Identification and Functional Analysis of MAPKAPK2 in Hyriopsis cumingii
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals, Preparation and Sample Collection
2.2. Immune Response
2.3. RNA Extraction and cDNA Synthesis
2.4. MK2 Full-Length Acquisition and Sequence Analysis
2.5. Quantitative Real-Time PCR
2.6. In Situ Hybridization
2.7. Data Processing, Statistical Analysis and Graph Production
3. Result
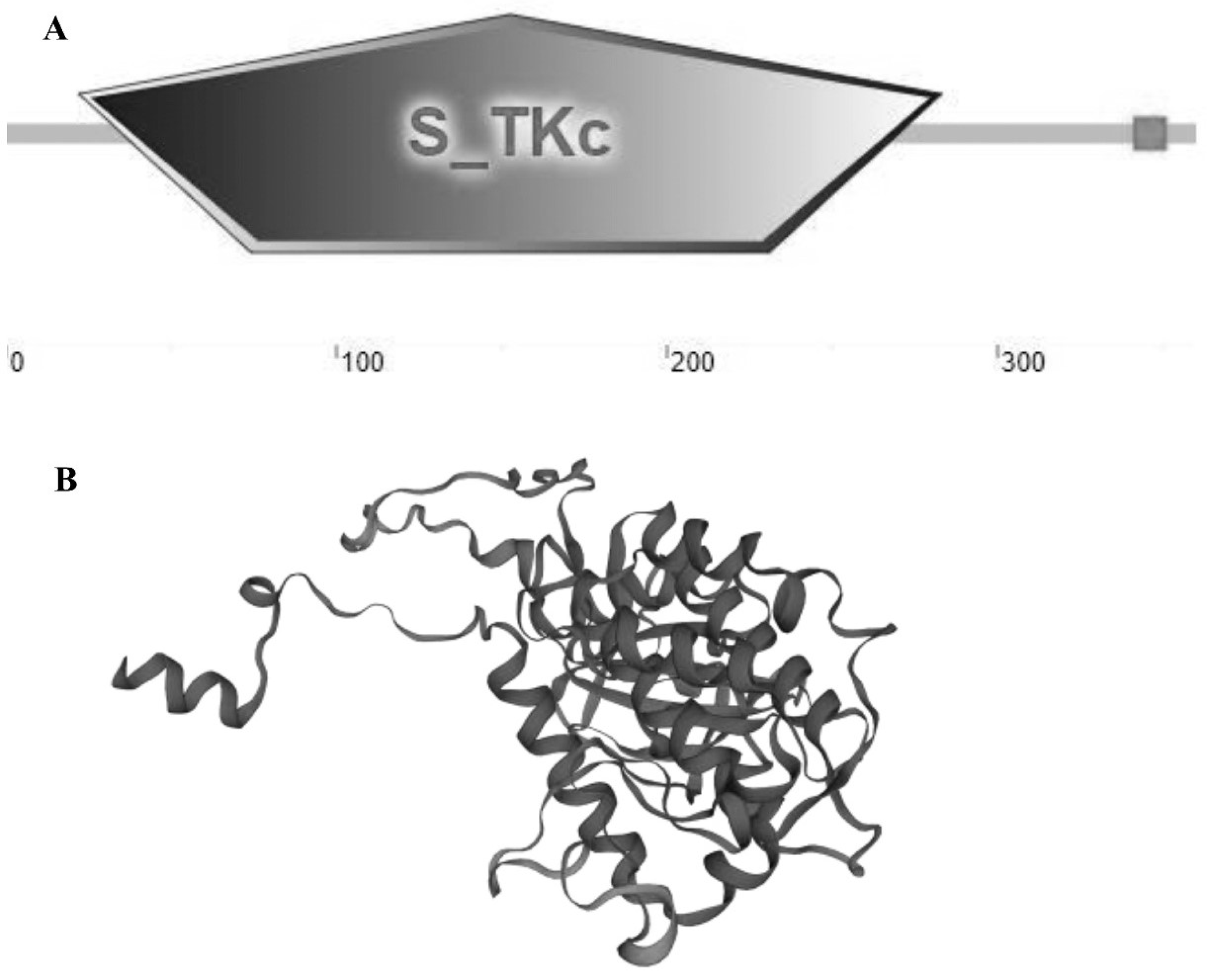
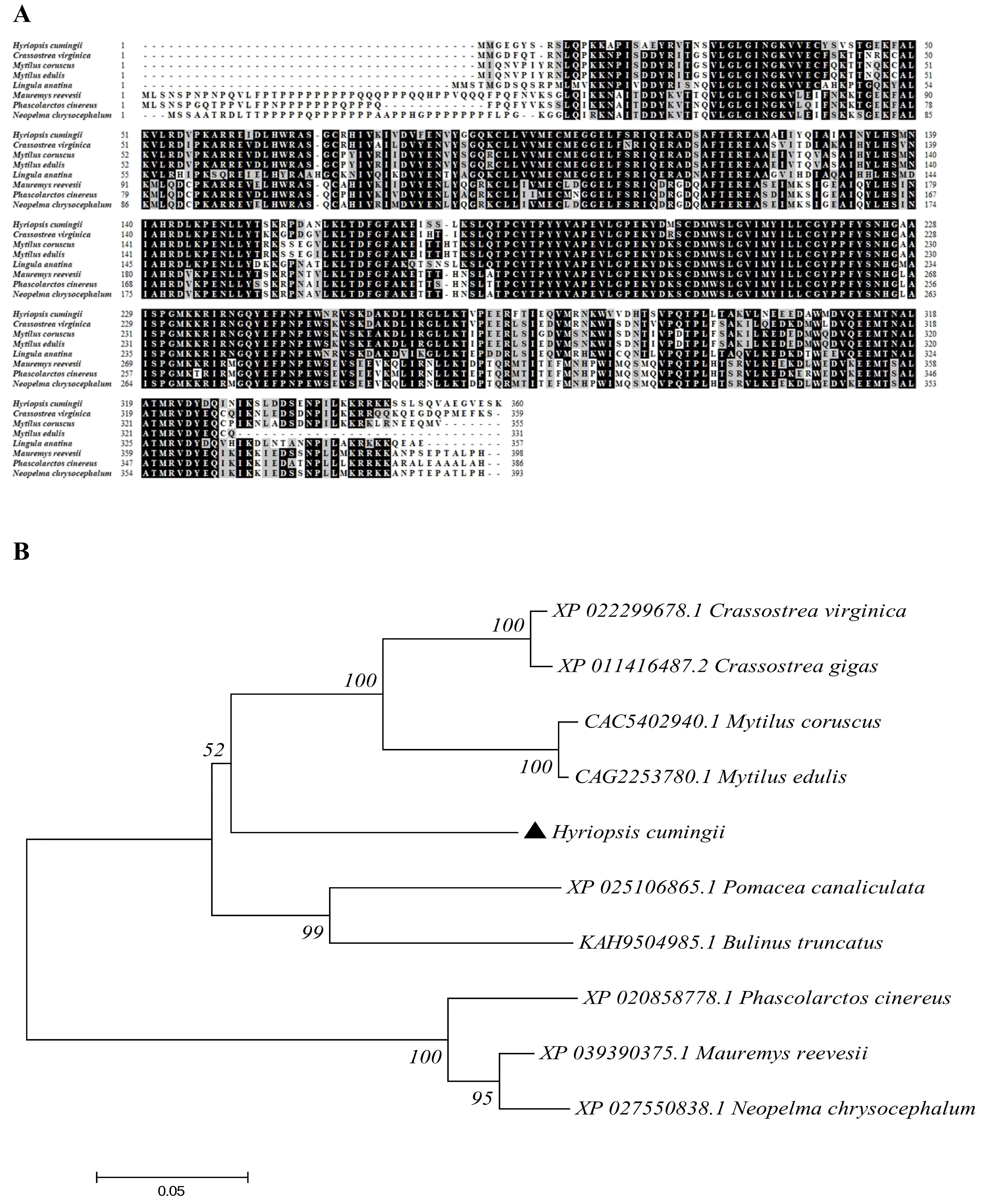
3.1. Full-Length Cloning and Sequence Characterization of MK2 cDNA in H. cumingii
3.2. Expression of MK2 in Various Tissues of the Mature H. cumingii
3.3. MK2 Responded to the Transcriptional Level after Infection by A. hydrophila and LPS in H. cumingii
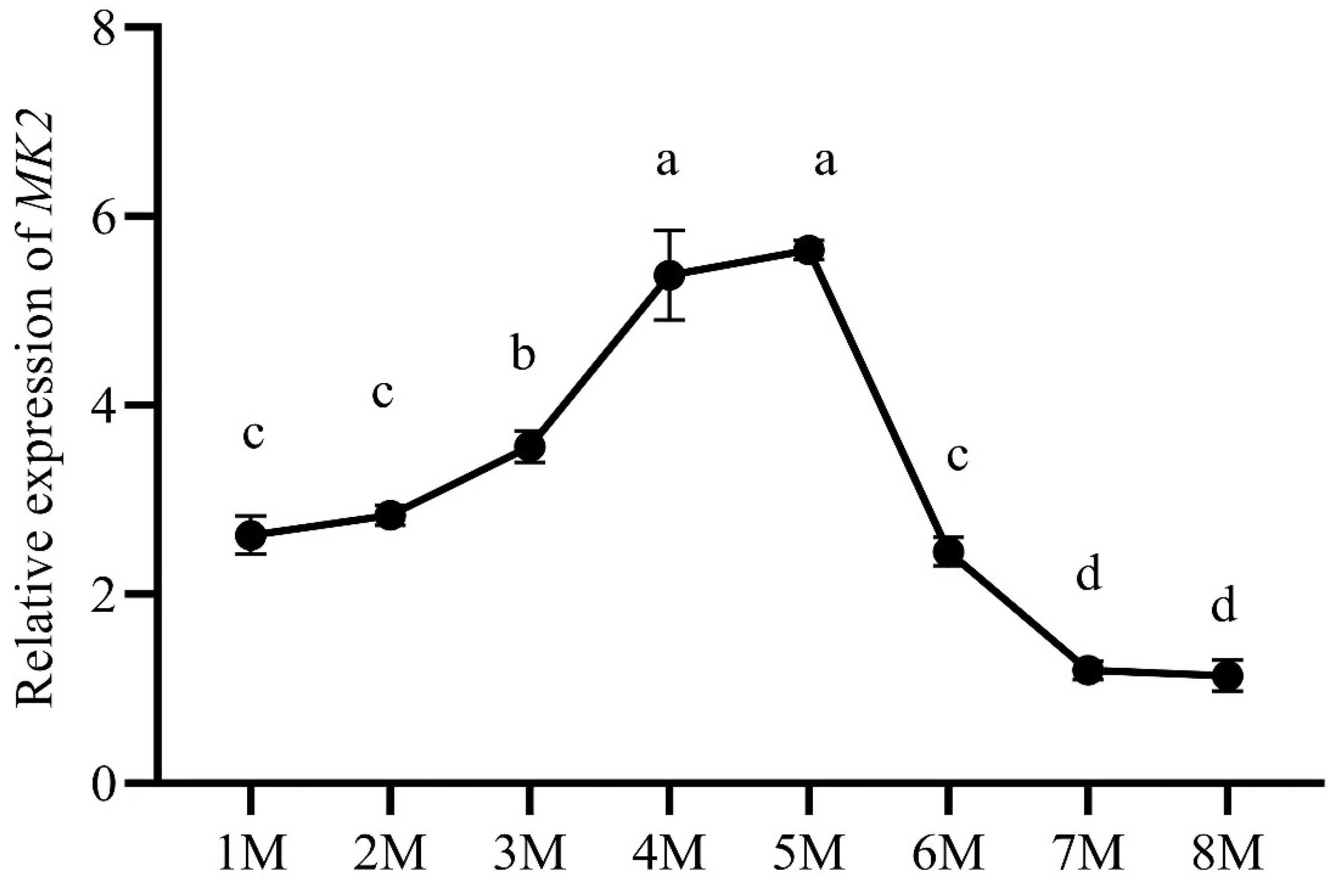
3.4. Expression of MK2 in Juvenile H. cumingii
3.5. Expression of MK2 in Adult H. cumingii
3.6. Localization of MK2 in H. cumingii
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zarubin, T.; Han, J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Hitti, E.; Iakovleva, T.; Brook, M.; Deppenmeier, S.; Gruber, A.D.; Radzioch, D.; Clark, A.R.; Blackshear, P.J.; Kotlyarov, A.; Gaestel, M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell Biol. 2006, 26, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.-H.; Li, S.; Xu, B.-Z.; Chen, L.-N.; Jiang, M.-X.; Chen, S.-Q.; Chen, N.-Q. Mitogen-activated protein kinase-activated protein kinase 2 is a critical regulator of pig oocyte meiotic maturation. Reprod. Fertil. Dev. 2017, 29, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Krug, M.S.; McMillan, E.A.; Peake, J.D.; Davis, T.L.; Cocklin, S.; Strochlic, T.I. Phosphorylation of the RNA-binding protein Dazl by MAPKAP kinase 2 regulates spermatogenesis. Mol. Biol. Cell 2016, 27, 2341–2350. [Google Scholar] [CrossRef] [PubMed]
- Holloway, B.A.; Gomez de la Torre Canny, S.; Ye, Y.; Slusarski, D.C.; Freisinger, C.M.; Dosch, R.; Chou, M.M.; Wagner, D.S.; Mullins, M.C. A novel role for MAPKAPK2 in morphogenesis during zebrafish development. PLoS Genet. 2009, 5, e1000413. [Google Scholar] [CrossRef] [PubMed]
- Holloway, B.; de la Torre Canny, S.; Ye, Y.; Slusarski, D.; Freisinger, C.; Dosch, R.; Chou, M.; Wagner, D.; Mullins, M. Sexual Dimorphism in MAPK-Activated Protein Kinase-2 (MK2) Regulation of RANKL-Induced Osteoclastogenesis in Osteoclast Progenitor Subpopulations. PLoS ONE 2015, 10, e0125387. [Google Scholar]
- Stokoe, D.; Campbell, D.; Nakielny, S.; Hidaka, H.; Leevers, S.; Marshall, C.; Cohen, P. MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992, 11, 3985–3994. [Google Scholar] [CrossRef]
- Kotlyarov, A.; Neininger, A.; Schubert, C.; Eckert, R.; Birchmeier, C.; Volk, H.-D.; Gaestel, M. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1999, 1, 94–97. [Google Scholar] [CrossRef]
- Winzen, R.; Kracht, M.; Ritter, B.; Wilhelm, A.; Chen, C.-Y.A.; Shyu, A.-B.; Müller, M.; Gaestel, M.; Resch, K.; Holtmann, H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999, 18, 4969–4980. [Google Scholar] [CrossRef]
- Su, X.; Ao, L.; Zou, N.; Song, Y.; Yang, X.; Cai, G.-Y.; Fullerton, D.A.; Meng, X. Post-transcriptional regulation of TNF-induced expression of ICAM-1 and IL-8 in human lung microvascular endothelial cells: An obligatory role for the p38 MAPK-MK2 pathway dissociated with HSP27. Biochim. Biophys. Acta 2008, 1783, 1623–1631. [Google Scholar] [CrossRef]
- Funding, A.T.; Johansen, C.; Gaestel, M.; Bibby, B.M.; Lilleholt, L.L.; Kragballe, K.; Iversen, L. Reduced oxazolone-induced skin inflammation in MAPKAP kinase 2 knockout mice. J. Investig. Dermatol. 2009, 129, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Iliev, D.B.; Hansen, T.; Jørgensen, S.M.; Krasnov, A.; Jørgensen, J.B. CpG- and LPS-activated MAPK signaling in in vitro cultured salmon (Salmo salar) mononuclear phagocytes. Fish Shellfish Immunol. 2013, 35, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Lehner, M.D.; Schwoebel, F.; Kotlyarov, A.; Leist, M.; Gaestel, M.; Hartung, T. Mitogen-activated protein kinase-activated protein kinase 2-deficient mice show increased susceptibility to Listeria monocytogenes infection. J. Immunol. 2002, 168, 4667–4673. [Google Scholar] [CrossRef]
- Bobo, L.D.; El Feghaly, R.E.; Chen, Y.-S.; Dubberke, E.R.; Han, Z.; Baker, A.H.; Li, J.; Burnham, C.-A.D.; Haslam, D.B. MAPK-activated protein kinase 2 contributes to Clostridium difficile-associated inflammation. Infect. Immun. 2013, 81, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Sarder, H.; Khan, T.; Saha, M.; Punom, N.; Mandal, S.; Rahman, M. Prevalence and antibiotic susceptibility of Aeromonas hydrophila isolated from freshwater fishes. J. Fish. 2016, 4, 411–419. [Google Scholar] [CrossRef]
- Liu, X.; Sun, W.; Zhang, Y.; Zhou, Y.; Xu, J.; Gao, X.; Zhang, S.; Zhang, X. Impact of Aeromonas hydrophila and infectious spleen and kidney necrosis virus infections on susceptibility and host immune response in Chinese perch (Siniperca chuatsi). Fish Shellfish Immunol. 2020, 105, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-D.; Li, J.-H.; Yao, Y.-Y.; Zhang, Y.-A. Aeromonas hydrophila suppresses complement pathways via degradation of complement C3 in bony fish by metalloprotease. Fish Shellfish Immunol. 2019, 94, 739–745. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, S.; Chu, W. Comparative proteomic analysis of sensitive and multi-drug resistant Aeromonas hydrophila isolated from diseased fish. Microb. Pathog. 2020, 139, 103930. [Google Scholar] [CrossRef]
- Wen, Z.-Y.; Liu, X.-Y.; Jin, X.-L. Study on Pathology of Hyriopsis cumingii Lea Disease Caused by Aeromonas hydrophila. J. Hunan Agric. Univ. 2001, 1, 56–59. [Google Scholar]
- Wang, G.L.; Yuan, Y.M.; Li, J.L. SSR analysis of genetic diversity and phylogenetic relationships among different populations of Hyriopsis cumingii from the five lakes of China. J. Fish. China 2007, 12, 12–18. [Google Scholar]
- Zhao, Y.; Bai, Z.; Fu, L.; Liu, Y.; Wang, G.; Li, J. Comparison of growth and pearl production in males and females of the freshwater mussel, Hyriopsis cumingii, in China. Aquac. Int. 2013, 21, 1301–1310. [Google Scholar] [CrossRef]
- Huang, D.; Bai, Z.; Shen, J.; Zhao, L.; Li, J. Identification of tumor necrosis factor receptor-associated factor 6 in the pearl mussel Hyriopsis cumingii and its involvement in innate immunity and pearl sac formation. Fish Shellfish Immunol. 2018, 80, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, M.; Zhang, J.; Bai, Z.; Li, J. miR-4504 is involved in nacre color formation in Hyriopsis cumingii. Biochem. Biophys. Res. Commun. 2019, 517, 210–215. [Google Scholar] [CrossRef]
- Bai, Z.; Zheng, H.; Lin, J.; Wang, G.; Li, J. Comparative Analysis of the Transcriptome in Tissues Secreting Purple and White Nacre in the Pearl Mussel Hyriopsis cumingii. PLoS ONE 2013, 8, e53617. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ren, Q. HcCUB-Lec, a newly identified C-type lectin that contains a distinct CUB domain and participates in the immune defense of the triangle sail mussel Hyriopsis cumingii. Dev. Comp. Immunol. 2019, 93, 66–77. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, J.; Wang, G.; Wu, C.; Li, J. Molecular cloning, sequencing, and expression profiles of heat shock protein 90 (HSP90) in Hyriopsis cumingii exposed to different stressors: Temperature, cadmium and Aeromonas hydrophila. Aquac. Fish. 2017, 2, 59–66. [Google Scholar] [CrossRef]
- Little, T.J.; Hultmark, D.; Read, A.F. Invertebrate immunity and the limits of mechanistic immunology. Nat. Immunol. 2005, 6, 651–654. [Google Scholar] [CrossRef]
- Chen, J.; Shao, B.; Wang, J.; Shen, Z.; Liu, H.; Li, S. Chlorpyrifos caused necroptosis via MAPK/NF-κB/TNF-α pathway in common carp (Cyprinus carpio L.) gills. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021, 249, 109126. [Google Scholar] [CrossRef]
- Ni, P.-J.; Feng, L.; Jiang, W.-D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.-Y.; Tang, L.; Tang, W.-N.; Zhou, X.-Q. Impairing of gill health through decreasing immune function and structural integrity of grass carp (Ctenopharyngodon idella) fed graded levels dietary lipids after challenged with Flavobacterium columnare. Fish Shellfish Immunol. 2019, 86, 922–933. [Google Scholar] [CrossRef]
- Shui, Y.; Xie, J.; Zhou, Y.; Li, J.; Gan, J. Molecular characterization of p38 MAPK and tissue-specific expression under cadmium stress in red swamp crayfish (Procambarus clarkii). Sci. Total Environ. 2020, 720, 137325.1–137325.11. [Google Scholar] [CrossRef]
- Huang, D.D. Differential expression analysis of genes in the development of pearl sac in Hyriopsis cumingii post mantle implantation and function analysis of IRAK4 and TRAF6. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2019. [Google Scholar]
- Fan, H.-Y.; Sun, Q.-Y. Involvement of Mitogen-Activated Protein Kinase Cascade during Oocyte Maturation and Fertilization in mammals. Biol. Reprod. 2004, 70, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.Y.; Breitbart, H.; Schatten, H. Role of the MAPK cascade in mammalian germ cells. Reprod. Fertil. Dev. 1999, 11, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.; Rulong, S.; Resau, J.; Fukasawa, K.; Matten, W.; Kuriyama, R.; Mansour, S.; Ahn, N.; Woude, G.F.V. Mos/mitogen-activated protein kinase can induce early meiotic phenotypes in the absence of maturation-promoting factor: A novel system for analyzing spindle formation during meiosis I. Proc. Natl. Acad. Sci. USA 1996, 93, 4730–4735. [Google Scholar] [CrossRef] [PubMed]
- Verlhac, M.-H.; De Pennart, H.; Maro, B.; Cobb, M.; Clarke, H. MAP kinase becomes stably activated at metaphase and is associated with microtubule-organizing centers during meiotic maturation of mouse oocytes. Dev. Biol. 1993, 158, 330–340. [Google Scholar] [CrossRef]
- Verlhac, M.; Kubiak, J.; Clarke, H.; Maro, B. Microtubule and chromatin behavior follow MAP kinase activity but not MPF activity during meiosis in mouse oocytes. Development 1994, 120, 1017–1025. [Google Scholar] [CrossRef]
- Xue, T. Study on DUI Occurring and Gonad Development of Freshwater Pearl Mussels. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2016. [Google Scholar]





| Primer Name | Sequences (5′—3′) | Usage |
|---|---|---|
| MK2-Outer | AGAGGTGGCGAAACCCGACAGGACT | 3′RACE outer |
| MK2-Inner | GGAACCGAGTCTCCAAGGATGCCA | 3′RACE inner |
| qPCRMK2-F | CTCGCTAAAATCATTACAGACCC | qPCR |
| qPCRMK2-R | GAATGGAGGATAGCCACATAACA | qPCR |
| EFl-αF | GGAACTTCCCAGGCAGACTGTGC | qPCR |
| EFl-αR | TCAAAACGGGCCGCAGAGAAT | qPCR |
| IMK2-F | CTCGCTAAAATCATTACAGACCC | ISH |
| IMK2-R | TAATACGACTCACTATAGGGGAATGG AGGATAGCCACATAACA | ISH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Liu, M.; Wang, Y.; Huo, Y.; Liu, Z.; Jin, W.; Wang, G. Identification and Functional Analysis of MAPKAPK2 in Hyriopsis cumingii. Genes 2022, 13, 2060. https://doi.org/10.3390/genes13112060
Gu Y, Liu M, Wang Y, Huo Y, Liu Z, Jin W, Wang G. Identification and Functional Analysis of MAPKAPK2 in Hyriopsis cumingii. Genes. 2022; 13(11):2060. https://doi.org/10.3390/genes13112060
Chicago/Turabian StyleGu, Yang, Meiling Liu, Yayu Wang, Yingduo Huo, Zongyu Liu, Wu Jin, and Guiling Wang. 2022. "Identification and Functional Analysis of MAPKAPK2 in Hyriopsis cumingii" Genes 13, no. 11: 2060. https://doi.org/10.3390/genes13112060
APA StyleGu, Y., Liu, M., Wang, Y., Huo, Y., Liu, Z., Jin, W., & Wang, G. (2022). Identification and Functional Analysis of MAPKAPK2 in Hyriopsis cumingii. Genes, 13(11), 2060. https://doi.org/10.3390/genes13112060




