percent (%) | Plasmid Number | Percent (%) | Plasmid Number | Percent (%) | |
| Escherichia | 5508 | 29.41 | 267 | 15.50 | 258 | 18.86 |
| Klebsiella | 4714 | 25.17 | 614 | 35.64 | 542 | 39.62 |
| Acinetobacter | 1438 | 7.68 | 58 | 3.37 | 19 | 1.39 |
| Salmonella | 1343 | 7.17 | 249 | 14.45 | 178 | 13.01 |
| Enterobacter | 1031 | 5.50 | 175 | 10.16 | 150 | 10.96 |
| Citrobacter | 655 | 3.50 | 139 | 8.07 | 75 | 5.48 |
| Xanthomonas | 398 | 2.12 | 1 | 0.06 | 1 | 0.07 |
| Pseudomonas | 385 | 2.06 | 75 | 4.35 | 38 | 2.78 |
| Shigella | 288 | 1.54 | 4 | 0.23 | 3 | 0.22 |
| Vibrio | 258 | 1.38 | 1 | 0.06 | 1 | 0.07 |
| Aeromonas | 225 | 1.20 | 29 | 1.68 | 25 | 1.83 |
| Pantoea | 197 | 1.05 | 4 | 0.23 | 4 | 0.29 |
| Yersinia | 187 | 1.00 | 1 | 0.06 | 1 | 0.07 |
| Serratia | 149 | 0.80 | 17 | 0.99 | 12 | 0.88 |
| Raoultella | 113 | 0.60 | 16 | 0.93 | 11 | 0.80 |
| Proteus | 81 | 0.43 | 10 | 0.58 | 10 | 0.73 |
| Providencia | 77 | 0.41 | 6 | 0.35 | 6 | 0.44 |
| Unclassed genera | 58 | 0.31 | 7 | 0.41 | 6 | 0.44 |
| Moraxella | 54 | 0.29 | 4 | 0.23 | ||
| Cronobacter | 52 | 0.28 | 2 | 0.12 | 2 | 0.15 |
| Shewanella | 47 | 0.25 | 8 | 0.46 | 6 | 0.44 |
| Edwardsiella | 38 | 0.20 | 1 | 0.06 | 1 | 0.07 |
| Leclercia | 37 | 0.20 | 12 | 0.70 | 12 | 0.88 |
| Pseudoalteromonas | 25 | 0.13 | 2 | 0.12 | ||
| Alteromonas | 19 | 0.10 | 3 | 0.17 | ||
| Halomonas | 14 | 0.07 | 4 | 0.23 | ||
| Marinobacter | 11 | 0.06 | 2 | 0.12 | ||
| Stenotrophomonas | 10 | 0.05 | 4 | 0.23 | 2 | 0.15 |
| Kluyvera | 7 | 0.04 | 2 | 0.12 | 1 | 0.07 |
| Phytobacter | 5 | 0.03 | 3 | 0.17 | 3 | 0.22 |
| Mixta | 3 | 0.02 | 1 | 0.06 | 1 | 0.07 |
| Marinomonas | 1 | 0.01 | 1 | 0.06 | ||
| Salinimonas | 1 | 0.01 | 1 | 0.06 | ||
| HRG Cluster Type | Incidence * | Percent of Incidences (%) |
|---|---|---|
| merR-merT-merP-X-merA-merD-merE | 543 | 32.32 |
| merR-merT-merP-merC-merA-merD-merE | 457 | 27.20 |
| merA-merD-merE | 96 | 5.71 |
| merR-merT-merP-merA-merB-merD-merE | 77 | 4.58 |
| merR-merT-merP-X-merA | 76 | 4.52 |
| merR-merT-merP-merF-merA-merD-merE | 62 | 3.69 |
| merD-merE | 48 | 2.86 |
| merR-merT-merP-merA-merD-merE | 39 | 2.32 |
| merR-merT-merP | 31 | 1.85 |
| merR-X-merB-merD-merE | 27 | 1.61 |
| merR-merT-merP-X-merA-merD | 16 | 0.95 |
| merR-X-X-merC-merA-merD | 16 | 0.95 |
| merR-merT-merP-merC-merA-merD | 14 | 0.83 |
| merR-merT | 11 | 0.65 |
| merR-merT-merP-merC-merA | 10 | 0.60 |
| merR-merT-merP-merA-merG-merB1 | 9 | 0.54 |
| merR-merT-merP-merA-merB-X-X-merE | 8 | 0.48 |
| merT-merP-X-merA-merD-merE | 7 | 0.42 |
| merP-X-merA | 6 | 0.36 |
| merR-X-X-merC-X-X-merD | 6 | 0.36 |
| merT-merP-merC-merA-merD-merE | 5 | 0.30 |
| merP-X-merA-merD-merE | 5 | 0.30 |
| merB-merD-merE | 5 | 0.30 |
| merR-X-merC-merA | 5 | 0.30 |
| ARG Class Nearest to HRGs | Plasmid Number | Percentage (%) |
|---|---|---|
| sulfonamide | 351 | 26.77 |
| macrolide-lincosamide-streptogramin | 332 | 25.32 |
| aminoglycoside | 195 | 14.87 |
| tetracycline | 154 | 11.75 |
| β-lactam | 126 | 9.61 |
| chloramphenicol | 53 | 4.04 |
| trimethoprim | 33 | 2.52 |
| quinolone | 26 | 1.98 |
| polymyxin | 20 | 1.53 |
| vancomycin | 11 | 0.84 |
| fosfomycin | 6 | 0.46 |
| rifamycin | 3 | 0.23 |
| kasugamycin | 1 | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, Z.; Zhang, G.; Si, S.; Wu, X.; Cai, L. Plasmid Genomes Reveal the Distribution, Abundance, and Organization of Mercury-Related Genes and Their Co-Distribution with Antibiotic Resistant Genes in Gammaproteobacteria. Genes 2022, 13, 2149. https://doi.org/10.3390/genes13112149
Li X, Yang Z, Zhang G, Si S, Wu X, Cai L. Plasmid Genomes Reveal the Distribution, Abundance, and Organization of Mercury-Related Genes and Their Co-Distribution with Antibiotic Resistant Genes in Gammaproteobacteria. Genes. 2022; 13(11):2149. https://doi.org/10.3390/genes13112149
Chicago/Turabian StyleLi, Xiangyang, Zilin Yang, Guohui Zhang, Shengli Si, Xianzhi Wu, and Lin Cai. 2022. "Plasmid Genomes Reveal the Distribution, Abundance, and Organization of Mercury-Related Genes and Their Co-Distribution with Antibiotic Resistant Genes in Gammaproteobacteria" Genes 13, no. 11: 2149. https://doi.org/10.3390/genes13112149
APA StyleLi, X., Yang, Z., Zhang, G., Si, S., Wu, X., & Cai, L. (2022). Plasmid Genomes Reveal the Distribution, Abundance, and Organization of Mercury-Related Genes and Their Co-Distribution with Antibiotic Resistant Genes in Gammaproteobacteria. Genes, 13(11), 2149. https://doi.org/10.3390/genes13112149






