Polymorphic Variants of AGT, ABCA1, and CYBA Genes Influence the Survival of Patients with Coronary Artery Disease: A Prospective Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample
2.2. Follow-Up and Events
2.3. Statistical Methods
3. Results
3.1. Endpoint and Traditional Risk Factors
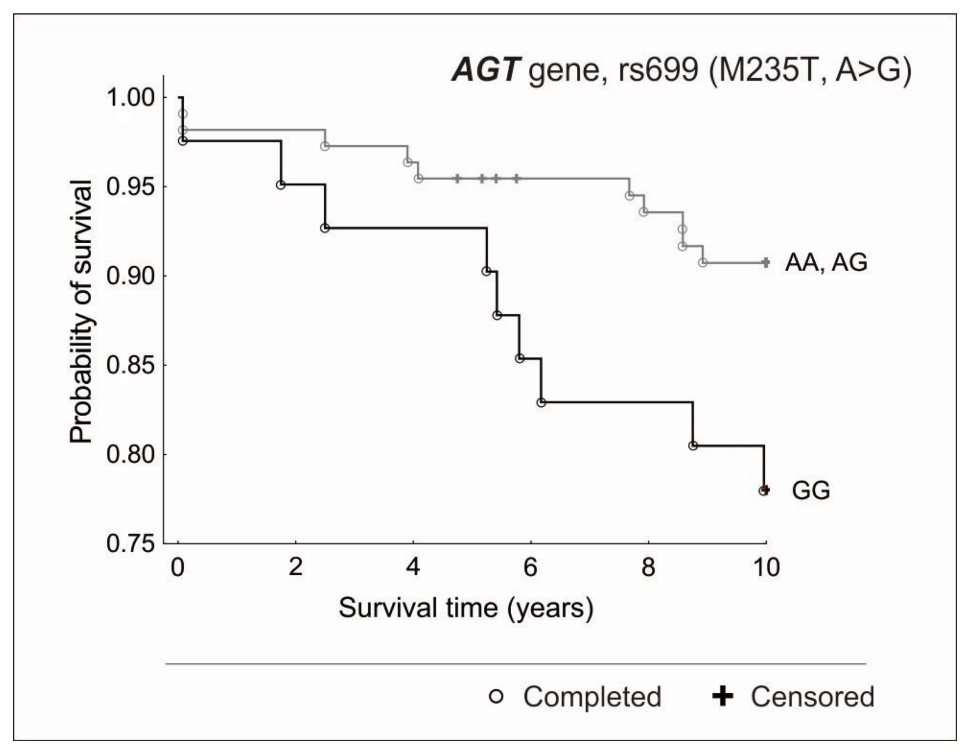
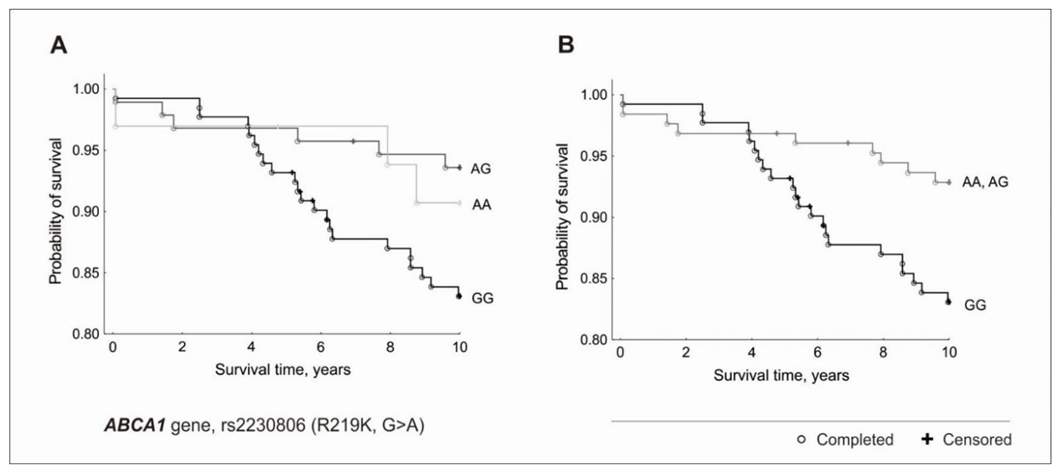
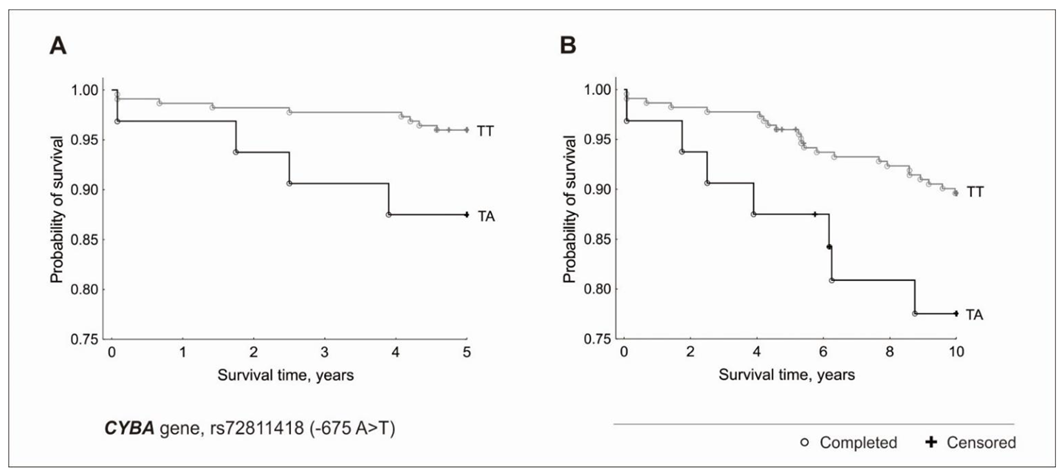
3.2. Genetic Factors and Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Schunkert, H. Genetics of coronary artery disease in the post-GWAS era. J. Intern. Med. 2021, 290, 980–992. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Ito, K.; Terao, C.; Akiyama, M.; Horikoshi, M.; Momozawa, Y.; Matsunaga, H.; Ieki, H.; Ozaki, K.; Onouchi, Y.; et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020, 52, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.; Izarraras-Gomez, A.; Ortega-Del Vecchyo, D. Populations, Traits, and Their Spatial Structure in Humans. Genome Biol. Evol. 2021, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Niemiec, P.; Nowak, T.; Iwanicki, T.; Gorczynska-Kosiorz, S.; Balcerzyk, A.; Krauze, J.; Grzeszczak, W.; Wiecha, M.; Zak, I. The rs2516839 Polymorphism of the USF1 Gene May Modulate Serum Triglyceride Levels in Response to Cigarette Smoking. Int. J. Mol. Sci. 2015, 16, 13203–13216. [Google Scholar] [CrossRef]
- Niemiec, P.; Zak, I.; Wita, K. The M235T polymorphism of the AGT gene modifies the risk of coronary artery disease associated with the presence of hypercholesterolemia. Eur. J. Epidemiol. 2008, 23, 349–354. [Google Scholar] [CrossRef]
- Iwanicka, J.; Iwanicki, T.; Balcerzyk, A.; Niemiec, P.; Nowak, T.; Krauze, J.; Trautsolt, W.; Ochalska-Tyka, A.; Grzeszczak, W.; Żak, I. Relationship between rs4674344 CYP27A1 gene polymorphism and coronary artery disease in a Polish population. Kardiol. Pol. 2020, 78, 65–67. [Google Scholar] [CrossRef]
- Niemiec, P.; Zak, I.; Wita, K. The risk of coronary artery disease associated with cigarette smoking and hypercholesterolemia is additionally increased by the presence of the AT1R gene 1166C allele. Biochem. Genet. 2008, 46, 799–809. [Google Scholar] [CrossRef]
- Balcerzyk, A.; Zak, I.; Krauze, J. Protective effect of R allele of PON1 gene on the coronary artery disease in the presence of specific genetic background. Dis. Markers. 2008, 24, 81–88. [Google Scholar] [CrossRef][Green Version]
- Iwanicka, J.; Iwanicki, T.; Niemiec, P.; Nowak, T.; Krauze, J.; Grzeszczak, W.; Górczyńska-Kosiorz, S.; Ochalska-Tyka, A.; Żak, I. Relationship between rs854560 PON1 Gene Polymorphism and Tobacco Smoking with Coronary Artery Disease. Dis. Markers. 2017, 2017, 1540949. [Google Scholar] [CrossRef]
- Iwanicki, T.; Balcerzyk, A.; Niemiec, P.; Trautsolt, W.; Grzeszczak, W.; Ochalska-Tyka, A.; Krauze, J.; Nowak, T.; Żak, I. The relationship between CYP7A1 polymorphisms, coronary artery disease & serum lipid markers. Biomark. Med. 2019, 13, 1199–1208. [Google Scholar] [CrossRef]
- Iwanicki, T.; Balcerzyk, A.; Niemiec, P.; Nowak, T.; Ochalska-Tyka, A.; Krauze, J.; Kosiorz-Gorczynska, S.; Grzeszczak, W.; Zak, I. CYP7A1 gene polymorphism located in the 5’ upstream region modifies the risk of coronary artery disease. Dis. Markers. 2015, 2015, 185969. [Google Scholar] [CrossRef]
- Niemiec, P.; Gorczynska-Kosiorz, S.; Iwanicki, T.; Krauze, J.; Trautsolt, W.; Grzeszczak, W.; Bochenek, A.; Zak, I. The rs10757278 polymorphism of the 9p21.3 locus is associated with premature coronary artery disease in Polish patients. Genet. Test. Mol. Biomarkers 2012, 16, 1080–1085. [Google Scholar] [CrossRef]
- Balcerzyk, A.; Zak, I.; Krauze, J. Synergistic effect between polymorphisms of PPARA and ABCA1 genes on the premature coronary artery disease. Acta. Cardiol. 2007, 62, 233–238. [Google Scholar] [CrossRef]
- Iwanicka, J.; Iwanicki, T.; Niemiec, P.; Balcerzyk, A.; Krauze, J.; Górczyńska-Kosiorz, S.; Ochalska-Tyka, A.; Grzeszczak, W.; Żak, I. Relationship between CETP gene polymorphisms with coronary artery disease in Polish population. Mol. Biol. Rep. 2018, 45, 1929–1935. [Google Scholar] [CrossRef]
- Niemiec, P.; Zak, I.; Wita, K. The 242T variant of the CYBA gene polymorphism increases the risk of coronary artery disease associated with cigarette smoking and hypercholesterolemia. Coron Artery Dis. 2007, 18, 339–346. [Google Scholar] [CrossRef]
- Niemiec, P.; Nowak, T.; Balcerzyk, A.; Krauze, J.; Zak, I. The CYBA gene A640G polymorphism influences predispositions to coronary artery disease through interactions with cigarette smoking and hypercholesterolemia. Biomarkers 2011, 16, 405–412. [Google Scholar] [CrossRef]
- Nowak, T.; Niemiec, P.; Górczyńska-Kosiorz, S.; Balcerzyk, A.; Iwanicki, T.; Krauze, J.; Grzeszczak, W.; Ochalska-Tyka, A.; Iwanicka, J.; Zak, I. The CYBA Gene (*)49A>G Polymorphism (rs7195830) Is Associated with Hypertension in Patients with Coronary Artery Disease. Biomed. Res. Int. 2016, 2016, 1539671. [Google Scholar] [CrossRef]
- Niemiec, P.; Nowak, T.; Iwanicki, T.; Krauze, J.; Gorczynska-Kosiorz, S.; Grzeszczak, W.; Ochalska-Tyka, A.; Zak, I. The -930A>G polymorphism of the CYBA gene is associated with premature coronary artery disease. A case-control study and gene-risk factors interactions. Mol. Biol. Rep. 2014, 41, 3287–3294. [Google Scholar] [CrossRef]
- Nowak, T.; Niemiec, P.; Iwanicki, T.; Balcerzyk, A.; Krauze, J.; Ochalska-Tyka, A.; Zak, I. Analysis of selected promoter polymorphisms and haplotypes of the CYBA gene encoding the p22phox, subunit of NADPH oxidases, in patients with coronary artery disease. Free Radic Res. 2018, 52, 1132–1139. [Google Scholar] [CrossRef]
- Zak, I.; Niemiec, P.; Sarecka, B.; Balcerzyk, A.; Ciemniewski, Z.; Rudowska, E.; Dylag, S. Carrier-state of D allele in ACE gene insertion/deletion polymorphism is associated with coronary artery disease, in contrast to the C677-->T transition in the MTHFR gene. Acta Biochim. Pol. 2003, 50, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Niemiec, P.; Zak, I.; Wita, K. Modification of the coronary artery disease risk associated with the presence of traditional risk factors by insertion/deletion polymorphism of the ACE gene. Genet. Test. 2007, 11, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Balcerzyk, A.; Zak, I., Krauze. Synergistic effects of apolipoprotein E gene epsilon polymorphism and some conventional risk factors on premature ischaemic heart disease development. Kardiol. Pol. 2007, 65, 1058–1065. [Google Scholar] [PubMed]
- Zak, I.; Niemiec, P.; Balcerzyk, A.; Krauze, J. Combined "pro-atherosclerotic" variants of the ACE and APOE genes increase the risk of the coronary artery disease associated with the presence of cigarette smoking. Acta Cardiol. 2008, 63, 741–747. [Google Scholar] [CrossRef]
- Jeunemaitre, X.; Soubrier, F.; Kotelevtsev, Y.V.; Lifton, R.P.; Williams, C.S.; Charru, A.; Hunt, S.C.; Hopkins, P.N.; Williams, R.R.; Lalouel, J.M. Molecular basis of human hypertension: Role of angiotensinogen. Cell 1992, 71, 169–180. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, R.; Hu, S.; Rong, J. Gene polymorphism associated with angiotensinogen (M235T), endothelial lipase (584C/T) and susceptibility to coronary artery disease: A meta-analysis. Biosci. Rep. 2020, 40, BSR20201414. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Wang, H.W.; Wang, X.Y.; Li, X.Q.; Zhang, L.L. The M235T polymorphism in the angiotensinogen gene and heart failure: A meta-analysis. J. Renin. Angiotensin Aldosterone Syst. 2014, 15, 190–195. [Google Scholar] [CrossRef]
- Zhen, Z.; Gao, L.; Wang, Q.; Chen, X.; Na, J.; Xu, X.; Yuan, Y. Angiotensinogen M235T polymorphism and susceptibility to hypertrophic cardiomyopathy in Asian population: A meta-analysis. J. Renin. Angiotensin Aldosterone Syst. 2020, 21, 1470320320978100. [Google Scholar] [CrossRef]
- Ma, X.Y.; Liu, J.P.; Song, Z.Y. Associations of the ATP-binding cassette transporter A1 R219K polymorphism with HDL-C level and coronary artery disease risk: A meta-analysis. Atherosclerosis 2011, 215, 428–434. [Google Scholar] [CrossRef]
- Shi, Z.; Tian, Y.; Zhao, Z.; Wu, Y.; Hu, X.; Li, J.; Chen, Q.; Wang, Y.; An, C.; Zhang, K. Association between the ABCA1 (R219K) polymorphism and lipid profiles: A meta-analysis. Sci. Rep. 2021, 11, 21718. [Google Scholar] [CrossRef]
- Fan, Q.; Zhu, Y.; Zhao, F. Association of rs2230806 in ABCA1 with coronary artery disease: An updated meta-analysis based on 43 research studies. Medicine 2020, 99, e18662. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhang, H.; Qin, X.Y.; Lu, X.Z.; Yang, B.; Chen, M.L. ATP-binding cassette transporter A1 R219K polymorphism and coronary artery disease in Chinese population: A meta-analysis of 5,388 participants. Mol. Biol. Rep. 2012, 39, 11031–11039. [Google Scholar] [CrossRef]
- Cenarro, A.; Artieda, M.; Castillo, S.; Mozas, P.; Reyes, G.; Tejedor, D.; Alonso, R.; Mata, P.; Pocoví, M.; Civeira, F.; et al. A common variant in the ABCA1 gene is associated with a lower risk for premature coronary heart disease in familial hypercholesterolaemia. J. Med. Genet. 2003, 40, 163–168. [Google Scholar] [CrossRef]
- Moreno, M.U.; José, G.S.; Fortuño, A.; Beloqui, O.; Redón, J.; Chaves, F.J.; Corella, D.; Díez, J.; Zalba, G. A novel CYBA variant, the -675A/T polymorphism, is associated with essential hypertension. J. Hypertens. 2007, 25, 1620–1626. [Google Scholar] [CrossRef]
- San José, G.; Fortuño, A.; Beloqui, O.; Díez, J.; Zalba, G. NADPH oxidase CYBA polymorphisms, oxidative stress and cardiovascular diseases. Clin. Sci. 2008, 114, 173–182. [Google Scholar] [CrossRef]
- Rabelo, F.; Stefano, J.T.; Cavaleiro, A.M.; Lima, R.; de Campos Mazo, D.F.; Carrilho, F.J.; Correa-Giannella, M.L.; Oliveira, C.P. Association between the CYBA and NOX4 genes of NADPH oxidase and its relationship with metabolic syndrome in non-alcoholic fatty liver disease in Brazilian population. Hepatobiliary Pancreat. Dis. Int. 2018, 17, 330–335. [Google Scholar] [CrossRef]
- Racis, M.; Sobiczewski, W.; Stanisławska-Sachadyn, A.; Wirtwein, M.; Bluj, E.; Nedoszytko, M.; Borzyszkowska, J.; Limon, J.; Rynkiewicz, A.; Gruchała, M. NADPH Oxidase Gene Polymorphism is Associated with Mortality and Cardiovascular Events in 7-Year Follow-Up. J. Clin. Med. 2020, 9, 1475. [Google Scholar] [CrossRef]
- Hai, Q.; Ritchey, B.; Robinet, P.; Alzayed, A.M.; Brubaker, G.; Zhang, J.; Smith, J.D. Quantitative Trait Locus Mapping of Macrophage Cholesterol Metabolism and CRISPR/Cas9 Editing Implicate an ACAT1 Truncation as a Causal Modifier Variant. Arterioscler Thromb. Vasc. Biol. 2018, 38, 83–91. [Google Scholar] [CrossRef]
- Ritchey, B.; Hai, Q.; Han, J.; Barnard, J.; Smith, J.D. Genetic variant in 3' untranslated region of the mouse pycard gene regulates inflammasome activity. Elife 2021, 10, e68203. [Google Scholar] [CrossRef]
- Ioannidis, J.P.; Ntzani, E.E.; Trikalinos, T.A. Contopoulos-Ioannidis, DG. Replication validity of genetic association studies. Nat. Genet. 2001, 29, 306–309. [Google Scholar] [CrossRef]
- Hai, Q.; Smith, J.D. Acyl-Coenzyme A: Cholesterol Acyltransferase (ACAT) in Cholesterol Metabolism: From Its Discovery to Clinical Trials and the Genomics Era. Metabolites 2021, 11, 543. [Google Scholar] [CrossRef] [PubMed]
| Locus | Gene | rs | Common Name | Risk Allele | Association with CAD | Gene-Gene or Gene-Traditional Risk Factor Interaction Increasing CAD Risk a | Ref. No |
|---|---|---|---|---|---|---|---|
| 1q23.3 | USF1 | 2516839 | T | - | + | [5] | |
| 3737787 | T | - | - | [5] | |||
| 1q42.2 | AGT | 699 | M235T (A > G) | G | + | + | [6] |
| 2q35 | CYP27A1 | 4674344 | T | - | - | [7] | |
| 3q24 | AGTR1 | 5186 | 1166 A > C | C | - | + | [8] |
| 7q21.3 | PON1 | 662 | Q192R | Q | + | + | [9] |
| 854560 | T | + | + | [10] | |||
| 8q12.1 | CYP7A1 | 3808607 | 278 A/C | C | - | - | [11] |
| 7833904 | A | + | - | [12] | |||
| 8192879 | G | - | - | [11] | |||
| 10504255 | G | - | - | [11] | |||
| 10957057 | C | - | - | [11] | |||
| 11786580 | C | - b | + | [11] | |||
| 9p21 | intergenic | 10757278 | G | + | - | [13] | |
| 9q31.1 | ABCA1 | 2230806 | R219K (G > A) | A | - | + | [14] |
| 16q13 | CETP | 247616 | C | + | + | [15] | |
| 708272 | C | - | + | [15] | |||
| 1532624 | C | + | + | [15] | |||
| 16q24.2 | CYBA | 4673 | 214C > T, C242T | T | - | + | [16] |
| 1049255 | * 24A > G, A640G | G | - | + | [17] | ||
| 7195830 | * 49 A > G | G | + | - | [18] | ||
| 9932581 | −930 A > G | G | + | + | [19] | ||
| 13306296 | −536 C > T | C | - | - | [20] | ||
| 16966671 | −852 C > G | C | - | + | [20] | ||
| 72811418 | −675 A > T | T | - | + | [20] | ||
| 17q23.3 | ACE | 1799752 | I/D, A287bpAlu | D | + | + | [21,22] |
| 19q13.32 | APOE | 7412 ε2 429358 ε4 | Epsilon1/2/3/4 | ε4 | - | + | [23,24] |
| 22q13.31 | PPARA | 4253778 | G > C, intron 7 | C | - | + | [14] |
| ICD-10 Code | Cause of Death | 5-Year Follow-Up | 10-Year Follow-Up | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| I20.8 | Ischaemic heart diseases. Other forms of angina pectoris. | 0 | 0.00 | 1 | 3.13 |
| I21.0 | Ischaemic heart diseases. Acute transmural myocardial infarction of anterior wall. | 0 | 0.00 | 1 | 3.13 |
| I21.9 | Ischaemic heart diseases. Acute myocardial infarction, unspecified. | 6 | 42.86 | 9 | 28.13 |
| I22.9 | Ischaemic heart diseases. Subsequent myocardial infarction of unspecified site. | 2 | 14.29 | 3 | 9.38 |
| I24.9 | Ischaemic heart diseases. Acute ischaemic heart disease, unspecified. | 0 | 0.00 | 1 | 3.13 |
| I25.0 | Ischaemic heart diseases. Atherosclerotic cardiovascular disease, so described. | 1 | 7.14 | 1 | 3.13 |
| I25.1 | Ischaemic heart diseases. Atherosclerotic heart disease. | 0 | 0.00 | 1 | 3.13 |
| I25.5 | Ischaemic heart diseases. Ischaemic cardiomyopathy. | 0 | 0.00 | 2 | 6.25 |
| I25.9 | Ischaemic heart diseases. Chronic ischaemic heart disease, unspecified. | 2 | 14.29 | 6 | 18.75 |
| I46.9 | Other forms of heart disease. Cardiac arrest, unspecified. | 0 | 0.00 | 1 | 3.13 |
| I50.1 | Other forms of heart disease. Left ventricular failure. | 1 | 7.14 | 1 | 3.13 |
| I61.6 | Cerebrovascular diseases. Intracerebral haemorrhage, multiple localized. | 0 | 0.00 | 1 | 3.13 |
| I63.5 | Cerebrovascular diseases. Cerebral infarction due to unspecified occlusion or stenosis of cerebral arteries. | 1 | 7.14 | 1 | 3.13 |
| I63.9 | Cerebrovascular diseases. Cerebral infarction, unspecified. | 1 | 7.14 | 1 | 3.13 |
| I70.2 | Diseases of arteries, arterioles and capillaries. Atherosclerosis of arteries of extremities. | 0 | 0.00 | 1 | 3.13 |
| I70.9 | Diseases of arteries, arterioles and capillaries. Generalized and unspecified atherosclerosis. | 0 | 0.00 | 1 | 3.13 |
| Σ | 14 | 100.00 | 32 | 100.00 | |
| Characteristics | 5-Year Follow-Up | 10-Year Follow-Up | ||||||
|---|---|---|---|---|---|---|---|---|
| Dead a (n = 14) | Alive b (n = 262) | OR (95%CI) | p | Dead a (n = 32) | Alive b (n = 244) | OR (95%CI) | p | |
| Age (SD) | 45.92 (4.55) | 45.33 (6.56) | - | 0.80 | 44.29 (5.53) | 45.49 (6.59) | - | 0.18 |
| Male, n (%) | 12 (85.71) | 178 (67.93) | 2.83 (0.62–12.94) | 0.16 | 25 (78.13) | 165 (67.62) | 1.71 (0.71–4.12) | 0.23 |
| BMI (SD) | 26.91 (4.94) | 27.16 (4.20) | - | 0.67 | 26.30 (4.32) | 27.25 (4.22) | - | 0.23 |
| Cigarette smoking, n (%) | 11 (78.57) | 150 (57.25) | 2.74 (0.75–10.04) | 0.11 | 21 (65.63) | 140 (57.38) | 1.42 (0.66–3.07) | 0.37 |
| Hypertension, n (%) | 5 (35.71) | 143 (54.58) | 0.46 (0.15–1.42) | 0.17 | 17 (53.13) | 131 (53.69) | 0.98 (0.47–2.05) | 0.95 |
| Diabetes mellitus, n (%) | 4 (28.57) | 19 (7.25) | 5.12 (1.47–17.86) | 0.02 * | 6 (18.75) | 17 (6.97) | 3.08 (1.17–8.51) | 0.05 |
| Hypercholesterolemia, n (%) | 4 (28.57) | 70 (26.72) | 1.10 (0.33–3.61) | 0.78 | 10 (31.25) | 64 (26.23) | 1.28 (0.57–2.85) | 0.55 |
| Hypertriglyceridemia, n (%) | 3 (21.43) | 24 (9.16) | 2.70 (0.71–10.37) | 0.13 | 4 (12.50) | 23 (9.43) | 1.37 (0.44–4.26) | 0.58 |
| Mixed hyperlipidemia, n (%) | 5 (35.71) | 101 (38.55) | 0.89 (0.29–2.71) | 0.83 | 11 (34.38) | 95 (38.93) | 0.82 (0.38–1.78) | 0.62 |
| Critical stenosis, n (%) | 11 (78,57) | 163 (62.21) | 2.23 (0.61–8.17) | 0.22 | 25 (78.16) | 149 (61.07) | 2.28 (0.95–5.47) | 0.06 |
| Multivessel disease, n (%) | 5 (35.71) | 47 (17.94) | 2.54 (0.81–7.93) | 0.10 | 8 (25.00) | 44 (18.03) | 1.51 (0.64–3.60) | 0.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balcerzyk-Matić, A.; Nowak, T.; Mizia-Stec, K.; Iwanicka, J.; Iwanicki, T.; Bańka, P.; Jarosz, A.; Filipecki, A.; Żak, I.; Krauze, J.; et al. Polymorphic Variants of AGT, ABCA1, and CYBA Genes Influence the Survival of Patients with Coronary Artery Disease: A Prospective Cohort Study. Genes 2022, 13, 2148. https://doi.org/10.3390/genes13112148
Balcerzyk-Matić A, Nowak T, Mizia-Stec K, Iwanicka J, Iwanicki T, Bańka P, Jarosz A, Filipecki A, Żak I, Krauze J, et al. Polymorphic Variants of AGT, ABCA1, and CYBA Genes Influence the Survival of Patients with Coronary Artery Disease: A Prospective Cohort Study. Genes. 2022; 13(11):2148. https://doi.org/10.3390/genes13112148
Chicago/Turabian StyleBalcerzyk-Matić, Anna, Tomasz Nowak, Katarzyna Mizia-Stec, Joanna Iwanicka, Tomasz Iwanicki, Paweł Bańka, Alicja Jarosz, Artur Filipecki, Iwona Żak, Jolanta Krauze, and et al. 2022. "Polymorphic Variants of AGT, ABCA1, and CYBA Genes Influence the Survival of Patients with Coronary Artery Disease: A Prospective Cohort Study" Genes 13, no. 11: 2148. https://doi.org/10.3390/genes13112148
APA StyleBalcerzyk-Matić, A., Nowak, T., Mizia-Stec, K., Iwanicka, J., Iwanicki, T., Bańka, P., Jarosz, A., Filipecki, A., Żak, I., Krauze, J., & Niemiec, P. (2022). Polymorphic Variants of AGT, ABCA1, and CYBA Genes Influence the Survival of Patients with Coronary Artery Disease: A Prospective Cohort Study. Genes, 13(11), 2148. https://doi.org/10.3390/genes13112148







