Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Processing
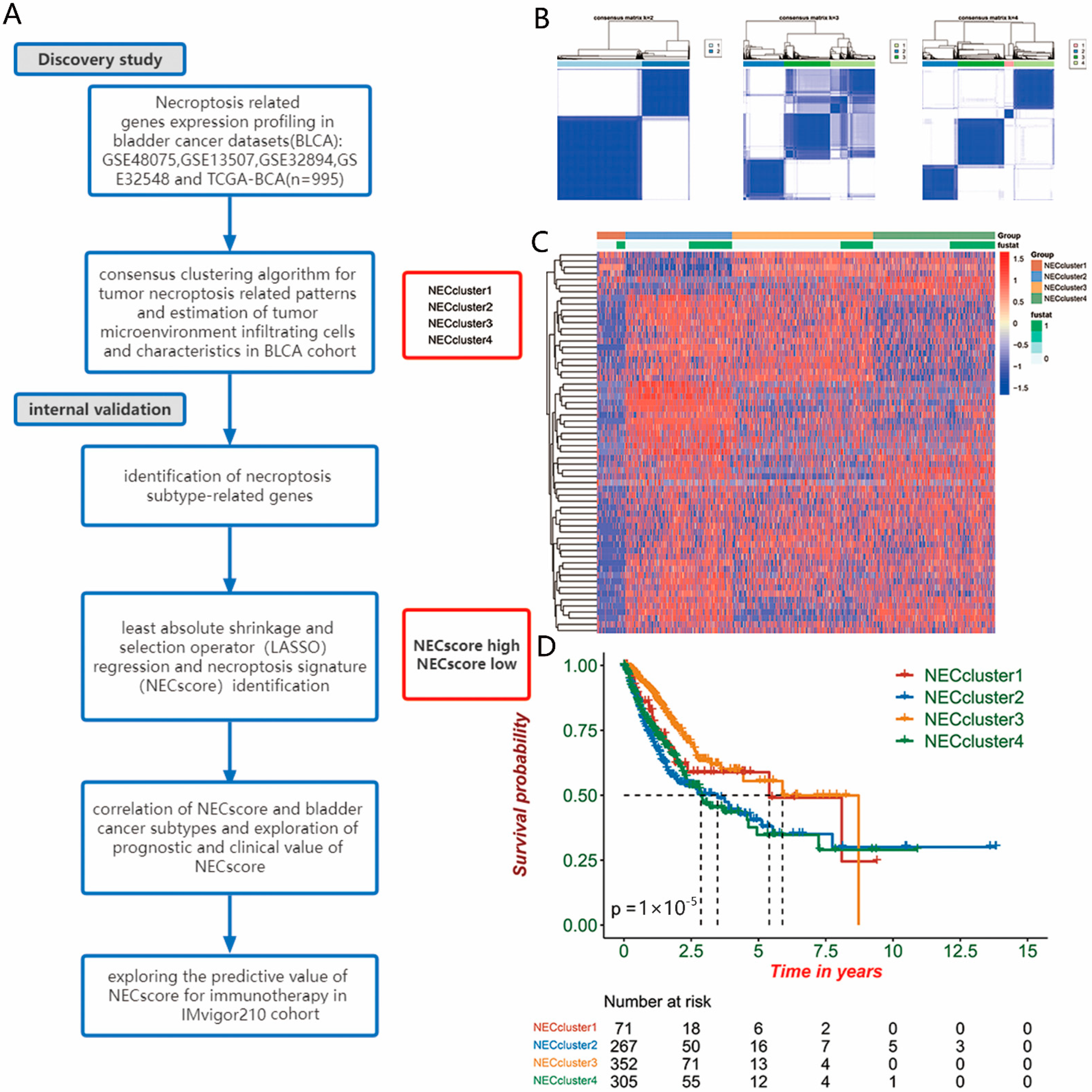
2.2. Consensus Clustering for Necroptosis-Related Patterns
2.3. Identification of Differentially Expressed Gene between Different Necroptosis-Related Patterns
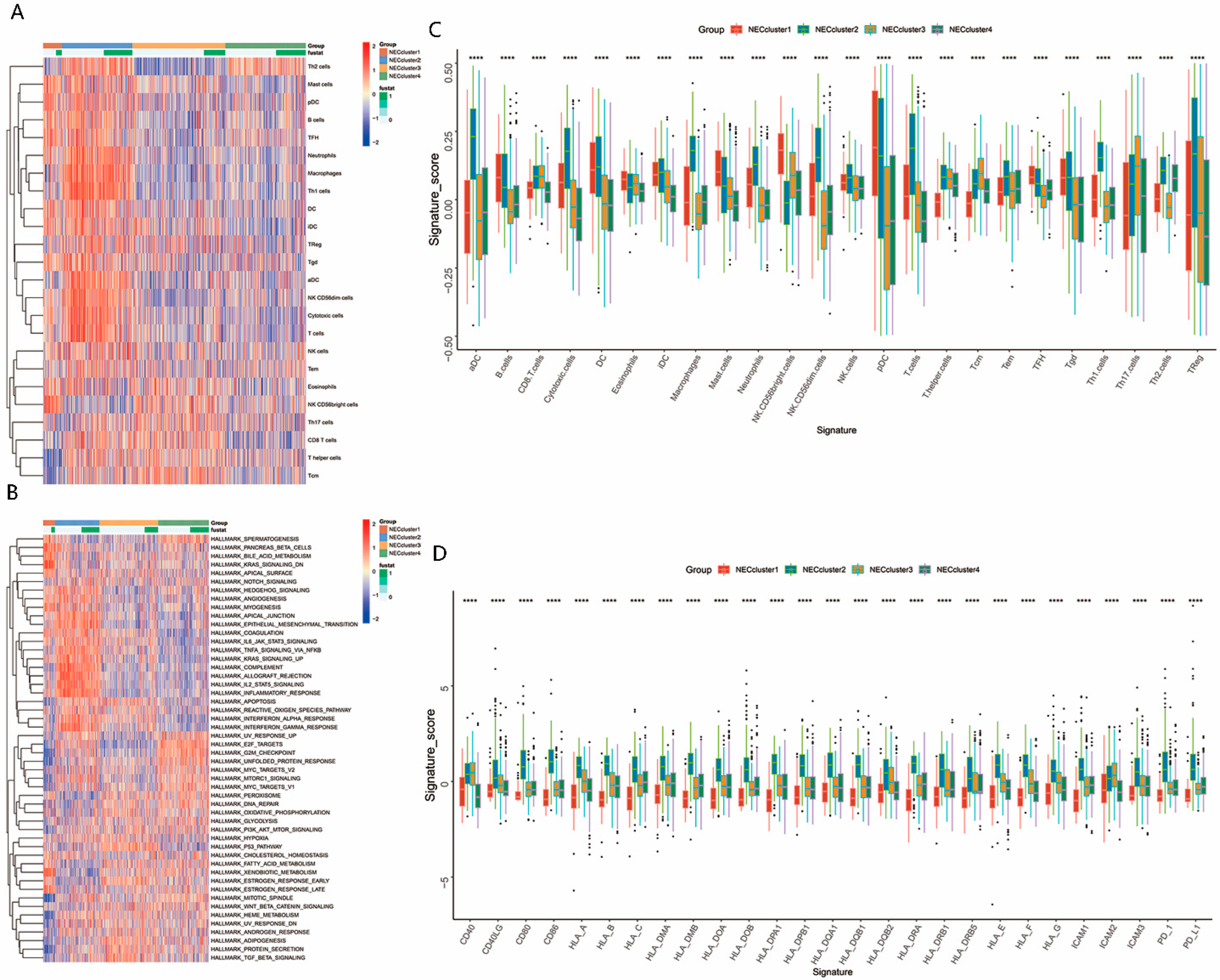
2.4. Evaluation of Infiltrating Immune Cells in the TME
2.5. Functional and Pathway Enrichment Analyses
2.6. Establishment of NEC-Cluster-Related Gene Signature
2.7. Tissue Specimens
2.8. RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR (qRT-PCR)
2.9. Statistical Analyses
3. Results
3.1. Characterization of Necroptosis-Related Genes Expression in BLCA
3.2. The TME Immune Cell Infiltration in Distinct NEC Cluster
3.3. Characteristics of the Biological Process in NEC Clusters
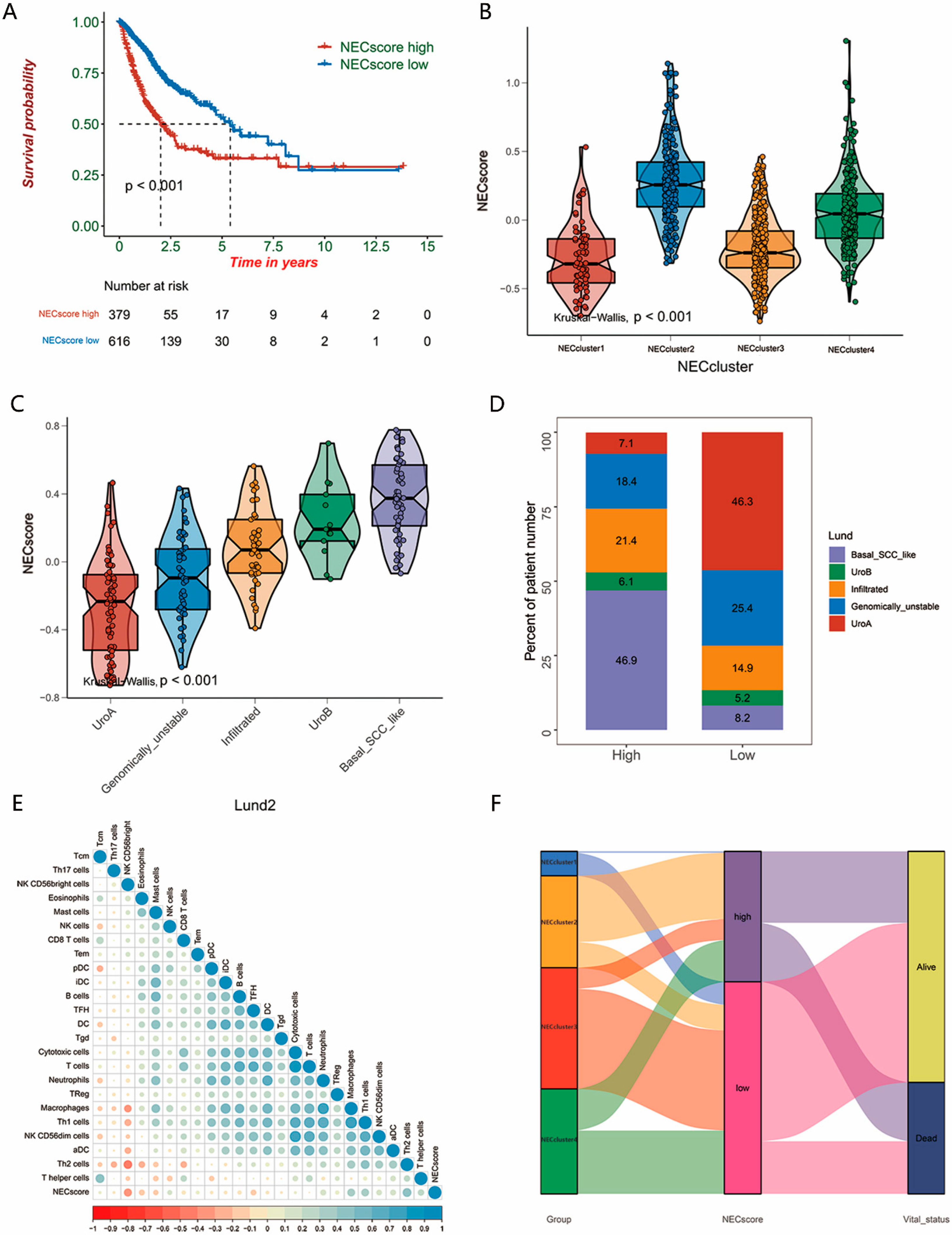
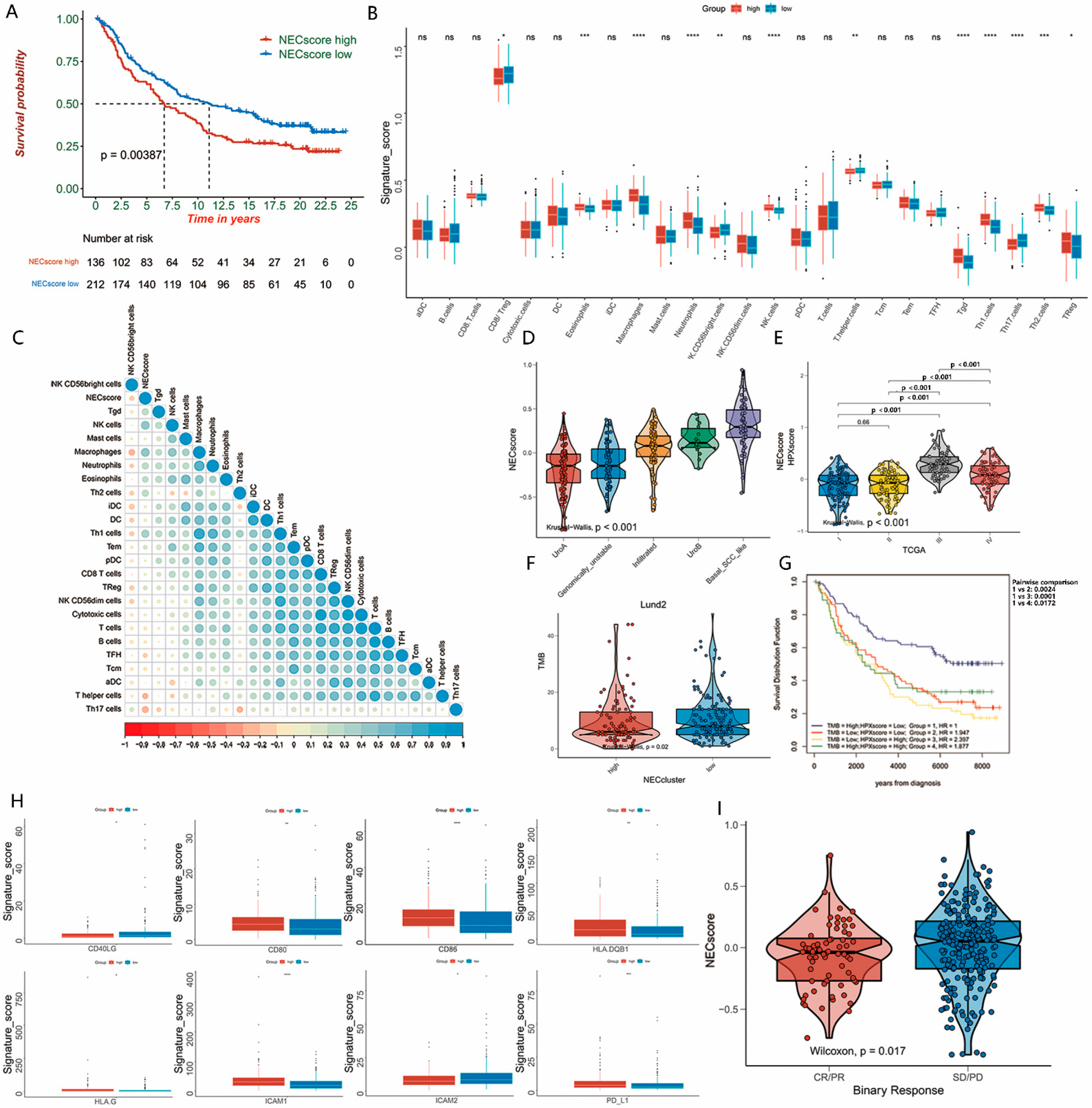
3.4. Establish of the NEC Score
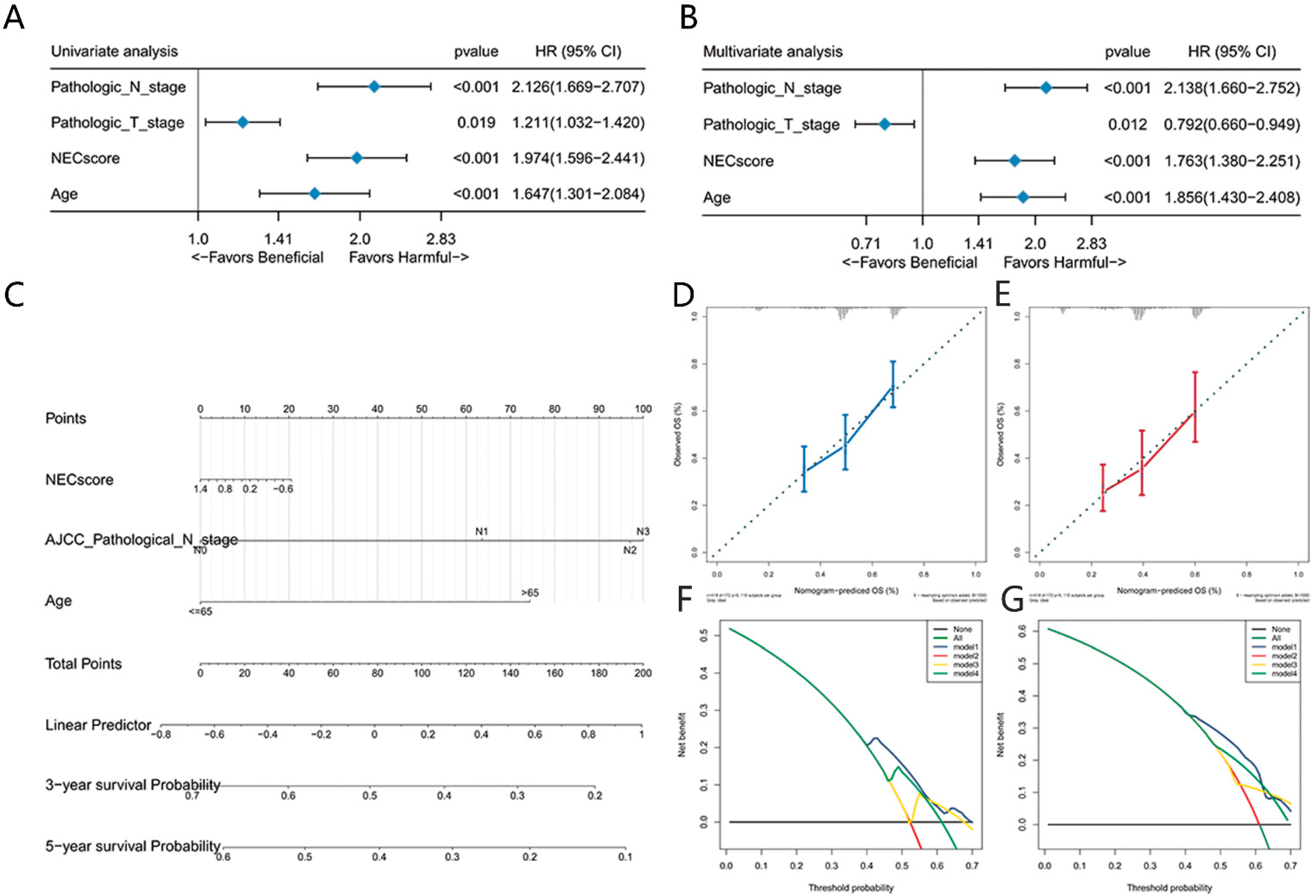
3.5. NEC Score Can Be Used as an Independent Prognostic Factor in BLCA
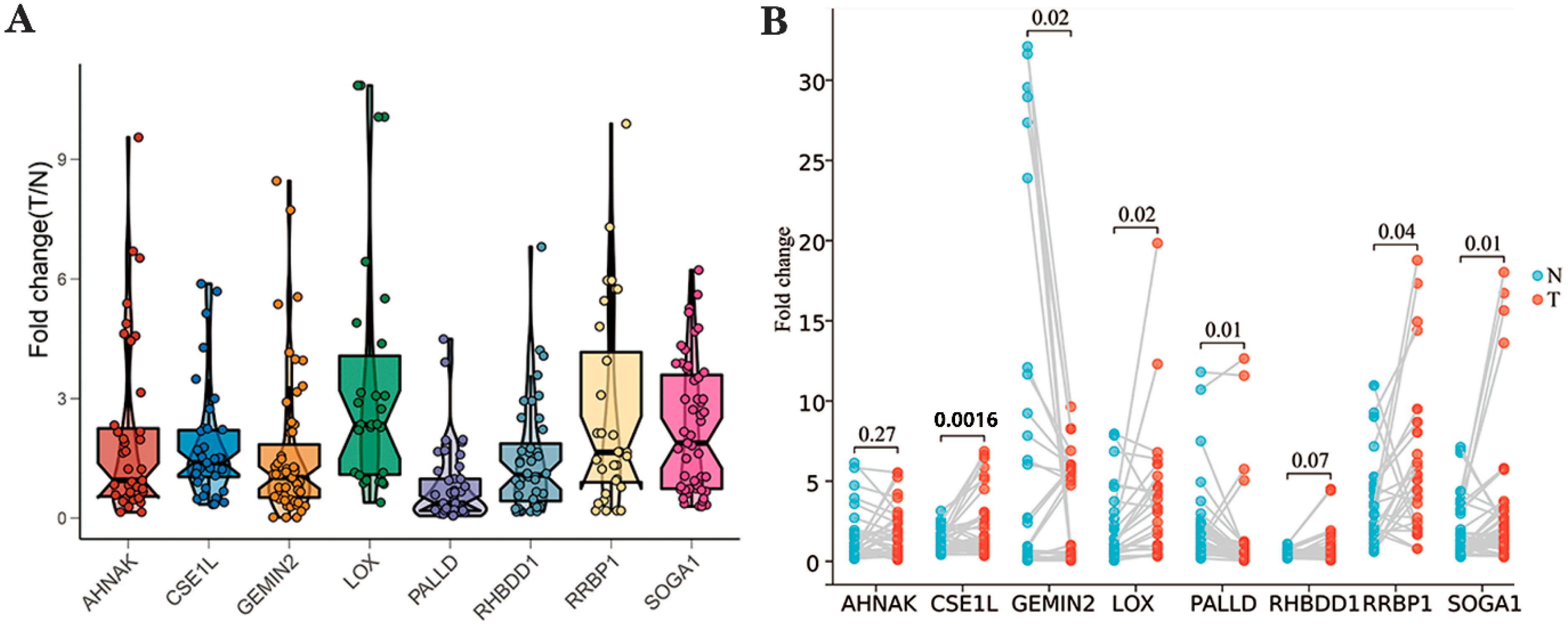
3.6. Expression Level of NEC Score-Related Candidate Genes in BLCA Patients’ Sample
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- He, Y.T.; Li, D.J.; Liang, D.; Zheng, R.S.; Zhang, S.W.; Zeng, H.M.; Chen, W.Q.; He, J. Incidence and mortality of bladder cancer in China, 2014. Zhonghua Zhong Liu Za Zhi 2018, 40, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.L.; Lee, C.T.; Montie, J.E. Bladder cancer in 2010: How far have we come? CA Cancer J. Clin. 2010, 60, 244–272. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.M.; Clark, P.E. Bladder cancer. Curr. Opin. Oncol. 2010, 22, 242–249. [Google Scholar] [CrossRef]
- Shariat, S.F.; Karakiewicz, P.I.; Palapattu, G.S.; Lotan, Y.; Rogers, C.G.; Amiel, G.E.; Vazina, A.; Gupta, A.; Bastian, P.J.; Sagalowsky, A.I.; et al. Outcomes of radical cystectomy for transitional cell carcinoma of the bladder: A contemporary series from the Bladder Cancer Research Consortium. J. Urol. 2006, 176, 2414–2422. [Google Scholar] [CrossRef]
- Lenis, A.T.; Lec, P.M.; Chamie, K.; Mshs, M.D. Bladder Cancer: A Review. JAMA 2020, 324, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.A.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Bell, B.D.; Leverrier, S.; Weist, B.M.; Newton, R.H.; Arechiga, A.F.; Luhrs, K.A.; Morrissette, N.S.; Walsh, C.M. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16677–16682. [Google Scholar] [CrossRef]
- Yatim, N.; Jusforgues-Saklani, H.; Orozco, S.; Schulz, O.; Barreira da Silva, R.; Reis e Sousa, C.; Green, D.R.; Oberst, A.; Albert, M.L. RIPK1 and NF-kappaB signaling in dying cells determines cross-priming of CD8(+) T cells. Science 2015, 350, 328–334. [Google Scholar] [CrossRef]
- Chan, F.K.; Luz, N.F.; Moriwaki, K. Programmed necrosis in the cross talk of cell death and inflammation. Annu. Rev. Immunol. 2015, 33, 79–106. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 2015, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Bridelance, J.; Roelandt, R.; Vandenabeele, P.; Takahashi, N. MLKL in cancer: More than a necroptosis regulator. Cell Death Differ. 2021, 28, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater 2020, 32, e2002054. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Lanitis, E.; Dangaj, D.; Irving, M.; Coukos, G. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann. Oncol. 2017, 28, xii18–xii32. [Google Scholar] [CrossRef]
- Tower, J. Programmed cell death in aging. Ageing Res. Rev. 2015, 23, 90–100. [Google Scholar] [CrossRef]
- Nicole, L.; Sanavia, T.; Cappellesso, R.; Maffeis, V.; Akiba, J.; Kawahara, A.; Naito, Y.; Radu, C.M.; Simioni, P.; Serafin, D.; et al. Necroptosis-driving genes RIPK1, RIPK3 and MLKL-p are associated with intratumoral CD3(+) and CD8(+) T cell density and predict prognosis in hepatocellular carcinoma. J. Immunother. Cancer 2022, 10, e004031. [Google Scholar] [CrossRef]
- Gaiha, G.D.; McKim, K.J.; Woods, M.; Pertel, T.; Rohrbach, J.; Barteneva, N.; Chin, C.R.; Liu, D.; Soghoian, D.Z.; Cesa, K.; et al. Dysfunctional HIV-specific CD8+ T cell proliferation is associated with increased caspase-8 activity and mediated by necroptosis. Immunity 2014, 41, 1001–1012. [Google Scholar] [CrossRef]
- Kang, Y.J.; Bang, B.R.; Han, K.H.; Hong, L.; Shim, E.J.; Ma, J.; Lerner, R.A.; Otsuka, M. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 2015, 6, 8371. [Google Scholar] [CrossRef]
- Lu, J.V.; Chen, H.C.; Walsh, C.M. Necroptotic signaling in adaptive and innate immunity. Semin. Cell Dev. Biol. 2014, 35, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Werthmoller, N.; Frey, B.; Wunderlich, R.; Fietkau, R.; Gaipl, U.S. Modulation of radiochemoimmunotherapy-induced B16 melanoma cell death by the pan-caspase inhibitor zVAD-fmk induces anti-tumor immunity in a HMGB1-, nucleotide- and T-cell-dependent manner. Cell Death Dis. 2015, 6, e1761. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Song, Y.K.; Burckart, G.J.; Ji, E.; Kim, I.W.; Oh, J.M. Regulation of Pharmacogene Expression by microRNA in The Cancer Genome Atlas (TCGA) Research Network. Biomol. Ther. 2017, 25, 482–489. [Google Scholar] [CrossRef]
- Cao, R.; Yuan, L.; Ma, B.; Wang, G.; Qiu, W.; Tian, Y. An EMT-related gene signature for the prognosis of human bladder cancer. J. Cell Mol. Med. 2020, 24, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Nidheesh, N.; Abdul Nazeer, K.A.; Ameer, P.M. An enhanced deterministic K-Means clustering algorithm for cancer subtype prediction from gene expression data. Comput. Biol. Med. 2017, 91, 213–221. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Iasonos, A.; Schrag, D.; Raj, G.V.; Panageas, K.S. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008, 26, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Qian, C.; Cao, X. Dendritic cells in the regulation of immunity and inflammation. Semin. Immunol. 2018, 35, 3–11. [Google Scholar] [CrossRef]
- Chamie, K.; Litwin, M.S.; Bassett, J.C.; Daskivich, T.J.; Lai, J.; Hanley, J.M.; Konety, B.R.; Saigal, C.S.; Urologic Diseases in America Project. Recurrence of high-risk bladder cancer: A population-based analysis. Cancer 2013, 119, 3219–3227. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Dolina, J.S.; Van Braeckel-Budimir, N.; Thomas, G.D.; Salek-Ardakani, S. CD8(+) T Cell Exhaustion in Cancer. Front. Immunol. 2021, 12, 715234. [Google Scholar] [CrossRef] [PubMed]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion during Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Yuan, L.; Ma, B.; Wang, G.; Tian, Y. Tumour microenvironment (TME) characterization identified prognosis and immunotherapy response in muscle-invasive bladder cancer (MIBC). Cancer Immunol. Immunother. 2021, 70, 1–18. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Lai, Y.; Tang, F.; Huang, Y.; He, C.; Chen, C.; Zhao, J.; Wu, W.; He, Z. The tumour microenvironment and metabolism in renal cell carcinoma targeted or immune therapy. J. Cell Physiol. 2021, 236, 1616–1627. [Google Scholar] [CrossRef]
- Toor, S.M.; Sasidharan Nair, V.; Decock, J.; Elkord, E. Immune checkpoints in the tumor microenvironment. Semin. Cancer Biol. 2020, 65, 1–12. [Google Scholar] [CrossRef]
- Choucair, K.; Morand, S.; Stanbery, L.; Edelman, G.; Dworkin, L.; Nemunaitis, J. TMB: A promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene 2020, 27, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.J.; Liao, C.F.; Su, T.C.; Shen, K.H.; Chang, C.C.; Lin, S.H.; Yeh, C.M.; Shen, S.C.; Lee, W.R.; Chiou, J.F.; et al. Presence of CSE1L protein in urine of patients with urinary bladder urothelial carcinomas. Int. J. Biol. Mrk. 2012, 27, e280–e284. [Google Scholar] [CrossRef]
- Chang, C.C.; Tai, C.J.; Su, T.C.; Shen, K.H.; Lin, S.H.; Yeh, C.M.; Yeh, K.T.; Lin, Y.M.; Jiang, M.C. The prognostic significance of nuclear CSE1L in urinary bladder urothelial carcinomas. Ann. Diagn. Pathol. 2012, 16, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Amendola, P.G.; Reuten, R.; Erler, J.T. Interplay Between LOX Enzymes and Integrins in the Tumor Microenvironment. Cancers 2019, 11, 729. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, M.; Yoshimura, R.; Tsuchida, K.; Takemoto, Y.; Segawa, Y.; Shinnka, T.; Kawahito, Y.; Sano, H.; Nakatani, T. Lipoxygenase inhibitors prevent urological cancer cell growth. Int. J. Mol. Med. 2004, 13, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Miao, F.; Huang, R.; Liu, W.; Zhao, Y.; Jiao, T.; Lu, Y.; Wu, F.; Wang, X.; Wang, H.; et al. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J. Exp. Clin. Cancer Res. 2018, 37, 22. [Google Scholar] [CrossRef]
- Wang, M.; Liu, S.; Zhou, B.; Wang, J.; Ping, H.; Xing, N. RRBP1 is highly expressed in bladder cancer and is associated with migration and invasion. Oncol. Lett. 2020, 20, 203. [Google Scholar] [CrossRef]
- Luo, H.L.; Liu, H.Y.; Chang, Y.L.; Sung, M.T.; Chen, P.Y.; Su, Y.L.; Huang, C.C.; Peng, J.M. Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma. Int. J. Mol. Sci. 2021, 22, 8761. [Google Scholar] [CrossRef]
- Lv, S.W.; Shi, Z.G.; Wang, X.H.; Zheng, P.Y.; Li, H.B.; Han, Q.J.; Li, Z.J. Ribosome Binding Protein 1 Correlates with Prognosis and Cell Proliferation in Bladder Cancer. OncoTargets Ther. 2020, 13, 6699–6707. [Google Scholar] [CrossRef]
- Pogue-Geile, K.L.; Chen, R.; Bronner, M.P.; Crnogorac-Jurcevic, T.; Moyes, K.W.; Dowen, S.; Otey, C.A.; Crispin, D.A.; George, R.D.; Whitcomb, D.C.; et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006, 3, e516. [Google Scholar] [CrossRef]
- Kim, J.; Kwiatkowski, D.; McConkey, D.J.; Meeks, J.J.; Freeman, S.S.; Bellmunt, J.; Getz, G.; Lerner, S.P. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur. Urol. 2019, 75, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Jalanko, T.; de Jong, J.J.; Gibb, E.A.; Seiler, R.; Black, P.C. Correction to: Genomic Subtyping in Bladder Cancer. Curr. Urol. Rep. 2020, 21, 25. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Cao, R.; Wang, R.; Wang, Y.; Shang, D.; Tian, Y. Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer. Genes 2022, 13, 2120. https://doi.org/10.3390/genes13112120
Chen Z, Cao R, Wang R, Wang Y, Shang D, Tian Y. Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer. Genes. 2022; 13(11):2120. https://doi.org/10.3390/genes13112120
Chicago/Turabian StyleChen, Zhenghao, Rui Cao, Ren Wang, Yichuan Wang, Donghao Shang, and Ye Tian. 2022. "Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer" Genes 13, no. 11: 2120. https://doi.org/10.3390/genes13112120
APA StyleChen, Z., Cao, R., Wang, R., Wang, Y., Shang, D., & Tian, Y. (2022). Prognosis Risk Model Based on Necroptosis-Related Signature for Bladder Cancer. Genes, 13(11), 2120. https://doi.org/10.3390/genes13112120





