From Recharge, to Groundwater, to Discharge Areas in Aquifer Systems in Quebec (Canada): Shaping of Microbial Diversity and Community Structure by Environmental Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Sampling
2.3. Geochemical Analyses
2.4. DNA Extraction, Illumina Sequencing and Sequence Analysis
2.5. Sequence Analyses
2.6. Statistical Analyses
3. Results
3.1. Groundwater Environmental Parameters
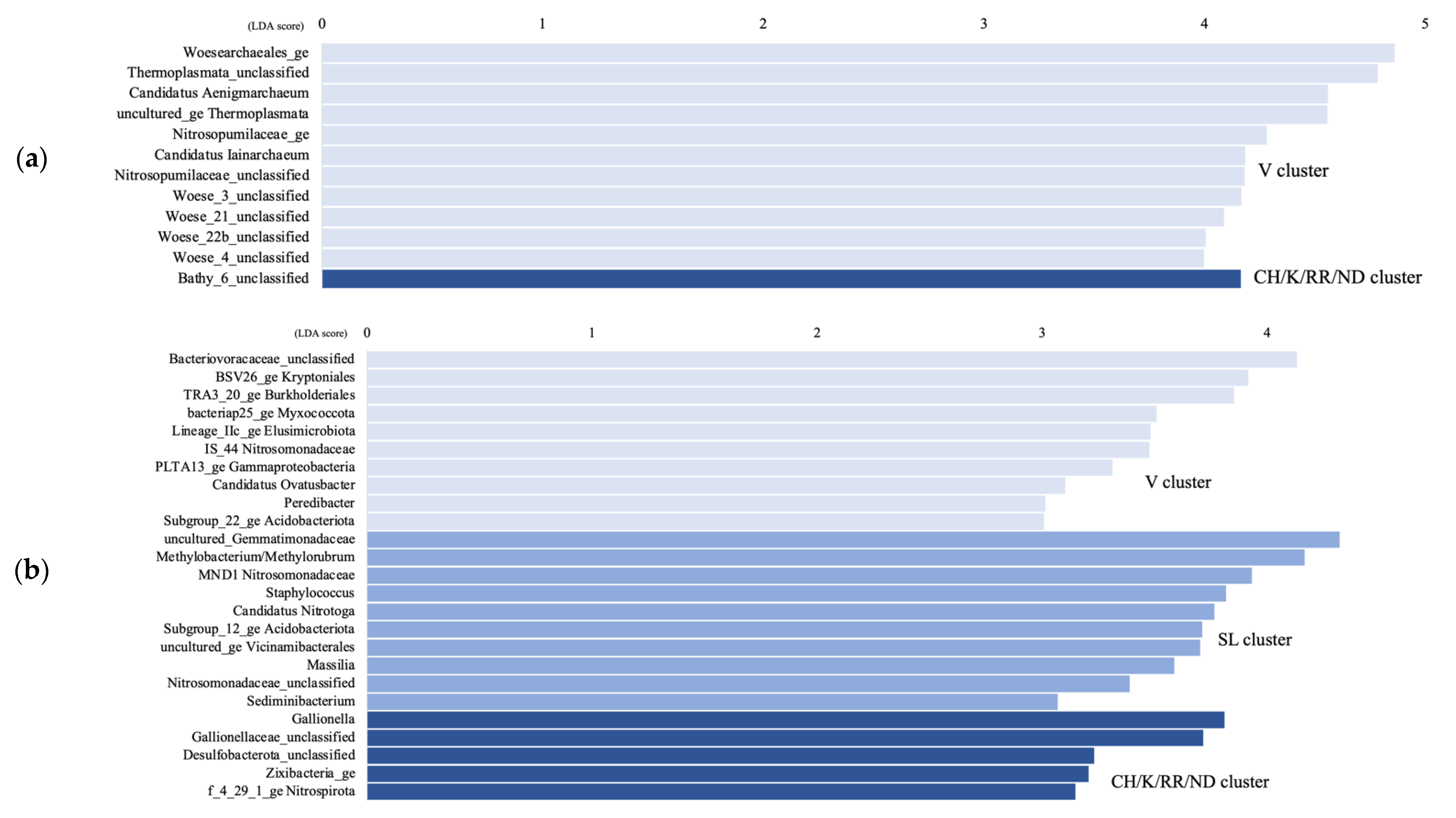
3.2. Archaeal and Bacterial 16S rRNA Gene Taxonomy
3.3. Alpha-Diversity Indices and Comparison between Habitats
3.4. Beta-Diversity Community Composition in Aquatic Habitats
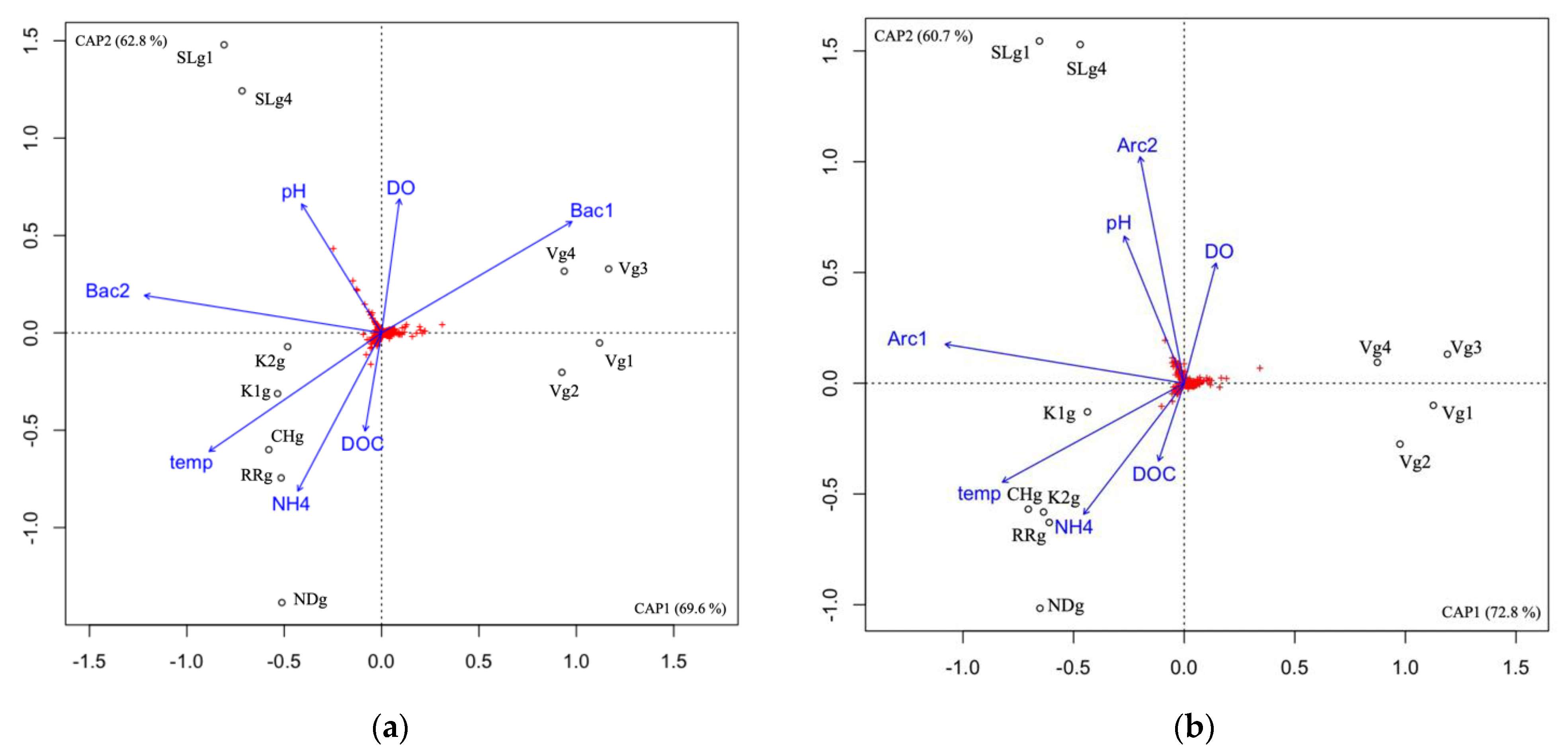
3.5. Microbial Community Correlation with Environmental Variables
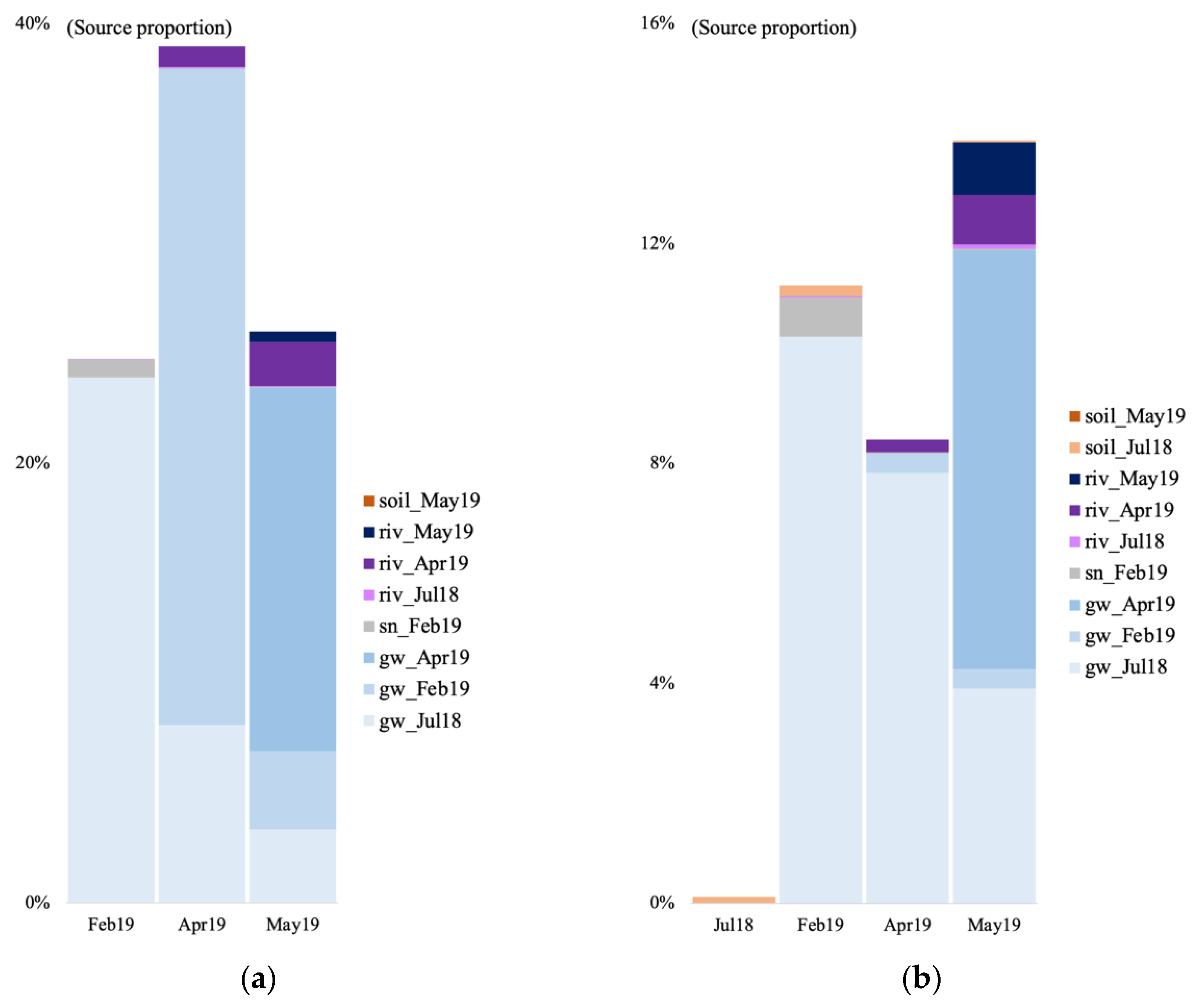
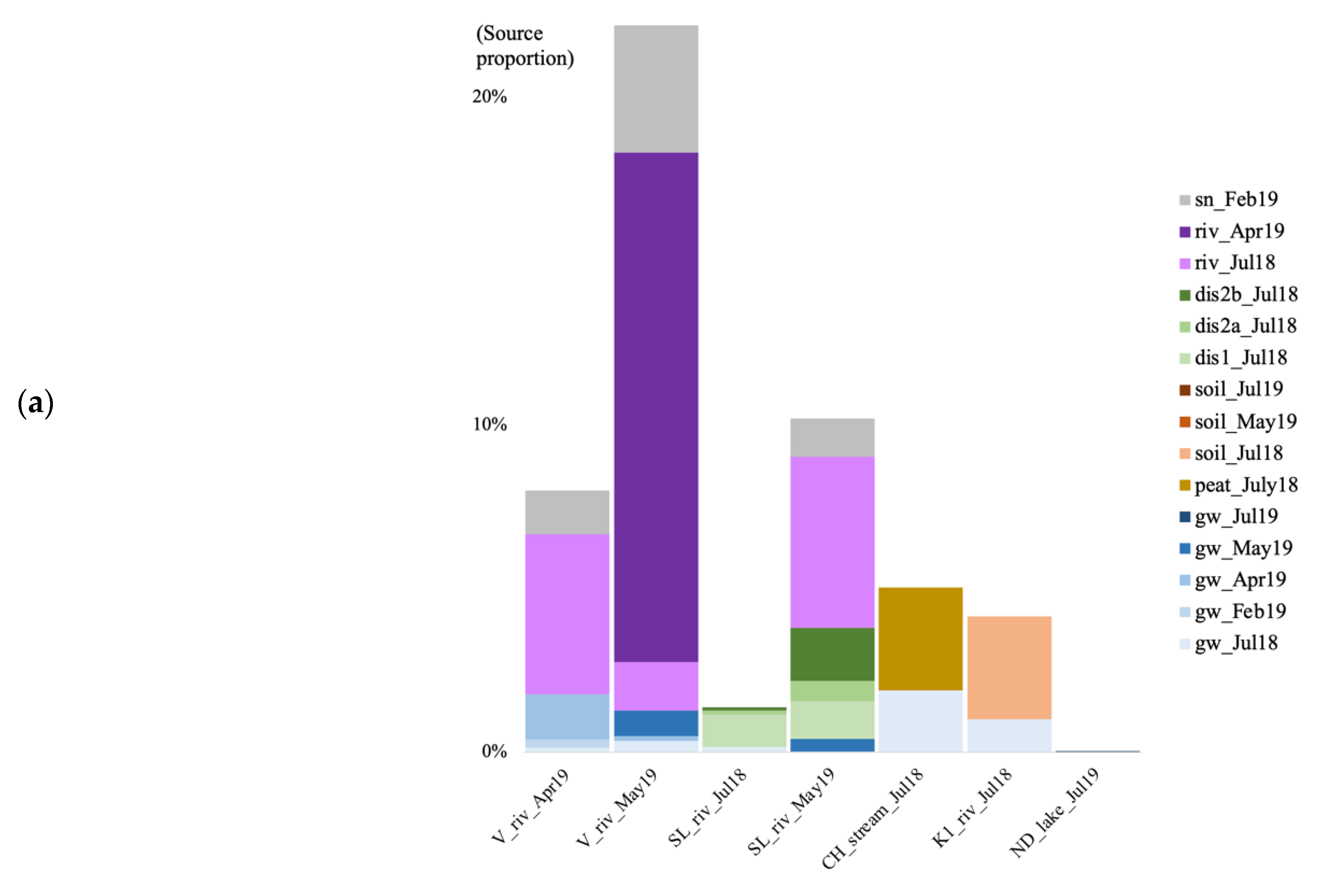
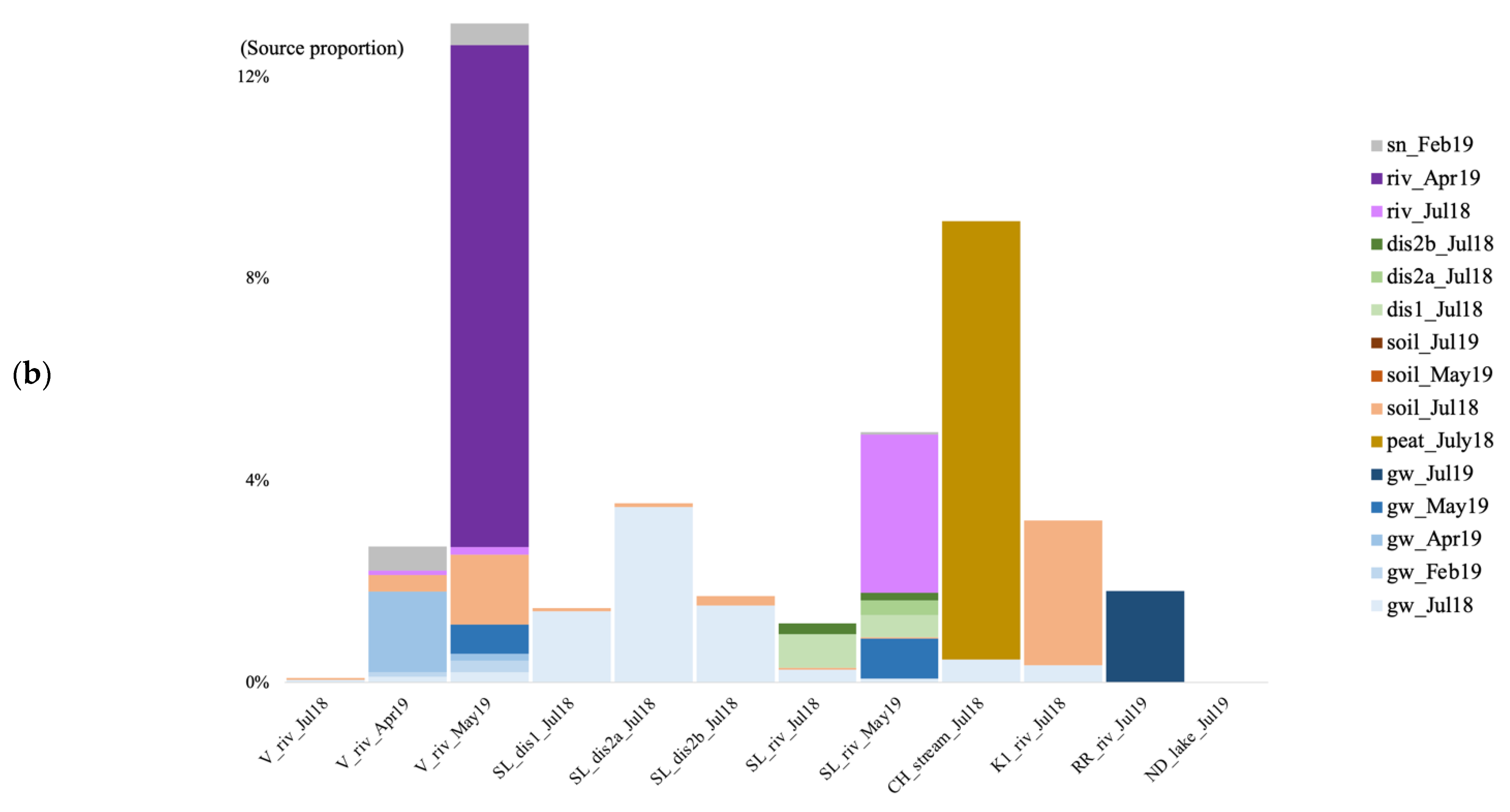
3.6. Microbial Source Tracking
4. Discussion
4.1. Microbial Communities Differ in Distinct Groundwater Ecosystems and Correlation with Environmental Parameters
4.2. Temporal Microbial Succession at the Vaudreuil Aquifer Site
4.3. Source of Groundwater Microbial Communities at the Vaudreuil Aquifer Site
4.4. Changes in the Microbial Community from Subsurface to the Surface Discharge Aquatic Ecosystems
4.5. Source of Microbial Communities in the Surface Discharge Aquatic Ecosystems
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fillinger, L.; Hug, K.; Griebler, C. Aquifer recharge viewed through the lens of microbial community ecology: Initial disturbance response, and impacts of species sorting versus mass effects on microbial community assembly in groundwater during riverbank filtration. Water Res. 2021, 189, 116631. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-Y.; Lo, M.-H.; Wada, Y.; Famiglietti, J.S.; Reager, J.T.; Yeh, P.J.F.; Ducharne, A.; Yang, Z.-L. Divergent effects of climate change on future groundwater availability in key mid-latitude aquifers. Nat. Commun. 2020, 11, 3710. [Google Scholar] [CrossRef]
- Wang, C.; Gomez-Velez, J.D.; Wilson, J.L. Dynamic coevolution of baseflow and multiscale groundwater flow system during prolonged droughts. J. Hydrol. 2022, 609, 127657. [Google Scholar] [CrossRef]
- Cardenas, M.B. Potential contribution of topography-driven regional groundwater flow to fractal stream chemistry: Residence time distribution analysis of Tóth flow. Geophys. Res. Lett. 2007, 34, L05403. [Google Scholar] [CrossRef]
- Merino, N.; Jackson, T.R.; Campbell, J.H.; Kersting, A.B.; Sackett, J.; Fisher, J.C.; Bruckner, J.C.; Zavarin, M.; Hamilton-Brehm, S.D.; Moser, D.P. Subsurface microbial communities as a tool for characterizing regional-scale groundwater flow. Sci. Total Environ. 2022, 842, 156768. [Google Scholar] [CrossRef]
- Retter, A.; Karwautz, C.; Griebler, C. Groundwater Microbial Communities in Times of Climate Change. Curr. Issues Mol. Biol. 2020, 41, 509–538. [Google Scholar] [CrossRef]
- Rivera, A. Canada’s Groundwater Resources; Fitzhenry & Whiteside: Toronto, ON, Canada, 2014. [Google Scholar]
- Zhang, Y.; Dekas, A.E.; Hawkins, A.J.; Parada, A.E.; Gorbatenko, O.; Li, K.; Horne, R.N. Microbial Community Composition in Deep-Subsurface Reservoir Fluids Reveals Natural Interwell Connectivity. Water Resour. Res. 2020, 56, e2019WR025916. [Google Scholar] [CrossRef]
- Yan, L.; Hermans, S.M.; Totsche, K.U.; Lehmann, R.; Herrmann, M.; Küsel, K. Groundwater bacterial communities evolve over time, exhibiting oscillating similarity patterns in response to recharge. bioRxiv 2021, 201, 117290. [Google Scholar] [CrossRef]
- Knobloch, S.; Klonowski, A.M.; Tómasdóttir, S.; Kristjánsson, B.R.; Guðmundsson, S.; Marteinsson, V.Þ. Microbial intrusion and seasonal dynamics in the groundwater microbiome of a porous basaltic rock aquifer used as municipal water reservoir. FEMS Microbiol. Ecol. 2021, 97, fiab014. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kellermann, C.; Griebler, C. Spatio-temporal patterns of microbial communities in a hydrologically dynamic pristine aquifer. FEMS Microbiol. Ecol. 2012, 81, 230–242. [Google Scholar] [CrossRef]
- Herrmann, M.; Wegner, C.-E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in Groundwater Is Caused by Their Preferential Mobilization from Soils and Flourishing Under Oligotrophic Conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef]
- Larocque, M.; Meyzonnat, G.; Ouellet, M.A.; Graveline, M.; Gagné, S.; Barnetche, D.; Dorner, S. Projet de connaissance des eaux souterraines de la zone de Vaudreuil- Soulanges. Rapp. Sci. 2015, 202. [Google Scholar]
- Girard, P.; Levison, J.; Parrott, L.; Larocque, M.; Ouellet, M.-A.; Green, D. Modeling cross-scale relationships between climate, hydrology, and individual animals: Generating scenarios for stream salamanders. Front. Environ. Sci. 2015, 3, 51. [Google Scholar] [CrossRef][Green Version]
- Levison, J.; Larocque, M.; Fournier, V.; Gagné, S.; Pellerin, S.; Ouellet, M.A. Dynamics of a headwater system and peatland under current conditions and with climate change. Hydrol. Process. 2014, 28, 4808–4822. [Google Scholar] [CrossRef]
- Meyer, J.; Zakhary, S.; Larocque, M.; Lazar, C.S. From Surface to Subsurface: Diversity, Composition, and Abundance of Sessile and Endolithic Bacterial, Archaeal, and Eukaryotic Communities in Sand, Clay and Rock Substrates in the Laurentians (Quebec, Canada). Microorganisms 2022, 10, 129. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Gantner, S.; Andersson, A.F.; Alonso-Sáez, L.; Bertilsson, S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 2011, 84, 12–18. [Google Scholar] [CrossRef]
- Stahl, D.A.; Amann, R.I. Development and Application of Nucleic Acid Probes. In Nucleic Acid Techniques in Bacterial Systematics; John Wiley & Sons: Hoboken, NJ, USA, 1991; pp. 205–248. [Google Scholar]
- Schloss, P.D. Reintroducing mothur: 10 Years Later. Appl. Environ. Microbiol. 2020, 86, e02343-19. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Zhou, Z.; Pan, J.; Wang, F.; Gu, J.-D.; Li, M. Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 2018, 42, 639–655. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Castelle, C.J.; Probst, A.J.; Zhou, Z.; Pan, J.; Liu, Y.; Banfield, J.F.; Gu, J.-D. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome 2018, 6, 102. [Google Scholar] [CrossRef]
- Pereira, M.B.; Wallroth, M.; Jonsson, V.; Kristiansson, E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genom. 2018, 19, 274. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.-C.; Astorg, L.; Derry, A.M.; Lazar, C.S. Response of Prokaryotic Communities to Freshwater Salinization. Appl. Microbiol. 2022, 2, 330–346. [Google Scholar] [CrossRef]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe’er, I.; Halperin, E. FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Baker, B.; Seitz, K.; Hyde, A.; Dick, G.; Hinrichs, K.-U.; Teske, A. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 2015, 18, 1200–1211. [Google Scholar] [CrossRef]
- Li, Y.-X.; Rao, Y.-Z.; Qi, Y.-L.; Qu, Y.-N.; Chen, Y.-T.; Jiao, J.-Y.; Shu, W.-S.; Jiang, H.; Hedlund, B.P.; Hua, Z.-S.; et al. Deciphering Symbiotic Interactions of “Candidatus Aenigmarchaeota” with Inferred Horizontal Gene Transfers and Co-occurrence Networks. mSystems 2021, 6, e00606-21. [Google Scholar] [CrossRef]
- Youssef, N.H.; Rinke, C.; Stepanauskas, R.; Farag, I.; Woyke, T.; Elshahed, M.S. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites’. ISME J. 2015, 9, 447–460. [Google Scholar] [CrossRef]
- Hu, W.; Pan, J.; Wang, B.; Guo, J.; Li, M.; Xu, M. Metagenomic insights into the metabolism and evolution of a new Thermoplasmata order (Candidatus Gimiplasmatales). Environ. Microbiol. 2021, 23, 3695–3709. [Google Scholar] [CrossRef]
- Murphy, C.L.; Biggerstaff, J.; Eichhorn, A.; Ewing, E.; Shahan, R.; Soriano, D.; Stewart, S.; VanMol, K.; Walker, R.; Walters, P.; et al. Genomic characterization of three novel Desulfobacterota classes expand the metabolic and phylogenetic diversity of the phylum. Environ. Microbiol. 2021, 23, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Castelle, C.J.; Hug, L.A.; Wrighton, K.C.; Thomas, B.C.; Williams, K.H.; Wu, D.; Tringe, S.G.; Singer, S.W.; Eisen, J.A.; Banfield, J.F. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat. Commun. 2013, 4, 2120. [Google Scholar] [CrossRef] [PubMed]
- Mujakić, I.; Piwosz, K.; Koblížek, M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms 2022, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.J.; Overmann, J. Vicinamibacteraceae fam. nov., the first described family within the subdivision 6 Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 2331–2334. [Google Scholar] [CrossRef]
- Kitzinger, K.; Koch, H.; Lücker, S.; Sedlacek, C.J.; Herbold, C.; Schwarz, J.; Daebeler, A.; Mueller, A.J.; Lukumbuzya, M.; Romano, S.; et al. Characterization of the First “Candidatus Nitrotoga” Isolate Reveals Metabolic Versatility and Separate Evolution of Widespread Nitrite-Oxidizing Bacteria. mBio 2018, 9, e01186-18. [Google Scholar] [CrossRef]
- Alessa, O.; Ogura, Y.; Fujitani, Y.; Takami, H.; Hayashi, T.; Sahin, N.; Tani, A. Comprehensive Comparative Genomics and Phenotyping of Methylobacterium Species. Front. Microbiol. 2021, 12, 740610. [Google Scholar] [CrossRef]
- Nercessian, O.; Noyes, E.; Kalyuzhnaya, M.G.; Lidstrom, M.E.; Chistoserdova, L. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 2005, 71, 6885–6899. [Google Scholar] [CrossRef]
- Taubert, M.; Stöckel, S.; Geesink, P.; Girnus, S.; Jehmlich, N.; Von Bergen, M.; Rösch, P.; Popp, J.; Küsel, K. Tracking active groundwater microbes with D 2 O labeling to understand their ecosystem function. Environ. Microbiol. 2017, 20, 369–384. [Google Scholar] [CrossRef]
- Wischer, D.; Kumaresan, D.; Johnston, A.; El Khawand, M.; Stephenson, J.; Hillebrand-Voiculescu, A.M.; Chen, Y.; Colin Murrell, J. Bacterial metabolism of methylated amines and identification of novel methylotrophs in Movile Cave. ISME J. 2015, 9, 195–206. [Google Scholar] [CrossRef]
- Davidov, Y.; Jurkevitch, E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax–Peredibacter clade as Bacteriovoracaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1439–1452. [Google Scholar] [CrossRef]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef]
- Dirren, S.; Posch, T. Promiscuous and specific bacterial symbiont acquisition in the amoeboid genus Nuclearia (Opisthokonta). FEMS Microbiol. Ecol. 2016, 92, fiw105. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Forchhammer, K. Bacterial Predation on Cyanobacteria. Microb. Physiol. 2021, 31, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, Z.; Pietrokovski, S.; Rotem, O.; Gophna, U.; Lurie-Weinberger, M.N.; Jurkevitch, E. By their genes ye all shall know them: Genomic signatures of predatory bacteria. ISME J. 2013, 7, 756–769. [Google Scholar] [CrossRef]
- Sirisena, K.A.; Daughney, C.J.; Moreau-Fournier, M.; Ryan, K.G.; Chambers, G.K. National survey of molecular bacterial diversity of New Zealand groundwater: Relationships between biodiversity, groundwater chemistry and aquifer characteristics. FEMS Microbiol. Ecol. 2013, 86, 490–504. [Google Scholar] [CrossRef]
- Thompson, P.L.; Guzman, L.M.; De Meester, L.; Horváth, Z.; Ptacnik, R.; Vanschoenwinkel, B.; Viana, D.S.; Chase, J.M. A process-based metacommunity framework linking local and regional scale community ecology. Ecol. Lett. 2020, 23, 1314–1329. [Google Scholar] [CrossRef]
- Li, J.; Wang, S. Seasonal variations and long-term trends of groundwater over the Canadian landmass. Hydrogeol. J. 2022, 30, 401–415. [Google Scholar] [CrossRef]
- Ruepp, A.; Graml, W.; Santos-Martinez, M.-L.; Koretke, K.K.; Volker, C.; Mewes, H.W.; Frishman, D.; Stocker, S.; Lupas, A.N.; Baumeister, W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 2000, 407, 508–513. [Google Scholar] [CrossRef]
- Lazar, C.S.; Baker, B.J.; Seitz, K.W.; Teske, A.P. Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J. 2017, 11, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Jiang, Y.; Liu, Q.; Wang, R.; Liu, X.; Liu, B. Enzymatic and regulatory properties of the trehalose-6-phosphate synthase from the thermoacidophilic archaeon Thermoplasma acidophilum. Biochimie 2014, 101, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, H.; Yang, R.; Li, S.; Zhou, M.; Gao, T.; An, L.; Chen, X.; Dyson, P. Complete genome sequence of a psychotrophic Pseudarthrobacter sulfonivorans strain Ar51 (CGMCC 4.7316), a novel crude oil and multi benzene compounds degradation strain. J. Biotechnol. 2016, 231, 81–82. [Google Scholar] [CrossRef]
- Choe, Y.-H.; Kim, M.; Woo, J.; Lee, M.J.; Lee, J.I.; Lee, E.J.; Lee, Y.K. Comparing rock-inhabiting microbial communities in different rock types from a high arctic polar desert. FEMS Microbiol. Ecol. 2018, 94, fiy070. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, M.; Ostrowski, M.; Fegatella, F.; Bowman, J.; Nichols, D.; Nishino, T.; Cavicchioli, R. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 2001, 67, 4945–4954. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, S.-I.; Lee, J.-C.; Ohta, H.; Whang, K.-S. Sphingomonas oligoaromativorans sp. nov., an oligotrophic bacterium isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 1679–1684. [Google Scholar] [CrossRef]
- Shen, Y.; Chapelle, F.H.; Strom, E.W.; Benner, R. Origins and bioavailability of dissolved organic matter in groundwater. Biogeochemistry 2015, 122, 61–78. [Google Scholar] [CrossRef]
- Valdivia-Anistro, J.A.; Eguiarte-Fruns, L.E.; Delgado-Sapién, G.; Márquez-Zacarías, P.; Gasca-Pineda, J.; Learned, J.; Elser, J.J.; Olmedo-Alvarez, G.; Souza, V. Variability of rRNA Operon Copy Number and Growth Rate Dynamics of Bacillus Isolated from an Extremely Oligotrophic Aquatic Ecosystem. Front. Microbiol. 2016, 6, 1486. [Google Scholar] [CrossRef]
- Harbison, A.B.; Price, L.E.; Flythe, M.D.; Bräuer, S.L. Micropepsis pineolensis gen. nov., sp. nov., a mildly acidophilic alphaproteobacterium isolated from a poor fen, and proposal of Micropepsaceae fam. nov. within Micropepsales ord. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 839–844. [Google Scholar] [CrossRef]
- Lehtovirta-Morley, L.E.; Sayavedra-Soto, L.A.; Gallois, N.; Schouten, S.; Stein, L.Y.; Prosser, J.I.; Nicol, G.W. Identifying Potential Mechanisms Enabling Acidophily in the Ammonia-Oxidizing Archaeon “Candidatus Nitrosotalea devanaterra”. Appl. Environ. Microbiol. 2016, 82, 2608–2619. [Google Scholar] [CrossRef]
- Fillinger, L.; Zhou, Y.; Kellermann, C.; Griebler, C. Non-random processes determine the colonization of groundwater sediments by microbial communities in a pristine porous aquifer. Environ. Microbiol. 2019, 21, 327–342. [Google Scholar] [CrossRef]
- Ben Maamar, S.; Aquilina, L.; Quaiser, A.; Pauwels, H.; Michon-Coudouel, S.; Vergnaud-Ayraud, V.; Labasque, T.; Roques, C.; Abbott, B.W.; Dufresne, A. Groundwater Isolation Governs Chemistry and Microbial Community Structure along Hydrologic Flowpaths. Front. Microbiol. 2015, 6, 1457. [Google Scholar] [CrossRef]
- Wen, Y. Impacts of Road De-Icing Salts on Manganese Transport to Groundwater in Roadside Soils. Master’s Thesis, KTH School of Architecture and the Built Environment, Stockholm, Sweden, 2012. [Google Scholar]
- Bushman, T.J.; Akob, D.M.; Bohu, T.; Beyer, A.; Woyke, T.; Shapiro, N.; Lapidus, A.; Klenk, H.-P.; Küsel, K. Draft Genome Sequence of Mn(II)-Oxidizing Bacterium Oxalobacteraceae sp. Strain AB_14. Microbiol. Resour. Announc. 2019, 8, e01024-19. [Google Scholar] [CrossRef] [PubMed]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef]
- Lin, X.; McKinley, J.; Resch, C.T.; Kaluzny, R.; Lauber, C.L.; Fredrickson, J.; Knight, R.; Konopka, A. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J. 2012, 6, 1665–1676. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally Rare Taxa Disproportionately Contribute to Temporal Changes in Microbial Diversity. mBio 2014, 5, e01371-14. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Frindte, K.; Dziallas, C.; Eckert, W.; Tang, K.W. Microbial methane production in oxygenated water column of an oligotrophic lake. Proc. Natl. Acad. Sci. USA 2011, 108, 19657–19661. [Google Scholar] [CrossRef]
- Bogard, M.J.; del Giorgio, P.A.; Boutet, L.; Chaves, M.C.G.; Prairie, Y.T.; Merante, A.; Derry, A.M. Oxic water column methanogenesis as a major component of aquatic CH4 fluxes. Nat. Commun. 2014, 5, 5350. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Hu, C.; Wang, Y.; Wu, J.; Hong, Y. Biogeographic Pattern of Methanogenic Community in Surface Water along the Yangtze River. Geomicrobiol. J. 2021, 38, 588–597. [Google Scholar] [CrossRef]
- Ortiz-Alvarez, R.; Casamayor, E.O. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ. Microbiol. Rep. 2016, 8, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Juottonen, H.; Fontaine, L.; Wurzbacher, C.; Drakare, S.; Peura, S.; Eiler, A. Archaea in boreal Swedish lakes are diverse, dominated by Woesearchaeota and follow deterministic community assembly. Environ. Microbiol. 2020, 22, 3158–3171. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Liu, Y.; Zhang, X.; Zhang, C.-J.; Zou, D.; Zheng, S.; Xu, W.; Luo, Z.; Liu, F.; Li, M. Comparative genomic analysis reveals metabolic flexibility of Woesearchaeota. Nat. Commun. 2021, 12, 5281. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.-H.; Yuan, H.-L. Sediminibacterium salmoneum gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from sediment of a eutrophic reservoir. Int. J. Syst. Evol. Microbiol. 2008, 58, 2191–2194. [Google Scholar] [CrossRef]
- Jogler, M.; Chen, H.; Simon, J.; Rohde, M.; Busse, H.-J.; Klenk, H.-P.; Tindall, B.J.; Overmann, J. Description of Sphingorhabdus planktonica gen. nov., sp. nov. and reclassification of three related members of the genus Sphingopyxis in the genus Sphingorhabdus gen. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 1342–1349. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, H.; Lee, B.-I.; Joung, Y.; Joh, K. Sediminibacterium goheungense sp. nov., isolated from a freshwater reservoir. Int. J. Syst. Evol. Microbiol. 2014, 64, 1328–1333. [Google Scholar] [CrossRef]
- Chen, L.-X.; Méndez-García, C.; Dombrowski, N.; Servín-Garcidueñas, L.E.; Eloe-Fadrosh, E.A.; Fang, B.-Z.; Luo, Z.-H.; Tan, S.; Zhi, X.-Y.; Hua, Z.-S.; et al. Metabolic versatility of small archaea Micrarchaeota and Parvarchaeota. ISME J. 2018, 12, 756–775. [Google Scholar] [CrossRef]
- Hahn, M.W.; Kasalický, V.; Jezbera, J.; Brandt, U.; Jezberová, J.; Šimek, K. Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2010, 60, 1358–1365. [Google Scholar] [CrossRef]
- Kasalický, V.; Jezbera, J.; Šimek, K.; Hahn, M.W. Limnohabitans planktonicus sp. nov. and Limnohabitans parvus sp. nov., planktonic betaproteobacteria isolated from a freshwater reservoir, and emended description of the genus Limnohabitans. Int. J. Syst. Evol. Microbiol. 2010, 60, 2710–2714. [Google Scholar] [CrossRef]
- Park, M.; Song, J.; Nam, G.G.; Cho, J.-C. Rhodoferax lacus sp. nov., isolated from a large freshwater lake. Int. J. Syst. Evol. Microbiol. 2019, 69, 3135–3140. [Google Scholar] [CrossRef]
- Zhou, D.; Tan, X.; Zhang, W.; Chen, H.-Y.; Fan, Q.-M.; He, X.-L.; Lv, J. Rhodoferax bucti sp. nov., isolated from fresh water. Int. J. Syst. Evol. Microbiol. 2019, 69, 3903–3909. [Google Scholar] [CrossRef] [PubMed]
- Ezzedine, J.A.; Jacas, L.; Desdevises, Y.; Jacquet, S. Bdellovibrio and Like Organisms in Lake Geneva: An Unseen Elephant in the Room? Front. Microbiol. 2020, 11, 98. [Google Scholar] [CrossRef]
- Bräuer, S.L.; Cadillo-Quiroz, H.; Ward, R.J.; Yavitt, J.B.; Zinder, S.H. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int. J. Syst. Evol. Microbiol. 2011, 61, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Savvichev, A.; Rusanov, I.; Dvornikov, Y.; Kadnikov, V.; Kallistova, A.; Veslopolova, E.; Chetverova, A.; Leibman, M.; Sigalevich, P.A.; Pimenov, N.; et al. The water column of the Yamal tundra lakes as a microbial filter preventing methane emission. Biogeosciences 2021, 18, 2791–2807. [Google Scholar] [CrossRef]
- Hahn, M.W.; Karbon, G.; Koll, U.; Schmidt, J.; Lang, E. Polynucleobacter sphagniphilus sp. nov. a planktonic freshwater bacterium isolated from an acidic and humic freshwater habitat. Int. J. Syst. Evol. Microbiol. 2017, 67, 3261–3267. [Google Scholar] [CrossRef] [PubMed]
- Eder, W.; Wanner, G.; Ludwig, W.; Busse, H.-J.; Ziemke-Kägeler, F.; Lang, E. Description of Undibacterium oligocarboniphilum sp. nov., isolated from purified water, and Undibacterium pigrum strain CCUG 49012 as the type strain of Undibacterium parvum sp. nov., and emended descriptions of the genus Undibacterium and the species Undibacterium pigrum. Int. J. Syst. Evol. Microbiol. 2011, 61, 384–391. [Google Scholar] [CrossRef][Green Version]
- Heckmann, K.; Schmidt, H.J. Polynucleobacter necessarius gen. nov., sp. nov., an Obligately Endosymbiotic Bacterium Living in the Cytoplasm of Euplotes aediculatus. Int. J. Syst. Evol. Microbiol. 1987, 37, 456–457. [Google Scholar] [CrossRef]
- González-Benítez, N.; Martín-Rodríguez, I.; Cuesta, I.; Arrayás, M.; White, J.F.; Molina, M.C. Endophytic Microbes Are Tools to Increase Tolerance in Jasione Plants Against Arsenic Stress. Front. Microbiol. 2021, 12, 664271. [Google Scholar] [CrossRef]
- Harrison, A.P. Acidiphilium cryptum gen. nov., sp. nov., Heterotrophic Bacterium from Acidic Mineral Environments. Int. J. Syst. Evol. Microbiol. 1981, 31, 327–332. [Google Scholar] [CrossRef]
- Jiang, Y.; Wiese, J.; Tang, S.-K.; Xu, L.-H.; Imhoff, J.F.; Jiang, C.-L. Actinomycetospora chiangmaiensis gen. nov., sp. nov., a new member of the family Pseudonocardiaceae. Int. J. Syst. Evol. Microbiol. 2008, 58, 408–413. [Google Scholar] [CrossRef]
- Borg, H.; Sundbom, M. Long-term trends of water chemistry in mountain streams in Sweden—Slow recovery from acidification. Biogeosciences 2014, 11, 173–184. [Google Scholar] [CrossRef]
- Høj, L.; Olsen, R.A.; Torsvik, V.L. Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2008, 2, 37–48. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, R.; Wang, H.; Gong, L.; Man, B.; Xu, Y. Distribution of Bathyarchaeota Communities Across Different Terrestrial Settings and Their Potential Ecological Functions. Sci. Rep. 2017, 7, 45028. [Google Scholar] [CrossRef]
- Marquart, K.A.; Haller, B.R.; Paper, J.M.; Flynn, T.M.; Boyanov, M.I.; Shodunke, G.; Gura, C.; Jin, Q.; Kirk, M.F. Influence of pH on the balance between methanogenesis and iron reduction. Geobiology 2019, 17, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Hua, B.; Huang, J.J.; Droste, R.L.; Zhou, Q.; Zhao, W.; Chen, L. Effect of different initial low pH conditions on biogas production, composition, and shift in the aceticlastic methanogenic population. Bioresour. Technol. 2019, 289, 121579. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Gu, J.-D. Ecological distribution and potential roles of Woesearchaeota in anaerobic biogeochemical cycling unveiled by genomic analysis. Comput. Struct. Biotechnol. J. 2021, 19, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Cruaud, P.; Vigneron, A.; Dorea, C.C.; Rodriguez, M.J.; Charette, S.J. Rapid Changes in Microbial Community Structures along a Meandering River. Microorganisms 2020, 8, 1631. [Google Scholar] [CrossRef] [PubMed]
- Balmonte, J.P.; Arnosti, C.; Underwood, S.; McKee, B.A.; Teske, A. Riverine Bacterial Communities Reveal Environmental Disturbance Signatures within the Betaproteobacteria and Verrucomicrobia. Front. Microbiol. 2016, 7, 1441. [Google Scholar] [CrossRef] [PubMed]
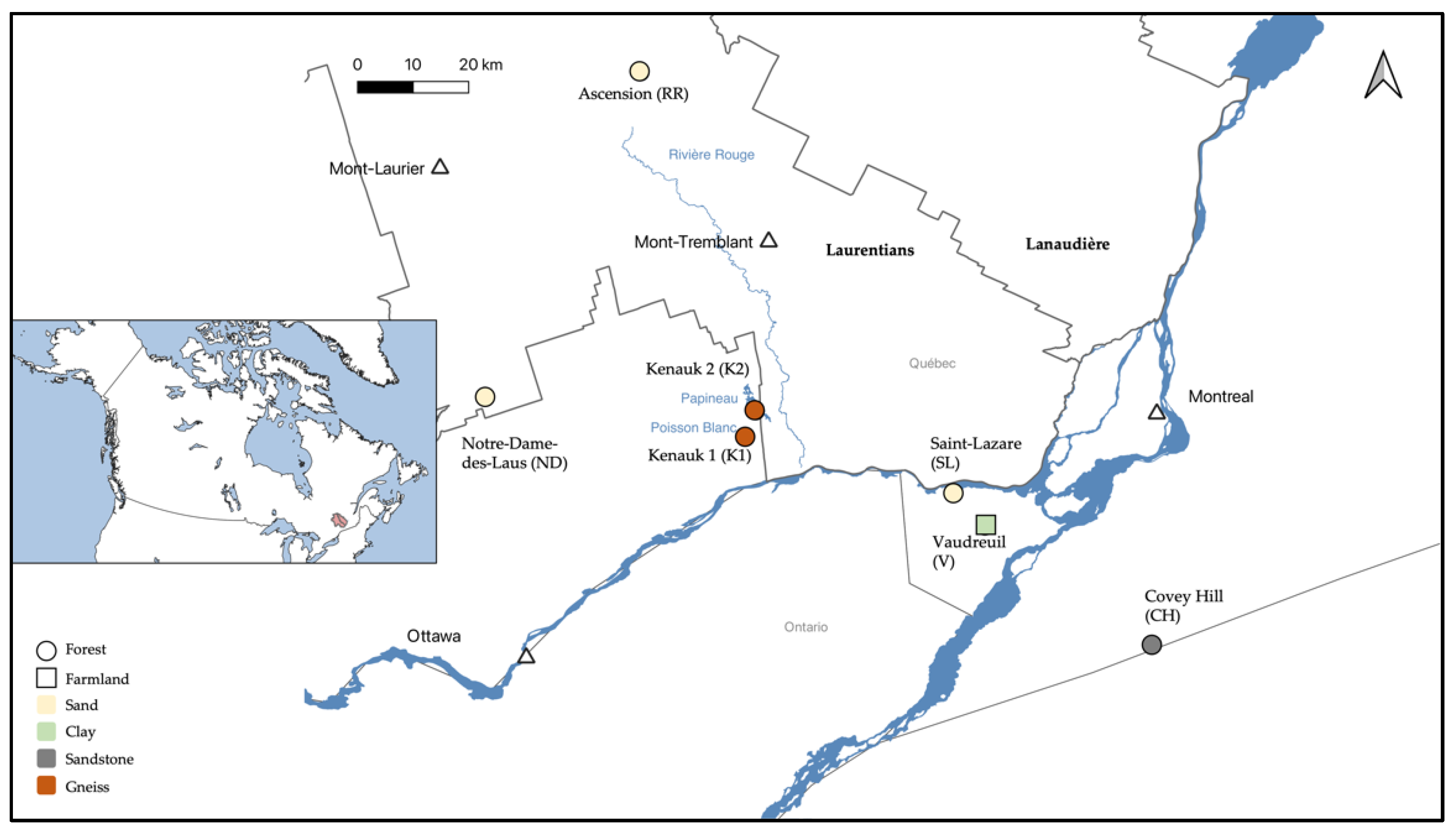
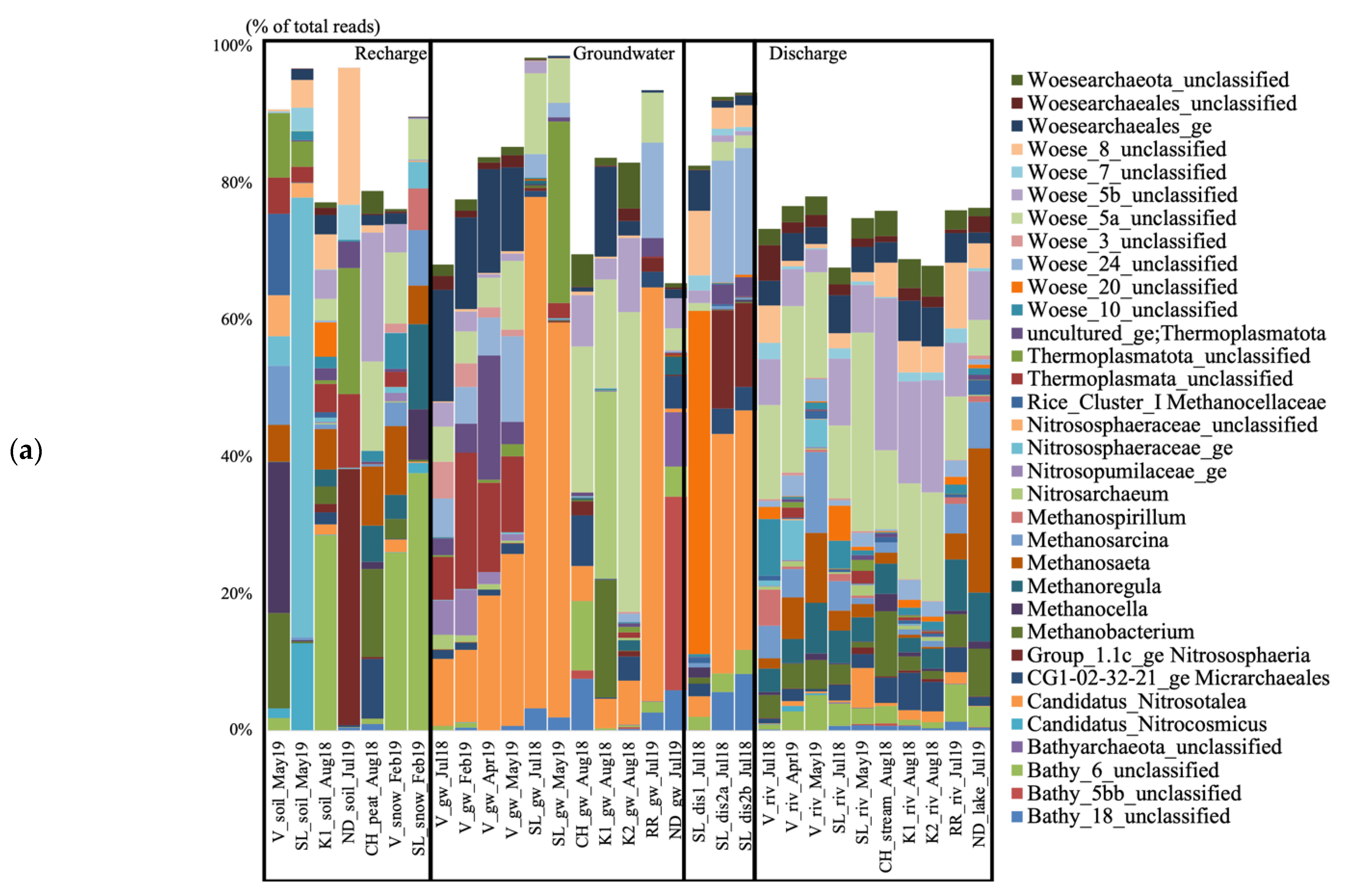
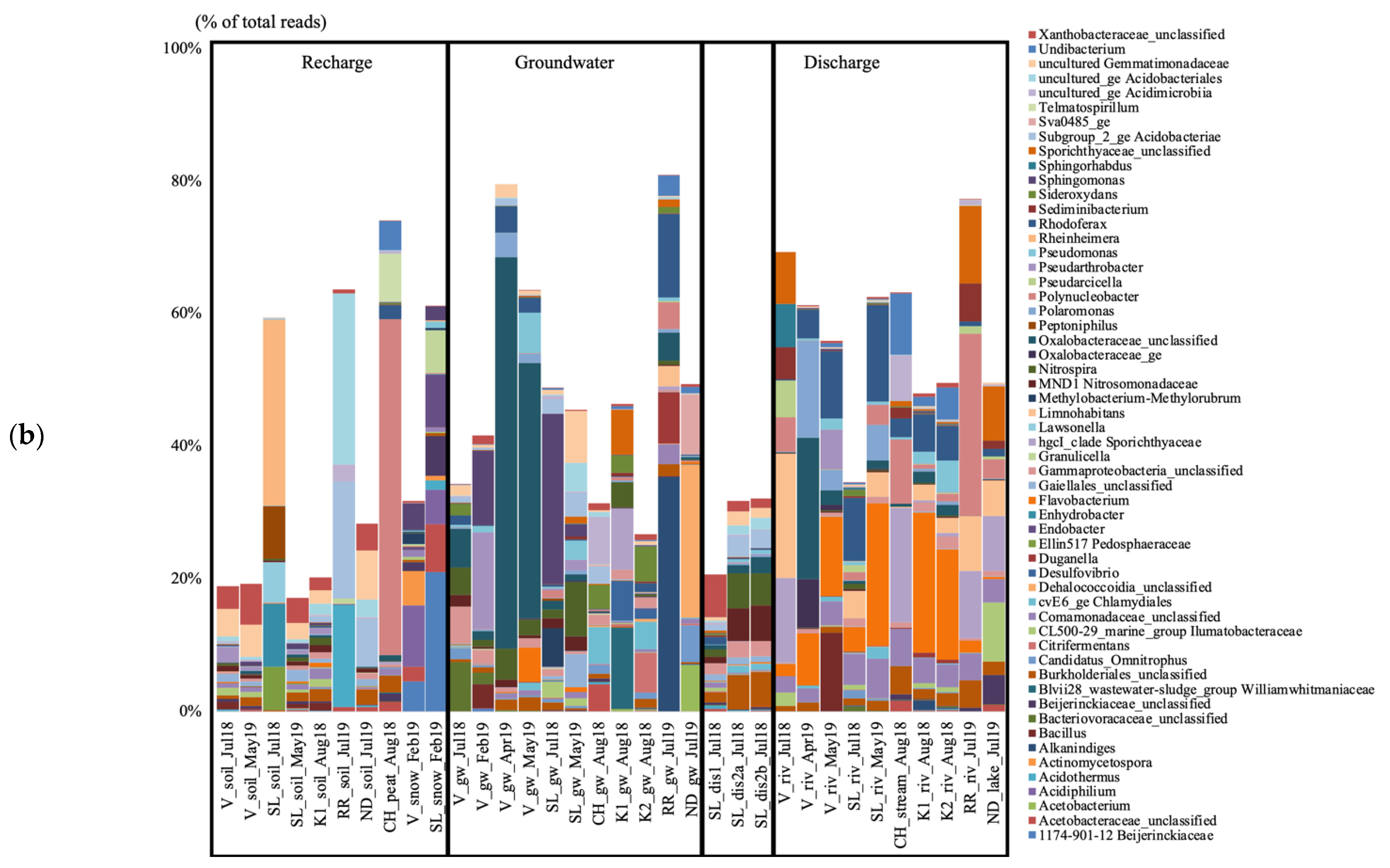

| Site | Surface Recharge | Groundwater | Surface Discharge |
|---|---|---|---|
| V | Soil (Jul 2018) Soil (May 2018) Snow (Feb 2019) | Jul 2018 Feb 2019 Apr 2019 May 2019 | River (Jul 2018) River (Apr 2019) River (May 2019) |
| SL | Soil (July 2018) Soil (May 2018) Snow (Feb 2019) | Jul 2018 May 2019 | Forest dis1 (Jul 2018) Forest dis2a (Jul 2018) Forest dis2b (Jul 2018) River (Jul 2018) River (May 2019) |
| CH | Peat (Aug 2018) | Aug 2018 | Stream (Aug 2018) |
| K1 | Soil (Aug 2018) | Aug 2018 | River (Aug 2018) |
| K2 | Soil (Aug 2018) | Aug 2018 | River (Aug 2018) |
| RR | Soil (Jul 2019) | Jul 2019 | River (Jul 2019) |
| ND | Soil (Jul 2019) | Jul 2019 | Lake (Jul 2019) |
| Archaea | |||||
| Df | SumOfSqs | R2 | F | Pr(>F) | |
| Habitat | 2 | 1.7638 | 0.29862 | 2.6004 | 0.001 |
| Site | 1 | 0.7514 | 0.12721 | 2.2155 | 0.002 |
| Residual | 10 | 3.3914 | 0.57417 | ||
| Total | 13 | 5.9066 | 1.00000 | ||
| Bacteria | |||||
| Df | SumOfSqs | R2 | F | Pr(>F) | |
| Habitat | 2 | 1.4322 | 0.23366 | 1.7254 | 0.001 |
| Site | 1 | 0.5467 | 0.08919 | 1.3172 | 0.049 |
| Residual | 10 | 4.1505 | 0.67714 | ||
| Total | 13 | 6.1294 | 1.00000 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villeneuve, K.; Violette, M.; Lazar, C.S. From Recharge, to Groundwater, to Discharge Areas in Aquifer Systems in Quebec (Canada): Shaping of Microbial Diversity and Community Structure by Environmental Factors. Genes 2023, 14, 1. https://doi.org/10.3390/genes14010001
Villeneuve K, Violette M, Lazar CS. From Recharge, to Groundwater, to Discharge Areas in Aquifer Systems in Quebec (Canada): Shaping of Microbial Diversity and Community Structure by Environmental Factors. Genes. 2023; 14(1):1. https://doi.org/10.3390/genes14010001
Chicago/Turabian StyleVilleneuve, Karine, Michel Violette, and Cassandre Sara Lazar. 2023. "From Recharge, to Groundwater, to Discharge Areas in Aquifer Systems in Quebec (Canada): Shaping of Microbial Diversity and Community Structure by Environmental Factors" Genes 14, no. 1: 1. https://doi.org/10.3390/genes14010001
APA StyleVilleneuve, K., Violette, M., & Lazar, C. S. (2023). From Recharge, to Groundwater, to Discharge Areas in Aquifer Systems in Quebec (Canada): Shaping of Microbial Diversity and Community Structure by Environmental Factors. Genes, 14(1), 1. https://doi.org/10.3390/genes14010001





