Abstract
Molecular biomarkers, such as IDH1/IDH2 mutations and 1p19q co-deletion, are included in the histopathological and clinical criteria currently used to diagnose and classify gliomas. IDH1/IDH2 mutation is a common feature of gliomas and is associated with a glioma-CpG island methylator phenotype (CIMP). Aberrant genomic methylation patterns can also be used to extrapolate information about copy number variation in a tumor. This project’s goal was to assess the feasibility of DNA methylation array for the simultaneous detection of glioma biomarkers as a more effective testing strategy compared to existing single analyte tests. Methods: Whole-genome methylation array (WGMA) testing was performed using 48 glioma DNA samples to detect methylation aberrations and chromosomal gains and losses. The analyzed samples include 39 tumors in the discovery cohort and 9 tumors in the replication cohort. Methylation profiles for each sample were correlated with IDH1 p.R132G mutation, immunohistochemistry (IHC), and previous 1p19q clinical testing to assess the sensitivity and specificity of the WGMA assay for the detection of these variants. Results: We developed a DNA methylation signature to specifically distinguish a IDH1/IDH2 mutant tumor from normal samples. This signature is composed of 11 CpG sites that were significantly hypermethylated in the IDH1/IDH2 mutant group. Copy number analysis using WGMA data was able to identify five of five positive samples for 1p19q co-deletion and was concordant for all negative samples. Conclusions: The DNA methylation signature presented here has the potential to refine the utility of WGMA to predict IDH1/IDH2 mutation status of gliomas, thus improving diagnostic yield and efficiency of laboratory testing compared to single analyte IDH1/IDH2 or 1p19q tests.
1. Introduction
Glioma is one of the most common primary brain tumors representing approximately 25% of all primary brain and other central nervous system tumors and 80% of malignant tumors [1]. Prior to the 2016 publication of the WHO classification of central nervous system tumors [2], classification of glioma tumors was primarily based on the histopathological and microscopic features of hematoxylin and eosin stained sections. According to histological criteria, glial-derived tumors could be classified as either diffuse astrocytoma, anaplastic astrocytoma, oligodendrogliomas, anaplastic oligodendrogliomas, oligoastrocytomas, or glioblastoma [3]. These classifications provided important information about the cellular etiology and developmental differentiation of brain malignancies and were the primary means for grading tumors. However, accurate histological classification of brain tumors is not always straightforward. Although histological evaluation has remained the mainstay for brain tumor classification and grading for decades, there is increasing evidence that molecular genetic testing can supplement histology by identifying genetic biomarkers that are more common in some glioma subtypes than others. The utility of molecular genetic testing to aid tumor classification is best exemplified by the incorporation of several genotype biomarkers in the most recent WHO classification systems [2,4]. The most significant of these genetic biomarkers for the classification of adult-type diffuse gliomas include IDH1 or IDH2 mutation, 1p19q co-deletion, TERT promoter mutations, chromosome 7 gains with chromosome 10 losses, and EGFR amplification, as well as various other aberrations, i.e., ATRX alteration, BRAF alteration, CDKN2A/B homozygous deletion, MYB or MYBL1 alteration, histone H3 mutation, and 10q23 (PTEN) losses [2]. Diffuse gliomas are the most prevalent form of gliomas in adults and are associated with a better overall prognosis. Molecular characterization of diffuse astrocytic and oligodendroglial tumors has led to the recognition of three molecular subtypes that better predict the survival of patients. The best survival is seen in Subtype 1, which is characterized by IDH1 or IDH2 mutation along with co-deletion of the chromosomal arms 1p and 19q. Intermediate survival is seen in Subtype 2, which is characterized by IDH1/IDH2 mutation but intact 1p19q. The least favorable survival is seen in Subtype 3, which is characterized by normal IDH1/IDH2 status and intact 1p19q [5,6]. The 2016 WHO CNS tumor classification included these molecular markers along with histological features to classify different subtypes of diffuse gliomas [2]. Three types of adult-type diffuse gliomas are also recognized in the most recent edition of the WHO CNS tumor classification (2021): astrocytoma, IDH-mutant; oligodendrogllioma, IDH-mutant and 1p19q-codeleted; and glioblastoma, IDH-wildtype [4].
Somatic IDH1/IDH2 mutations occur in approximately 80% of gliomas and secondary glioblastomas but in less than 10% of primary glioblastomas [7]. Two hotspots for IDH1/IDH2 mutation have been described in gliomas: IDH1 codon 132 (p.R132) and IDH2 codon 172 (p.R172). IDH1 and IDH2 normally catalyze the oxidative decarboxylation of isocitrate to alpha-ketoglutarate. Mutations at IDH1 codon 132 and IDH2 codon 172 enhance affinity for alpha-ketoglutarate (α-KG) which is then converted to D-2-hydroxyglutarate (D2HG), a competitive inhibitor of several α-KG-dependent dioxygenases (such as histone demethylases and the TET family of methylcytosine hydroxylases). The enzymatic inhibition induced by the accumulation of D2HG results in increased levels of histone H3 lysine methylation levels, global DNA hypermethylation (CpG island methylator phenotype, CIMP) and altered epigenetic regulation of cellular functions [8,9].
Immunohistochemistry (IHC) represents a common methodology for the direct detection of the most common IDH1 mutation in brain tumors, IDH1 p.R132H. However, a negative IHC result does not rule out the possibility of other IDH1 or IDH2 variants. Given the association between IDH1/IDH2 mutations and CIMP, it has been demonstrated that methylation patterns may be useful as a surrogate marker for mutations of the IDH1/IDH2-associated epigenetic pathway, particularly in cases with less common IDH1/IDH2 variants [8,10]. WGMA testing has the added potential for the concurrent detection of CIMP and relevant copy number variations (i.e., 1p19q co-deletion) in a single assay. In this study, we examined the use of whole-genome DNA methylation array (WGMA) testing to identify IDH1/IDH2 pathway disruption, through the detection of CIMP, in addition to the characterization of additional molecular alterations that may assist in the classification of glioma tumors. In addition, this study identified novel methylation loci associated with glioma CIMP that have the potential to refine the utility of WGMA assessment to detect CIMP and predict IDH1/IDH2 mutation status.
2. Methods
2.1. Sample Selection
This study included a discovery cohort of 39 glioma tumors from the Hamilton region collected between 2010 and 2013. An additional replication cohort of 9 blinded glioma tissue DNA samples was included for WGMA testing. These samples were used to assess the feasibility of our methylation array analysis pipeline for assessment of the IDH1/IDH2 mutation status in a blinded cohort. All samples from the discovery and replication cohort were anonymized and deidentified prior to testing. The study and use of tissues was reviewed and approved by the Hamilton Integrated Research Ethics Board.
Tumor histology was reviewed and interpreted by two senior neuropathologists as part of routine clinical care. Original tumor classifications were based on standard histopathology criteria regarding WHO grading at the time, including cellular morphology, mitotic activity, microvascular hyperplasia, and necrosis.
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor samples. Briefly, tissue curls from each specimen were deparaffinized using xylene and then washed with alcohol to enable DNA extraction using a QIAmp DNA Mini Kit (QIAGEN).
2.2. Immunohistochemistry for IDH1 R132H
Immunohistochemistry was performed on all retrospective samples using a standardized protocol and an anti-IDH1 R132H antibody. All immunohistochemistry results were reviewed by a senior neuropathologist.
2.3. 1p19q LOH Testing
Microsatellite markers on the short arm of chromosome 1 (D1S468 (1p36.3), 2D1S214 (1p36.31), D1S1161 (1p35.2)) and on the long arm of chromosome 19 (D19S559 (19q13.32), D19S412 (19q13.32), D19S601 (19q13.41)) were subjected to PCR amplification using fluorescently labeled primers and visualized by capillary electrophoresis. DNA from peripheral blood was compared with DNA extracted from tumor tissue. For informative markers, loss of heterozygosity (LOH) was calculated from the fluorescence capillary electrophoresis using the formula (B2/B1)/(T2/T), where B is the peak height of allele fragments amplified from blood and T is a peak height in tumor. Each marker is classified into one of the three categories: (1) LOH-positive (LOH value: ≥1.5 or ≤0.65), (2) inconclusive (LOH value: 0.66–0.79 or 1.21–1.49), or (3) no significant loss (LOH value: 0.80–1.20). 1p19q LOH was defined when at least 1 out of 3 markers in each arm (1p and 19q) were LOH-positive (Category 1) and there were no Category 3 markers.
2.4. Whole-Genome Methylation Array and CIMP Analysis
WGMA testing was performed on glioma tissue DNA of 48 individuals using the Infinium MethylationEPIC BeadChip (Illumina) according to a standard protocol. Briefly, FFPE DNA was bisulfite-treated using a Zymo Research EZ DNA Methylation Kit. Due to the degraded nature of FFPE DNA, the DNA was pre-processed using an Illumina Infinium HD FFPE Restore Kit, allowing for the samples to be suitable for amplification in the Illumina Infinium Methylation protocol. During the Infinium methylation protocol, samples underwent a whole-genome amplification, fragmentation, and purification prior to being hybridized to the EPIC array. Fluorescent probes hybridized to the array were detected using the Illumina iScan System. Sample quality was based on unmethylated and methylated median signal intensity as well as the number of probes with detection p-values above background (p < 0.05).
Beta values (estimate of methylation level using ratio of intensities between methylated and unmethylated alleles) were generated using Illumina Genome Studio Software and IDAT files were imported to Partek Genomic Suite software (PGS). An ANOVA test was performed using PGS to compare probe methylation levels between retrospective glioma samples with positive IHC for IDH1 R132H (13 individuals) and retrospective glioma samples with negative IHC for IDH1 R132H (26 individuals). We identified genomic regions (probes) with significant DNA methylation changes that met the following statistical criteria: (1) significant methylation change p < 0.01, (2) F-value (signal to noise) > 20, and (3) methylation estimate value (E, net methylation difference in IDH1-IHC-positive individuals as compared to IDH1-IHC-negative) > 15%. CpG probes with the most significant methylation changes were used to create a hierarchical clustering of differentially methylated genomic regions (epi-signature) in IDH1-p.R132H-positive versus IDH1-p.R132H-negative samples. Replication cohort samples were then plotted together in the hierarchical clustering of the epi-signature to classify their IDH status. A cutoff for average methylation beta values and Z-score was determined to classify CIMP status based on the selected CpG epi-signature and then applied to test results from each specimen within the replication cohort.
2.5. Copy Number Analysis
The 1p19q deletion analysis from the EPIC array data was performed according to published bioinformatics tools for copy number calling using the Conumee package [11]. Briefly, copy number calling consists of the following principal steps: normalization of probe intensities, calculation of the log R ratios (LRRs), and segmentation and determination of copy number status for each segment based on a chosen cut-off level or a p-value threshold. Baseline normalization was performed using blinded 1p19q non-deleted samples (previously tested by LOH). The cutoff for copy number determination was set at >±0.3.
2.6. Brain Tumor Methylation Classifier
IDAT files generated by Illumina Genome Studio Software were uploaded to v11b4 version of the online CNS tumor methylation classifier (https://www.molecularneuropathology.org (accessed on 27 July 2020)) and reports were produced as previously described [10].
2.7. Next-Generation Sequencing
Next-generation sequencing (NGS) was performed using Ion AmpliSeq™ Cancer Hotspot Panel v2, which targets hotspot regions of 50 cancer genes, including IDH1 (NM_005896.2) and IDH2 (NM_002168.2), on the Ion GeneStudio S5 sequencer (ThermoFisher, Waltham, MA, USA). Torrent Suite Software (version 5.4) was used for variant detection and Ion Reporter™Software (version 5.6, Waltham, MA, USA) was then used for the annotation of variants. Quality control parameters include a minimum coverage per base of 200×, and a minimum of 500× average coverage for IDH1 and IDH2. All discovery and replication samples with sufficient DNA remaining were tested by NGS. This assay was used to test discovery samples with a discordant IDH status between IHC and DNA methylation array; and to confirm the IDH status of the replication samples after DNA methylation analysis. All samples met the quality control cutoff, except 1 sample (Sample 20) that had an IDH2 coverage of 332×.
3. Results
3.1. Cohort Demographics
Tissues from thirty-nine tumors were included in the discovery cohort (refer to Supplemental Table S1). The average age of participants was 54 years (ranging from 27–82 years of age). Twenty-seven (69%) participants were male. Based on original histopathological classifications, 15 (38%), 14 (36%), and 10 (26%) samples were classified as oligodendroglioma (5 grade II, 10 grade III), oligoastrocytoma (7 grade II, 7 grade III), and glioblastoma, respectively.
The replication cohort included tissues from nine tumors (refer to Supplemental Table S1). The average age of participants in the replication cohort was 58 years (ranging from 32–71 years of age) and included four males (44%). Based on original histopathological classifications, five samples were classified as oligodendroglioma (two grade II, three grade III) and three were classified as glioblastoma. An original histopathological classification of the ninth sample was not available.
3.2. WGMA and CIMP
Quality control parameters: WGMA testing of the 48 samples yielded an average number of detected CpG (detection p-value < 0.05) of 846,023 (min = 746,229; max = 864,115), which demonstrates an adequate coverage of the total probes tested by the assay (over 850,000). For the selected 11 CpG sites (described below), the detection p-value was below 0.005 for all probes in all samples, except for one probe with p = 0.01 in one sample (refer to Supplemental Table S2).
The CIMP epi-signature was created using the discovery cohort of 39 tissue samples obtained from patients with diffuse glioma and was analyzed according to their IDH1 immunocytochemistry status. Various methylation changes at a single probe level were identified across the genome in the IDH1-IHC-positive group when compared to the IDH1-IHC-negative group. Using a less stringent cutoff of p < 0.01 and E > 10%, 851 CpG sites showed significant methylation change, the majority consisting of hypermethylation (844 CpG sites). A more stringent cutoff of p < 0.01, E > 15%, and F-value > 20 revealed 11 CpG sites, significantly hypermethylated, in the IDH-positive group (refer to Table 1 and Supplementary Table S3) compared to the IDH-negative group. The top 11 CpG sites comprise our epi-signature and were then used to create a hierarchical clustering of differentially methylated genomic regions, which demonstrated a unique methylation profile and sub-clustering of individuals positive for IDH1 p.R132H compared with individuals negative for IDH p.R132H (Figure 1). To identify the biological processes and molecular functions most enriched within our data set, we analyzed the 10 unique genes that overlapped with the 11 CpG sites that were differentially methylated in IDH-positive cases, using Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/ (accessed on 6 October 2022)). Due to a small number of genes, there were no significantly enriched categories after correction for multiple testing. However, interestingly, 5 out of 10 genes have roles in metal ion binding (p-value, 0.017, refer to Supplemental Table S4).

Table 1.
CIMP epi-signature: top 11 CG sites that were differentially methylated in IDH-positive group versus IDH-negative group.
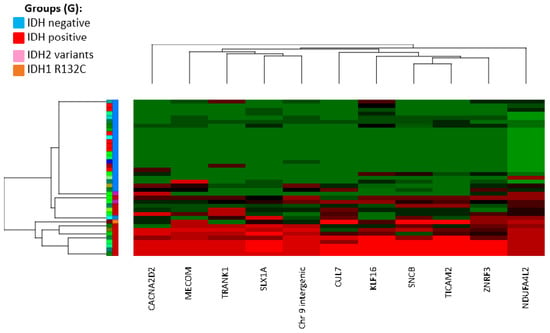
Figure 1.
Hierarchical clustering of the top 11 CG sites (column) that were differentially methylated (epi-signature) in IDH-positive group versus IDH-negative group. Row; S: individual samples. G: group (blue: IDH-negative, red: IDH-positive, pink: IDH2 variants by NGS, orange: IDH1 variant by NGS).
Comparison of the CIMP status by EPIC methylation array, the IDH1 mutation status tested by IHC and NGS is shown in Table 2. Initial classification of CIMP-positive versus CIMP-negative samples was based on hierarchical clustering using the 11 CpG sites. For the discovery cohort (n = 39), the average methylation (beta value; 0 = unmethylated; 1 = fully methylated) across those 11 CpG sites in CIMP-positive samples was significantly higher (mean = 0.63, SD = 0.25) than CIMP-negative samples (mean = 0.079, SD = 0.078) (refer to Supplemental Table S1). Three samples (Samples 21, 30, 33) with previous negative immunohistochemistry for IDH1 p.R132H were clustered in the IDH-positive group (Figure 1) and showed an average methylation of 0.60, 0.40, and 0.40. Sequencing analysis of two of those samples revealed a less common oncogenic IDH1 variant c.394C>T (HGVS: NM_005896.3(IDH1):c.394C>T, p.(Arg132Cys)) in Sample 21, and an IDH2 oncogenic variant, c.515G > A (HGVS: NM_002168.3(IDH2):c.515G>A, p.(Arg172Lys) in Sample 30. However, we could not obtain enough DNA to confirm the IDH1/IDH2 mutation status of the third discordant sample (Sample 33).

Table 2.
Tumor reclassification based on histological, molecular, and epigenetic findings. Molecular testing results for each case are provided, including IDH1 p.R132H immunohistochemistry, IDH1/IDH2 sequencing, CIMP positivity (as determined by hierarchical clustering, using the top 11 CpG sites that were differentially methylated) and 1p/19q status (based on single analyte testing and WGMA). Replication cohort samples are underlined. Tumor classifications as assessed using the MolecularNeuro-pathology.org Classifier (v11b4, accessed on 27 July 2020) are provided; calibrated scores for class and subclass are shown in brackets (recommended score class: >0.90, subclass > 0.5). Original tumor classifications as assessed by two senior neuropathologists are also provided. Histological criteria used for the original classifications include: oligodendroglioma grade II—no necrosis or microvascular hyperplasia (mvh); oligodendroglioma grade III—necrosis/mvh, increased mitotic activity; astrocytoma grade II—no necrosis/mvh or increased mitotic activity; astrocytoma grade III (anaplastic astrocytoma)—increased mitotic activity/anaplasia, no necrosis/mvh; glioblastoma—morphologically astrocytic with necrosis/mvh; oligoastrocytoma—morphologically mixed oligo/astrocytic, no necrosis/mvh or increased mitotic activity; oligoastrocytoma grade III—with anaplasia, increased mitotic activity.
One sample (Sample 9) was positive for an IDH2 c.476G>A (HGVS: NM_002168.3(IDH2):c.476G>A, p.(Arg159His)) variant detected by NGS. This variant was interpreted as a variant of uncertain clinical significance (VUS) based on limited information in either public tumor and control population databases or in the available literature. Our CIMP analysis revealed that this sample clustered with the IDH-negative group and had an average methylation of 0.20, suggesting that this IDH2 variant does not significantly affect methylation status and is therefore likely benign.
Following the primary analysis with the discovery cohort, a small replication cohort (n = 9) with unknown IDH1/IDH2 mutation status, was also examined (Figure 2). Analysis of additional replication cohort samples was not possible due to limited tissue and resources.
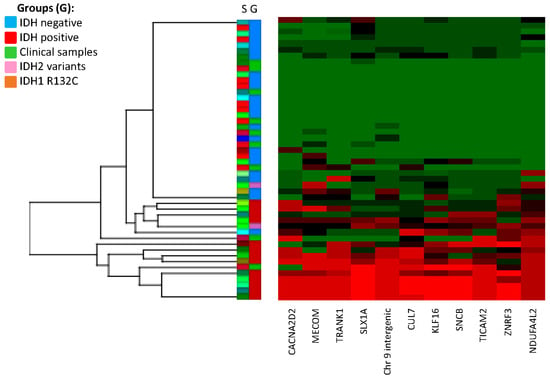
Figure 2.
Hierarchical clustering of discovery and replication cohorts. The top 11 CG sites (column) that were differentially methylated (epi-signature) in IDH-positive versus IDH-negative group. Row; S: individual samples. G: group (blue: IDH-negative, red: IDH-positive, pink: IDH2 variants by NGS, orange: IDH1 variant by NGS, green: replication cohort samples).
Hierarchical clustering analysis identified two samples (Sample 44 and Sample 45) that clustered with the hypermethylated IDH-positive group and seven samples that clustered with the IDH-negative group. The samples clustering with the IDH-positive group (CIMP-positive) had an average methylation beta value for the 11 CpG epi-signature sites of 0.42 and 0.75, whereas the remaining seven CIMP-negative samples had an average beta value ranging from 0.08 to 0.21. Subsequent sequencing analysis was able to confirm the presence of the common IDH1 p.R132H (HGVS: NM_005896.3(IDH1):c.395G>A, p.(Arg132His)) mutation on Samples 44 and 45. All of the remaining samples were negative for either IDH1 or IDH2 oncogenic variants by NGS.
The hierarchical clustering data from the discovery and replication cohorts demonstrate suitable separation of IDH-positive versus IDH-negative samples and suggest that the top 11 epi-signature targets may be used to identify tumors with IDH1 or IDH2 methylation defects. Using a calculated Z-score from the average methylation from CIMP-negative samples (Supplementary Table S3), we observed that the majority of CIMP-positive samples (15 out of 17) have a score > 4 (average methylation value is 4 standard deviations higher than the CIMP-negative average), whereas CIMP-negative samples have a Z-score < 2. Using a stringent average methylation (beta value) cutoff of ≥0.4 and Z-score ≥ 4 for CIMP-positive prediction, and an average beta value ≤ 0.2 and Z-score ≤ 2 for CIMP-negative prediction, we could accurately predict the IDH status of 44 out of 48 samples. Analysis of additional samples is required to further refine these cutoffs and verify the sensitivity and specificity of this test.
3.3. 1p19q Co-Deletion Analysis
Because CNV analysis relies on the probe intensity measurement across all chromosomes, only samples with high overall intensity (unmethylated and methylated intensity (log2) > 9.5) were included in the CNV analysis. Using this quality cutoff, 18 samples were excluded from further copy number analysis (refer to Table 2; excluded samples are indicated by “—” in the “1p del”, “19q del”, and “additional CNV” columns). Methylation array data from the remaining 30 samples were evaluated for copy number variation and assessment of 1p19q co-deletion status.
For the previous 1p19q PCR testing, 12 samples had the 1p19q status classified as inconclusive (refer to Table 2; inconclusive previous 1p19q co-deletion results are highlighted). These inconclusive reports resulted from a lack of available testing, lack of informative markers in each chromosomal region, or segmental loss involving LOH in only one of the chromosomal arms. Thus, for concordance between methylation array and LOH testing, only samples that had LOH patterns consistent with 1p19q co-deletion or with no LOH detected (total 18) were included. For these 18 samples, LOH testing and DNA methylation array analysis were concordant for all 18 samples (Table 2). Five out of the eighteen samples were positive for 1p19q loss by LOH; DNA methylation array was able to confirm the loss in all five cases, although only weak 19q loss was noted in one sample (Sample 30). A plot for the normalized copy number analysis for one sample is represented in Figure 3A.
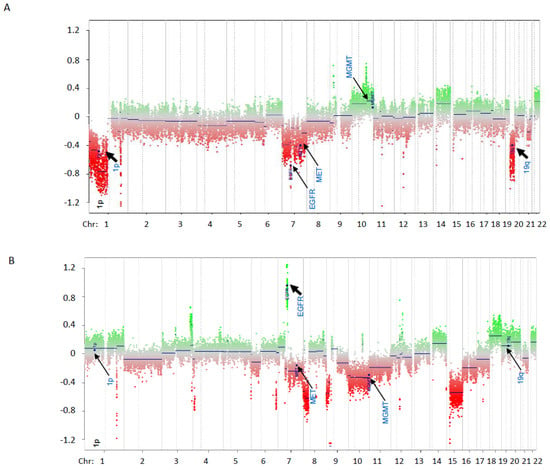
Figure 3.
Genome-wide copy number analysis from methylation array data (Infinium MethylationEPIC BeadChip). All chromosomes are represented in the X-axis. Normalized copy number is represented in Y-axis (baseline = 0). Green dots represent probes with normalized copy number above 0 while red dots represent probes with normalized copy number below 0. Positions of probes for 1p, 19q, EGFR, MGMT and MET are indicated by arrows. (A) Example of genome-wide copy number analysis with 1p19q co-deletion (Sample 44). Instances of 1p19q co-deletion are highlighted by black arrows. (B) Example of genome-wide copy number analysis with EGFR amplification. EGFR amplification is highlighted by a black arrow.
Copy number analysis of WGMA also demonstrated copy number variation in regions other than 1p and 19q (refer to Table 2) that may be relevant for tumor classification, including EGFR amplification (refer to Table 2 and Figure 3B). Further validation of the WGMA to reliably detect copy number variants of chromosomal segments other than 1p and 19q was beyond the scope of this project but are included in Table 2 for reference.
3.4. Brain Tumor Methylation Classifier
To compare the performance of the epi-signature against a pre-existing brain tumor methylation classifier system, methylation array data were assessed using a brain tumor methylation classifier developed by the German Cancer Consortium (DKTK) (https://www.molecularneuropathology.org (accessed on 27 July 2020)). The DKTK methylation classifier was chosen for analysis based on public availability at the time of the study and previous evidence demonstrating utility in the clinical setting [10,12,13]. Using the DKTK methylation classifier, the majority of our samples did not reach the optimal calibrated score threshold of ≥0.9 for methylation class and a threshold value of ≥0.5 for subclasses within methylation class families [10], probably due to suboptimal DNA quality (low intensity). Of those cases that reached the recommended scores (≥0.9 for methylation class; 0.5 for methylation subclasses; 15 samples), the IDH status and 1p19q co-deletion status were 100% concordant with our findings.
3.5. Refinement of Glioma Classification Using Genomic Information
According to the 2016 and 2021 World Health Organization Classification of Tumors of the Central Nervous System, molecular features should be incorporated in the definition of glioma tumors in addition to histological features [2,4,14]. Using the information obtained for IDH status and 1p19q co-deletion from WGMA, we could refine the classification of many tumor entities from the original histological classification (Table 2). In our cohort, a large number of oligoastrocytomas were observed in the histological classification. The 2016 WHO CNS classification strongly discourages the diagnosis of oligoastrocytoma, claiming that nearly all tumors with histological features suggesting both an astrocytic and an oligodendroglial component can be classified as either astrocytoma or oligodendroglioma using genetic testing for IDH mutation and 1p19q loss [2,4]. Following the current guideline, of the 14 oligoastrocytomas initially observed, 5 were reclassified to diffuse astrocytoma, IDH mutant; 5 were diffuse astrocytoma, IDH-wild-type; and 4 were oligodendrogliomas, IDH mutant, 1p19q co-deleted. In addition, our cohort seems to have an over-classification for oligodendrogliomas. Of the 20 initially classified oligodendrogliomas, only 4 met the genetic determinants for this entity (IDH mutation and 1p19q loss). Three histologically classified oligodendrogliomas had IDH mutation but intact 1p19q, being reclassified either to astrocytoma or diffuse astrocytoma, IDH mutant. Seven of the histologically classified oligodendrogliomas were reclassified as glioblastoma, IDH-wild-type. The remaining six samples designated as oligodendroglioma, NOS, were IDH-wild-type with 1p19q intact. However, careful examination of these genetic inconclusive specimens should be undertaken for features of glioblastoma or other tumors of similar histology.
4. Discussion
The advent of high throughput molecular techniques enabled the comprehensive characterization of genetic and epigenetic alterations in diffuse gliomas. Over the past few decades, several biomarkers of glioma have been identified, including IDH1/IDH2 mutations, 1p19q co-deletion, CDKN2A/B homozygous deletion, chromosome 7 gains/chromosome 10 losses, PTEN deletion, monosomy 6, and genetic alterations in ATRX, BRAF, and EGFR, as well as promoter methylation of MGMT [8,15,16,17]. Many of these biomarkers are currently being clinically tested to aid in the diagnosis of glioma tumors as part of the revised 2021 WHO classification system of CNS tumors, as well as to provide a better prognosis and treatment choice for patients with these tumors. The current 2021 WHO classification system also introduced methylome profiling as an effective ancillary method for brain and spinal cord tumor classification when used alongside other, standard technologies, including histology.
In this study, we evaluated the use of a DNA methylation array to simultaneously identify the two most important biomarkers of glioma: IDH1/IDH2 status and 1p19q co-deletion. It has been shown that IDH1/IDH2 mutations are associated with a global change in the DNA methylation levels, known as a CpG-island methylator phenotype (CIMP), which can be used to characterize the IDH1/IDH2 status of these tumors [8,12,18,19]. Initial studies using methylation data from the Illumina GoldenGate array had characterized CIMP in glioma tumors, which was associated with both glioma histological subtype and IDH1/IDH2 mutations [8,19]. In 2017, Paul and colleagues developed a DNA methylation signature to identify glioma subtypes. Using different sets of CpG methylation biomarkers from the 450K array, Paul et al. were able to classify IDH1/IDH2 mutant tumors versus IDH1/IDH2 wild-type tumors, as well as oligodendroglioma, astrocytoma, and glioblastoma [18], proving to be a powerful tool for glioma classification. Using the EPIC methylation array, which is a more robust array covering > 850,000 CpG sites, and a discovery cohort of 39 glioma tissue samples, we successfully developed a DNA methylation (CIMP) signature to specifically distinguish IDH1/IDH2-positive samples from IDH1/IDH2-negative samples. This signature is composed of 11 CpG sites that were significantly hypermethylated in the IDH-positive group. It is unclear whether the hypermethylation of these 11 CpG sites directly impacts oncogenic processes. Intriguingly, 5 of the 10 corresponding genes have roles in metal ion binding, although the significance of this observation is uncertain. The 11 CpG sites were not included in previous published studies and may further refine the predictive value of methylation analysis to identify aberrant IDH1/IDH2 pathways. Using this CIMP signature, we were able to correctly assign the IDH1/IDH2 status of an additional nine samples from the replication cohort. Although further studies with a larger number of replication cohort samples are required to fully validate this epi-signature, the preliminary findings from these samples suggest the utility of the signature to differentiate between IDH1/IDH2 mutant and normal tissues. This represents an important finding that has the potential to refine the utility of WGMA to predict the IDH1/IDH2 mutation status of gliomas, thus improving diagnostic yield and efficiency of laboratory testing compared to single analyte IDH1/IDH2 tests. In addition to correctly identifying the IDH1/IDH2 status of tumors with the common IDH1 p.Arg132His variant, the reported epi-signature also enabled the identification of tumors with less common pathogenic IDH1 and IDH2 mutations, including one tumor with an IDH1 p.Arg132Cys variant and one tumor with an IDH2 p.Arg172Lys variant. The IDH1/IDH2 mutation status would have been incorrectly assigned in these tumors if relying on immunohistochemistry for the common IDH1 variant as an isolated test. The CIMP signature may also provide additional information to better understand the functional consequences of IDH1 or IDH2 variants of unknown significance detected by NGS. As we presented here, NGS identified an IDH2 variant (c.476G>A, p.(Arg159His)) that had been initially interpreted as a variant of uncertain clinical significance. The absence of significant hypermethylation at the examined loci and clustering of this variant with the IDH-negative group, suggests that the variant is unlikely to disrupt normal DNA-methylation-based epigenetic processes in this tumor and is therefore is likely benign. Nevertheless, it is important to note that CIMP is not a specific phenotype of IDH1/IDH2 oncogenic variants but could also be detected in the presence of mutations in other regulators of the IDH-epigenetic pathway, such as TET2 mutations in leukemia [20] and the BRAF p.V600E mutation in colorectal cancer [21,22]. Recent evidence has also identified a new group of IDH-positive brain tumors with hypomethylated genomes rather than genomic hypermethylation that is characteristic of most IDH mutant gliomas [23]. Although it has been suggested that the CIMP-specific DNA methylation targets may be non-overlapping and manifested by several molecular pathways across different tumor types [22], testing the CIMP status of glioma tumors only provides a subjective assessment of IDH mutation status.
In addition to IDH status, WGMAs can be used to evaluate chromosome copy number. Of particular interest herein are the chromosomes 1p and 19q, which are co-deleted in the oligodendrogliomas with concurrent IDH1/IDH2 mutations. Using the R package Conumee for normalization and copy number calling, we detected 1p19q co-deletions as well as other relevant copy number variants, including chromosome 7 gains and chromosome 6 and 10 losses. The raw data also suggest possible detection of other clinically relevant molecular biomarkers, including EGFR amplification (refer to Figure 3B), MET (chromosome region 7q31.2) amplification, MDM2 (chromosome region 12q15) amplification, or CDKN2A/B (chromosome region 9p21.3) deletion. Detection of these molecular changes by WGMA can further streamline tumor classification by simultaneously screening for multiple relevant molecular markers in a single assay and limiting the number of tests required for tumors with limited sample quantity. Use of the EPIC WGMA for copy number assessment of these additional gene regions requires further validation against samples with a known copy number status of the relevant loci. An additional limitation of this study was the quality of the DNA extracted, presumably due to older tissue FFPE from our discovery samples (collected 5–8 years prior to WGMA testing and with either tumor heterogeneity or contamination with normal tissue). While copy number analysis was possible in all of the samples examined, poor overall WGMA array signal intensities in some samples made it clear that identification of subtle chromosome changes was more difficult than in other samples. More recently collected samples and microdissection of relevant tumor material in specimens with considerable tumor and normal tissue admixture may result in lower failure rates but we were unable to test this in the current study. Accordingly, the DNA quality was adequate and copy number calls were successful for the nine more recent (collected within 1 year) replication cohort samples that were included in the study, further highlighting the importance of specimen age as a pre-analytic consideration for FFPE tumor assessment using whole-genome methylation profiling.
Recently, a brain tumor methylation classifier was developed by the German Cancer Consortium (DKTK) to identify distinct DNA methylation classes of CNS tumors (free online tool: www.molecularneuropathology.org (accessed on 27 July 2020)) [10]. This classifier comprises 82 CNS tumor methylation classes and uses the information on methylation status (e.g., IDH status) as well as copy number changes (e.g., 1p19q co-deletion) to classify the tumors. In 2019, two studies evaluating the use of this classifier in glioma testing in a clinical laboratory showed that the classifier improved the diagnostic approach, especially for cases with ambiguous histology or non-informative molecular profiles [12,13]. However, these studies also highlight some limitations of this classifier in that a subset of samples did not reach the optimal calibrated score for use of this tool. Tumor cellularity and heterogeneity may represent additional limitations for the use of methylation-based classification tools. A study evaluating spatial heterogeneity showed that, while IDH-mutant gliomas have homogeneous methylation-based classification, intratumor variability in DNA methylation within glioblastomas, IDH-wild-type, and meningiomas can potentially affect tumor classification of these methylation-based biomarkers [24]. Applying the DKTK classifier to our study samples, only 15 out of 48 samples met the recommended calibrated scores to achieve optimal sensitivity and specificity for methylation class prediction [10]. Among these samples, IDH1/IDH2 mutation status and 1p19 co-deletion were concordant with results obtained by our in-house protocol. However, for the remaining samples, the IDH1/IDH2 status was not reliably assigned, likely due to suboptimal quality of DNA extracted from FFPE material. Thus, our in-house protocol for EPIC DNA methylation analysis of IDH1/IDH2 status and 1p19q co-deletion showed a better diagnostic yield than the other classifier used. Additional DNA methylation signatures for classification and/or prognostication of gliomas as well as other tumors have been developed in the last year and have been shown to be cost-effective and improve diagnostics over current clinical assays [25,26,27,28]. Further assessment of the utility of these classifiers for the detection of CIMP versus the CIMP epi-signature reported herein is beyond the scope of this project but represents an important area for future study.
The technical advantages of the EPIC DNA methylation array compared to IHC, targeted sequencing analysis, and copy number assay testing are grounded in three reasons: the first is the analytical advantage of using a single test to detect both IDH and 1p19q status, in addition to other copy number changes and promoter DNA methylation changes; the second is when there is a need to obtain a more accurate diagnosis for tumors with unusual or non-specific histology (i.e., oligoastrocytic tumors), and the third is when the molecular testing does not yield diagnostically informative results, such as in the case of IDH1 VUS identification. Finally, the implementation of robust molecular techniques, such as EPIC array and next-generation sequencing, will allow for a more precise categorization of glioma tumors according to the 2021 WHO classification of CNS tumors, and ultimately improve the diagnosis and tailoring of patient therapy.
5. Conclusions
Accurate detection of IDH1/IDH2 mutation and 1p19q co-deletion status is important for appropriate tumor classification of gliomas under the most recent 2021 WHO classification of central nervous tumors. We describe a novel DNA methylation epi-signature that specifically distinguishes brain tumors having oncogenic IDH1/IDH2 variants from tumors with normal IDH1/IDH2. Copy number assessment of WGMA data further enabled accurate detection of tumors having 1p19q co-deletion. The ability to accurately detect IDH1/IDH2 mutation status and 1p19q co-deletion in a single test enhances lab efficiency and reduces the need for additional testing of samples having limited material, Although further validation of this DNA methylation epi-signature is required, it has the potential to refine the utility of WGMA to predict IDH1/IDH2 mutations status in gliomas, thus enhancing patient care through improved diagnostic yield and accuracy compared to single analyte IDH1/IDH2 or 1p19q tests.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13112075/s1, Table S1: Demographic and histopathological information of glioma tissues samples; Table S2: Detection p-value per probe per sample for 11 differentially methylatied CpG sites; Table S3: Methylation beta values of each sample for 11 differentially methylated CpG sites; Table S4: DAVID (Database for Annotation, Visualization and Integrated Discovery) functional annotation for 10 genes overlapping 11 differentially methylated CpG sites.
Author Contributions
Conceptualization, E.M., D.G., J.P. and H.H; methodology, L.C.S., J.M. and M.C.; formal analysis, L.C.S., J.M., M.C. and D.G.; data curation, L.C.S., J.M., E.M., D.G. and J.P.; writing—original draft preparation, L.C.S., J.M., E.M. and D.G.; writing—review and editing, L.C.S., J.M., H.H., J.P., G.P., M.C., D.G. and E.M.; visualization, L.C.S., M.C. and E.M.; supervision, E.M., J.P. and G.P.; project administration, E.M.; funding acquisition: E.M., L.C.S. and J.P. All authors have read and agreed to the published version of the manuscript.
Funding
Ontario Molecular Pathology Research Network (OMPRN; Award number CPTRG-006), McMaster University Pathology and Molecular Medicine Resident Research Grant.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Hamilton Integrated Research Ethics Board (Approval Code: 3190, Approval Date: 18 July 2017, renewed 16 July 2019).
Informed Consent Statement
Patient consent was waived. All samples were retrospective, anonymized and deidentitifed.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21 (Suppl. 5), v1–v100. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Weller, M.; Weber, R.; Willscher, E.; Riehmer, V.; Hentschel, B.; Kreuz, M.; Felsberg, J.; Beyer, U.; Löffler-Wirth, H.; Kaulich, K.; et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015, 129, 679–693. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Marzouka, N.-A.; Nordlund, J.; Bäcklin, C.L.; Lönnerholm, G.; Syvänen, A.-C.; Almlöf, J.C. CopyNumber450kCancer: Baseline correction for accurate copy number calling from the 450k methylation array. Bioinformatics 2016, 32, 1080–1082. [Google Scholar] [CrossRef]
- Jaunmuktane, Z.; Capper, D.; Jones, D.T.W.; Schrimpf, D.; Sill, M.; Dutt, M.; Suraweera, N.; Pfister, S.M.; von Deimling, A.; Brandner, S. Methylation array profiling of adult brain tumours: Diagnostic outcomes in a large, single centre. Acta Neuropathol. Commun. 2019, 7, 24. [Google Scholar] [CrossRef]
- Karimi, S.; Zuccato, J.A.; Mamatjan, Y.; Mansouri, S.; Suppiah, S.; Nassiri, F.; Diamandis, P.; Munoz, D.G.; Aldape, K.D.; Zadeh, G. The central nervous system tumor methylation classifier changes neuro-oncology practice for challenging brain tumor diagnoses and directly impacts patient care. Clin. Epigenet. 2019, 11, 185. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Brandner, S.; Brat, D.J.; Ellison, D.W.; Giangaspero, F.; Hattab, E.M.; Hawkins, C.; Judge, M.J.; Kleinschmidt-DeMasters, B.; et al. Data Sets for the Reporting of Tumors of the Central Nervous System: Recommendations From The International Collaboration on Cancer Reporting. Arch. Pathol. Lab. Med. 2020, 144, 196–206. [Google Scholar] [CrossRef]
- Riemenschneider, M.J.; Hegi, M.E.; Reifenberger, G. MGMT promoter methylation in malignant gliomas. Target. Oncol. 2010, 5, 161–165. [Google Scholar] [CrossRef]
- Reifenberger, J.; Reifenberger, G.; Liu, L.; James, C.D.; Wechsler, W.; Collins, V.P. Molecular genetic analysis of oligoden-droglial tumors shows preferential allelic deletions on 19q and 1p. Am. J. Pathol. 1994, 145, 1175–1190. [Google Scholar] [PubMed]
- Wiestler, B.; Capper, D.; Holland-Letz, T.; Korshunov, A.; Von Deimling, A.; Pfister, S.M.; Platten, M.; Weller, M.; Wick, W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013, 126, 443–451. [Google Scholar] [CrossRef]
- Paul, Y.; Mondal, B.; Patil, V.; Somasundaram, K. DNA methylation signatures for 2016 WHO classification subtypes of diffuse gliomas. Clin. Epigenetics 2017, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.C.; Smith, A.A.; Zheng, S.; Koestler, D.C.; Houseman, E.A.; Marsit, C.; Wiemels, J.L.; Nelson, H.; Karagas, M.R.; Wrensch, M.R.; et al. DNA Methylation, Isocitrate Dehydrogenase Mutation, and Survival in Glioma. JNCI J. Natl. Cancer Inst. 2011, 103, 143–153. [Google Scholar] [CrossRef]
- Ichimura, N.; Shinjo, K.; An, B.; Shimizu, Y.; Yamao, K.; Ohka, F.; Katsushima, K.; Hatanaka, A.; Tojo, M.; Yamamoto, E.; et al. Aberrant TET1 Methylation Closely Associated with CpG Island Methylator Phenotype in Colorectal Cancer. Cancer Prev. Res. 2015, 8, 702–711. [Google Scholar] [CrossRef]
- Yagi, K.; Akagi, K.; Hayashi, H.; Nagae, G.; Tsuji, S.; Isagawa, T.; Midorikawa, Y.; Nishimura, Y.; Sakamoto, H.; Seto, Y.; et al. Three DNA Methylation Epigenotypes in Human Colorectal Cancer. Clin. Cancer Res. 2010, 16, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J. Clin. Investig. 2014, 124, 17–23. [Google Scholar] [CrossRef]
- Li, K.K.-W.; Shi, Z.-F.; Malta, T.M.; Chan, A.K.-Y.; Cheng, S.; Kwan, J.S.H.; Yang, R.R.; Poon, W.S.; Mao, Y.; Noushmehr, H.; et al. Identification of subsets of IDH-mutant glioblastomas with distinct epigenetic and copy number alterations and stratified clinical risks. Neuro Oncol. Adv. 2019, 1, vdz015. [Google Scholar] [CrossRef]
- Vega, S.F.; Wenger, A.; Kling, T.; Bontell, T.O.; Jakola, A.S.; Carén, H. Spatial heterogeneity in DNA methylation and chromosomal alterations in diffuse gliomas and meningiomas. Mod. Pathol. 2022, 35, 1551–1561. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Q.; Zhang, Z.-Y.; Long, L.; Ezhilarasan, R.; Karp, J.M.; Tsirigos, A.; Snuderl, M.; Wiestler, B.; Wick, W.; et al. DNA methylation-based epigenetic signatures predict somatic genomic alterations in gliomas. Nat. Commun. 2022, 13, 4410. [Google Scholar] [CrossRef]
- Liu, Q.; Reed, M.; Zhu, H.; Cheng, Y.; Almeida, J.; Fruhbeck, G.; Ribeiro, R.; Hu, P. Epigenome-wide DNA methylation and transcriptome profiling of localized and locally advanced prostate cancer: Uncovering new molecular markers. Genomics 2022, 2022, 110474. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, H.; An, K.; Liu, Z.; Hai, L.; Li, R.; Zhou, Y.; Zhao, W.; Jia, Y.; Wu, N.; et al. Whole-genome bisulfite sequencing analysis of circulating tumour DNA for the detection and molecular classification of cancer. Clin. Transl. Med. 2022, 12, e1014. [Google Scholar] [CrossRef]
- Gu, X.; Huang, X.; Zhang, X.; Wang, C. Development and Validation of a DNA Methylation-related Classifier of Circulating Tumour Cells to Predict Prognosis and to provide a therapeutic strategy in Lung Adenocarcinoma. Int. J. Biol. Sci. 2022, 18, 4984–5000. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).