Abstract
Pleurotomarioidea represents a truly isolated and basally diverging lineage in Vetigastropoda (Mollusca: Gastropoda) whose fossil record can date back to the late Cambrian, thus providing rare insights into the evolutionary history of molluscs. Here, we sequenced and assembled the complete mitochondrial genome of one representative species from Pleurotomarioidea—Entemnotrochus rumphii (Schepman, 1879)—of which the mitogenome is 15,795 bp in length, including 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. The nucleotide composition was biased toward AT, and A + T content reached 65.2%. E. rumphii was recovered as sister to all other living vetigastropods according to mitogenome-based phylogenetic analysis. The mitochondrial gene order was consistent with major vetigastropods and the hypothetical ancestral gastropoda, suggesting the deep conservation of mitogenome arrangement in Vetigastropoda.
1. Introduction
Entemnotrochus rumphii (Schepman, 1879) belongs to the superfamily Pleurotomarioidea, subclass Vetigastropoda, which is regarded as one of the most deeply diverging lineages among Gastropoda [1,2]. Pleurotomarioids can be distinguished from other gastropods based on their special morphological characteristics such as the coiled, conispiral shells and a slit along outer lips [3]. Pleurotomarioids are distributed in tropical and temperate latitudes [4]. Two species of Entemnotrochus are widely distributed, and one is E. rumphii, which is known from the southern islands of Japan to the central Philippines and Indonesia [3]. According to museum records, the bathymetric ranges of living E. rumphii range from about 80–280 m, and they inhabit sandy, coarse bottoms [5]. Tan [6] reported that the gut content and feces of E. rumphii consisted of sponge spicules, which indicated that this species lived on a diet of sponges.
Pleurotomarioidea is the only Gastropoda superfamily with fossil records from the Cambrian to the present and they are called “living fossils” [7,8,9,10]. The family Pleurotomariidae originated in the lower Triassic and is the only family of Pleurotomarioidea that survives now. Of 1500 described species of Pleurotomarioidea, only 25 are living today [4]. Their symmetrical organs and asymmetrical shells are regarded as transitional between symmetrical ancestors and asymmetrical modern gastropods [11,12]. Overall, these species have significance for understanding the early evolution of Gastropoda.
The mitochondrion is an important organelle in eukaryotes, and the mitochondrial genome has nearly the same structure in all animals, that is, a circular double-stranded DNA molecule, almost always including 13 protein-coding genes (PCGs), two ribosomal RNAs, and 22 transfer RNAs [13]. The conservation of its composition and structure makes the mitochondrial genome a good material for studying evolution and phylogenetic relationships [14,15]. Even as mitochondrial genome data are increasing for Gastropoda, few have been published yet for Vetigastropoda, especially within Pleurotomariida. To provide more molecular information on this order, we described a complete mitochondrial genome of E. rumphii, a representative species of Pleurotomarioidea. We also conducted a phylogenetic analysis and compared gene arrangements within Vetigastropoda.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
A specimen of E. rumphii was collected in 2020 from the East China Sea, near Japan (30.9° N, 129.9° E). The entire individual was preserved at −80 °C in the laboratory at the Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China. Total genomic DNA was extracted from its foot using CTAB methods.
2.2. Mitogenome Sequencing and Assembly
The complete mitochondrial genome was sequenced and assembled by Huitong Biotechnology, Shenzhen. Genomic libraries with an average length of 350 bp were constructed using the NexteraXT DNA Library Preparation Kit and then sequenced as 150 bp paired-end runs on the Illumina Novaseq platform (Total Genomics Solution Limited, SZHT) (San Diego, CA, USA). Raw data were filtered using the NGS QC Toolkit v2.3.3 [16] for quality control. De novo assembling was performed with SPAdes 3.11.0 [17].
2.3. Mitogenome Annotation, Sequence Analysis, and Data Acquisition
The mitochondrial genome of E. rumphii was annotated using MITOS Web Server (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 13 July 2021). Manual corrections of genes for start/stop codons were performed in SnapGene Viewer. The position and size of all protein-coding genes were corrected using Open Reading Frame Finder (ORFfinder Home–NCBI (nih.gov)). Finally, a mitochondrial genome map was visualized with the online tool Proksee (https://proksee.ca/, accessed on 3 August 2022). The nucleotide composition, composition skew, codon usage of PCGs, and relative synonymous codon usage (RSCU) were analyzed using PhyloSuite v1.2.2 [18]. The secondary structures of 22 tRNAs were predicted using tRNAscan-Se [19,20] and MITOS, and drawn with the ViennaRNA Web Services (http://rna.tbi.univie.ac.at/, accessed on 21 September 2022).
To explore the phylogenetic relationship and gene arrangement of Vetigastropoda, 21 complete mitochondrial genomes were obtained from the NCBI database (Table 1). Of these, 19 species belong to the subclass Vetigastropoda, while the other two species from the subclass Neritimorpha were viewed as outgroups.

Table 1.
Information of species for phylogenetic analysis.
2.4. Phylogenetic Analyses
To indicate the phylogenetic position of E. rumphii among Vetigastropoda, phylogenetic analysis was performed. Thirteen protein-coding genes were obtained using PhyloSuite v1.2.2 [18]. Amino acid sequences were aligned with Mafft v7 [21], and the misaligned positions were removed using Gblocks 0.91b [22] with default settings. All PCGs were concatenated using PhyloSuite v1.2.2 [18]. The best-fitting partition model was found by ModelFinder [23] using the Corrected Akaike Information Criterion (AICc). A maximum-likelihood (ML) tree was reconstructed by IQ-Tree v2.1.2 [24] with 1000 ultrafast bootstrap replications. Finally, the phylogenetic tree was shown by FigTree v1.4.4.
3. Results and Discussion
3.1. Mitogenome Characteristic
The circular mitogenome of E. rumphii was 15,795 bp in length, containing 13 protein-coding genes (PCGs), two ribosomal RNA genes (rrnS and rrnL), and 22 transfer RNA genes (Figure 1; Table 2). The length of this mitogenome was smaller than other vetigastropods except for Lepetodrilus schrolli (Table 3). Seven PCGs (cox1, cox2, cox3, nad2, nad3, atp6, atp8) and eight tRNAs (tRNA-SerAGC, tRNA-Ile, tRNA-Asn, tRNA-Arg, tRNA-Ala, tRNA-Lys, tRNA-Thr, tRNA-Asp) were encoded on the heavy strand, while the other genes, including two ribosomal RNAs, were encoded on the light strand. Twenty-two intergenic regions were found ranging from 1 to 364 bp, while the largest one was located between tRNA-Glu and cox3. Moreover, two overlaps were detected. Nad4 overlapped nad4l by seven bp, and tRNA-Thr overlapped tRNA-SerUCA by one bp.
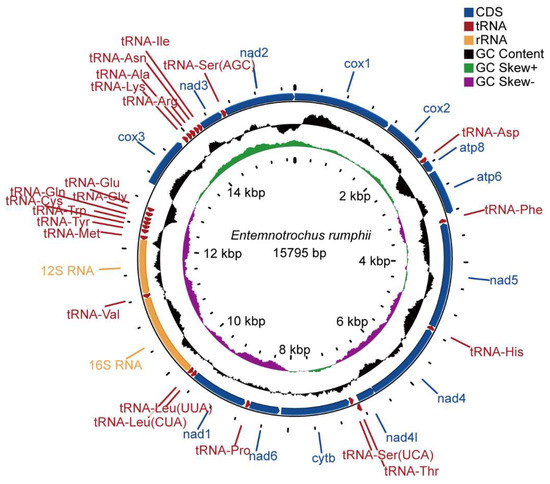
Figure 1.
The mitochondrial genome map of Entemnotrochus. rumphii. From outside to inside, the first circle represents gene arrangement, the second represents the GC content, the third represents the GC skew, and the innermost circle represents the scale. The GC content circus was centered at 50% and the GC skew was centered at zero.

Table 2.
Organization of the mitochondrial genome of E. rumphii.

Table 3.
Nucleotide composition of mitochondrial genomes of vetigastropods.
The nucleotide composition was 35.21% for A, 29.98% for T, 20.39% for C, and 14.43% for G, indicating it was biased toward A+T content at 65.19%, which was similar to other vetigastropods (Table 3). The AT skew of E. rumphii was positive, while the GC skew was negative. Among 20 vetigastropods analyzed in our study, the AT skews of eight species were positive, and the GC skews of 13 species were negative, not showing any apparent consistency.
3.2. Protein-Coding Genes
The total length of all PCGs was 11,310 bp, accounting for 71.6% of the complete mitochondrial genome. The size of the 13 PCGs ranged from 165 (atp8) to 1731 bp (nad5). All PCGs used ATG as the start codon. Among the 20 vetigastropods we studied, ATG was the most frequently used start codon and 11 species used it for all PCGs. Two different kinds of stop codons were found in the 13 PCGs; TAA in nine PCGs (cox1, cox2, cox3, cytb, nad2, nad3, nad4, nad5, atp8) and TAG in the remaining four PCGs (nad1, nad4l, nad6, atp6). The other 19 vetigastropods also used TAA and TAG as stop codons.
The base composition of the PCGs was 25.2% for A, 40% for T, 15.4% for C, and 19.4% for G. The A + T content of each individual PCG and the concatenated data all exceeded 60%, which showed that the PCGs preferred AT. Except for atp8, all PCGs had a negative AT skew, suggesting they were biased toward T. As for GC skew, eight PCGs had a positive GC skew, while the other five PCGs had a negative one.
The relative synonymous codon usage (RSCU) of E. rumphii mitogenome was presented in Figure 2, indicating Leu, Phe, and Val were the three most frequently used amino acids. UUA-Leu, UUU-Phe, and AUU-Ile were the three most frequently utilized codons. Among 22 amino acids, nine (Ala, Arg, Gly, Leu1, Pro, Ser1, Ser2, Thr, and Val) had four codons, and others had two codons.
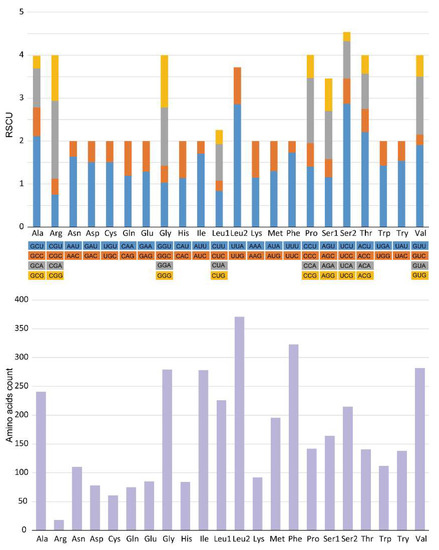
Figure 2.
Relative synonymous codon usage (RSCU) and amino acids count of E. rumphii mitochondrial genome.
3.3. Transfer RNAs and Ribosomal RNAs
The size of 22 transfer RNA genes ranged from 64 (tRNA-Cys) to 70 bp (tRNA-Gly). The AT content of total tRNAs was 62.6%, also showing an AT bias. Almost all tRNAs of E. rumphii had typical clover-leaf secondary structures with four arms, except for trnS1 (Figure 3). This phenomenon where this specific tRNA does not form a four-arms clover-leaf structure can be observed in most metazoan mitogenomes [25].
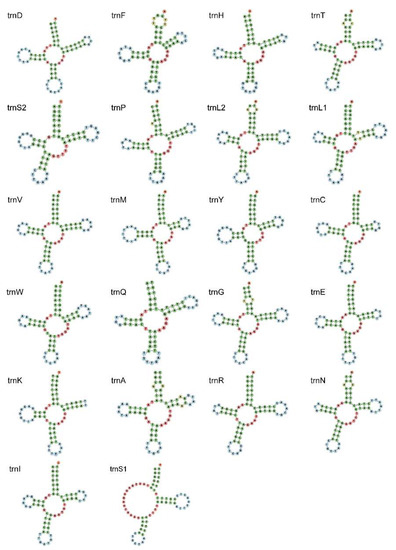
Figure 3.
Secondary structure of 22 tRNAs of E. rumphii mitochondrial genome.
The 12S ribosomal RNA (rrnS) had a length of 894 bp, and the 16S ribosomal RNA (rrnL) had a length of 1386 bp. The 12S ribosomal RNA was located between tRNA-Val and tRNA-Met, while the 16S ribosomal RNA gene was located between tRNA-LeuCUA and tRNA-Val. For rrnL, the AT skew and GC skew values were −0.118 and 0.321; for rrnS, they were −0.054 and 0.321. Both of these rRNAs were biased toward T and G.
3.4. Phylogenetic Analysis
In our study, phylogenetic analysis of Vetigastropoda was conducted based on 13 protein-coding genes (Figure 4A). Based on two Neritimorpha species Clithon retropictus and Nerita albicilla as outgroups, the vetigastropods grouped together in a single clade. Within Vetigastropoda, E. rumphii was recovered as sister to all other vetigastropods (BS = 100). Other vetigastropods were divided into three major clades. Fissurella volcano and Diodora graeca, representing Fissurelloidea, formed the first clade with full support. Lepetodriloidea, Haliotidea, and Seguenzioidea formed the second clade, though without significant support (BS = 50). Five families of Trochidea formed the third clade (BS = 98). However, the relationship of these clades remained unresolved because all bootstrap support values at deep nodes were lower than 95. Among Trochidea, all families were recovered as monophyletic groups with full support. Phasianellidae was resolved as the sister group to the remaining Trochidea species (BS = 98). However, phylogenetic relationships among the other four families were unstable. Turbinidae, Tegulidae, and Trochidae grouped together with weak support (BS = 66). This clade grouped with Angariidae, also without strong support (BS = 83).
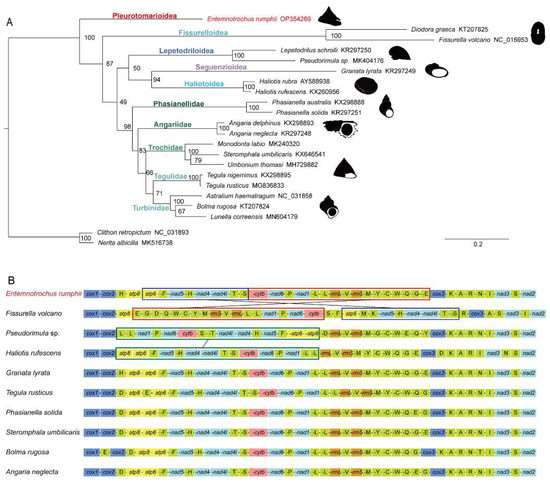
Figure 4.
Phylogenetic tree of Entemnotrochus rumphii, 20 species of Vetigastropoda, and 2 outgroup species based on the amino acid sequences of 13 protein-coding genes (A), and protein-coding gene arrangement of 10 species in Vetigastropoda (B). In the phylogenetic tree, bootstrap support values (BS) are shown on each node. The arrangement graph is shown with iTOL (https://itol.embl.de/, accessed on 15 October 2022) and the boxes in the same color show identical gene blocks.
Major phylogenetic relationships recovered in this study were consistent with previous studies based on Vetigastropoda mitogenomes [26,27]. However, bootstrap support values at some nodes were not high. The current matrix may not have enough phylogenetic information to resolve these nodes, so more data and research are needed for phylogenetic analyses about Vetigastropoda. E. rumphii was recovered as sister clade to all other vetigastropods, indicating that Pleurotomarioidea may be the most deeply diverging lineage of Vetigastropoda. This view was supported by some phylogenetic analyses based on morphology [28,29], or short molecular sequences [12,30], but first was proved by the mitochondrial genome data, as mitogenomes of Pleurotomarioidea were not included in other mitogenome-based studies. Moreover, a phylogenetic study of Vetigastropoda based on transcriptomes has been published recently [31]. Pleurotomarioidea was also recovered as sister group to all other vetigastropods in all their analyses.
3.5. Gene Arrangement
Nine vetigastropods of nine different families were chosen to explore Vetigastropoda mitochondrial gene arrangements (Figure 4B).
Mitochondrial gene arrangements of Vetigastropoda are mostly conserved. Five species of the order Trochida, Granata lyrata (Choristellidae), and Haliotis rufescens (Haliotidae) shared the same gene order except for the position of a very few tRNAs. The organization of their protein-coding genes was identical. As for E. rumphii, it showed the same gene order as G. lyrata and Angaria neglecta, and shared almost the same gene arrangement with most vetigastropods. However, Pseudorimula sp. (Lepetodrilidae) and Fissurella volcano (Fissurellidae) presented different gene arrangements. In F. volcano, the gene block cytb-nad6-trnP-nad1-trnL-trnL-16S-trnV-12S-trnM-trnY-trnC-trnW-trnQ-trnG-trnE reversed, and exchanged its position with the gene block atp6-trnF-nad5-trnH-nad4-nad4l-trnT-trnS. In P. sp, the gene block atp8-atp6-trnF-nad5-trnH-nad4-nad4l-trnT-trnS-cytb-nad6-trnP-nad1-trnL-trnL reversed, but did not change its position.
Stöger and Schrödl [32] compared mitogenome arrangements of different mollusca taxa. They found that most vetigastropods had the same gene order as Octopus and Katharina, so this arrangement may be plesiomorphic for gastropods. Gene arrangement of E. rumphii was completely consistent with the hypothetical ancestral gastropoda mitochondrial gene order, indicating its deep conservation.
Within Vetigastropoda, most species have the same gene order, except for Lepetodrilidae and Fissurellidae. Uribe [26] indicated that the high number of gene rearrangements is correlated with the high evolutionary rates of mitogenomes, and this correlation has been found in many molluscs [33,34]. This may have a negative effect on phylogenetic analyses. Therefore, mitogenomes may not be appropriate for phylogenetic reconstruction at deep nodes, compared with genome and transcriptome data.
4. Conclusions
The complete mitochondrial genome of E. rumphii was 15,795 bp in length, containing 13 PCGs, two rRNAs, and 22 tRNAs. Phylogenetic analysis based on complete mitogenomes recovered E. rumphii as sister group to all other vetigastropods. The arrangement of mitochondrial genes was the same as most vetigastropods as well as the inferred ancestral gastropoda. This suggested that E. rumphii was a deeply diverging lineage of Vetigastropoda, and its mitogenome arrangement was very conserved. This study provided a mitochondrial genome of Pleurotomarioidea, which could be helpful for understanding phylogenetic relationships and the early evolution of Vetigastropoda.
Author Contributions
H.W. and H.S. designed this experiment. P.M., Z.Z., Y.L., Y.C., C.L. and J.W. provided technical and sample assistance. The first draft of the manuscript was written by Y.W. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly supported by the National Key R&D Program of China (2019YFD0901303), the National Science Foundation of China (Nos. 42006080, 42076092, 41776179), the Strategic Priority Research Program of CAS (XDB42000000), the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-47), the Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), and the National Science & Technology Fundamental Resources Investigation Program of China (2022FY100304).
Institutional Review Board Statement
All procedures involving the care and use of animals conformed to the measures for the administration of animal experiment safety of the Institute of Oceanology, Chinese Academy of Science, and were approved by the Institutional Animal Care and Ethics Committee of the Institute of Oceanology, Chinese Academy of Science (Approval Code: Haifa 2020 [37], Approval Date: 17 June 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The mitochondrial genome of E. rumphii is available from GeneBank under the accession OP354269.
Acknowledgments
We appreciate the Oceanographic Data Center, IOCAS, for providing a computing server.
Conflicts of Interest
The authors have no relevant financial or non-financial interest to disclose.
References
- Rupert, E.E.; Barnes, R.D. Invertebrate Zoology, 6th ed.; Saunders College Publishing: New York, NY, USA, 1994; pp. 1–928. [Google Scholar]
- Bieler, R. Gastropod phylogeny and systematics. Annu. Rev. Ecol. Syst. 1992, 23, 311–338. [Google Scholar] [CrossRef]
- Harasewych, M.G. Pleurotomaroidean gastropods. Adv. Mar. Biol. 2002, 42, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Anseeuw, P.; Goto, Y. The Living Pleurotomariidae: A Synopsis of the Recent Pleurotomariidae Including Color Plates of All Extant Type Specimens; Elle Scientific Publications: Osaka, Japan, 1996; pp. 1–202. [Google Scholar]
- Okutani, T.; Hasegawa, K. Superfamily Pleurotomarioidea. In Marine Mollusks in Japan; Okutani, T., Ed.; Tokai University Press: Tokyo, Japan, 2000; pp. 1–374. [Google Scholar]
- Tan, T.H. A preliminary study on the anatomy of Pleurotoniaria (Eittemnotrochus) rumphii Schepman. Bull. Malacol. Soc. China 1974, 1, 15–20. [Google Scholar]
- Woodward, H. On Recent and fossil Pleurotomariae. Geol. Mag. 1885, 2, 433–439. [Google Scholar] [CrossRef]
- Wenz, W. Gastropoda. Allgemeiner Teil und Prosobranchia. In Handbuch der Paläozoologie; Schindewolf, O.H., Ed.; Gebrüder Borntraeger: Berlin, Germany, 1938; pp. 1–948. [Google Scholar]
- Cox, L.R. Thoughts on the classification of the Gastropoda. Proc. Malacol. Soc. Lond. 1960, 33, 239–261. [Google Scholar] [CrossRef]
- Knight, J.B.; Cox, L.R.; Keen, A.M.; Batten, R.L.; Yochelson, E.L.; Robertson, R. Gastropoda, Systematic Descriptions. In Treatise of Invertebrate Paleontology. I—Mollusca 1; Moore, R.C., Ed.; University of Kansas Press: Lawrence, KS, USA, 1960; pp. 1169–1331. [Google Scholar]
- Graham, A. Evolution within the Gastropoda: Prosobranchia. In The Mollusca. Volume 10—Evolution; Trueman, E.R., Clarke, M.R., Eds.; Academic Press: Orlando, FL, USA, 1985; pp. 151–186. [Google Scholar]
- Harasewych, M.G.; Adamkewicz, S.L.; Blake, J.A.; Saudek, D.; Spriggs, T.; Bult, C.J. Phylogeny and relationships of pleurotomariid gastropods (Mollusca: Gastropoda): An assessment based on partial 18S rDNA and cytochrome coxidase I sequences. Mol. Mar. Biol. Biotechnol. 1997, 6, 1–20. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Bernt, M.; Braband, A.; Middendorf, M.; Misof, B.; Rota-Stabelli, O.; Stadler, P.F. Bioinformatics methods for the comparative analysis of metazoan mitochondrial genome sequences. Mol. Phylogenet. Evol. 2013, 69, 320–327. [Google Scholar] [CrossRef]
- Kern, E.M.A.; Kim, T.; Park, J.K. The Mitochondrial Genome in Nematode Phylogenetics. Front. Ecol. Evol. 2020, 8, 250. [Google Scholar] [CrossRef]
- Ravi, K.P.; Mukesh, J. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Search and Contextual Analysis of Transfer RNA Genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Garey, J.R.; Wolstenholme, D.R. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989, 28, 374–387. [Google Scholar] [CrossRef]
- Uribe, J.E.; Kano, Y.; Templado, J.; Zardoya, R. Mitogenomics of Vetigastropoda: Insights into the evolution of pallial symmetry. Zool. Scr. 2016, 45, 145–159. [Google Scholar] [CrossRef]
- Lee, H.; Samadi, S.; Puillandre, N.; Tsai, M.H.; Dai, C.F.; Chen, W.J. Eight new mitogenomes for exploring the phylogeny and classification of Vetigastropoda. J. Molluscan Stud. 2016, 82, 534–541. [Google Scholar] [CrossRef]
- Boss, K.J. Mollusca. In Synopsis and Classification of Living Organisms; Parker, S.P., Ed.; McGraw-Hill: New York, NY, USA, 1982; Volume 2, pp. 1092–1096. [Google Scholar]
- Vaught, K.C. A Classification of Living Mollusca; American Malacologists: Melbourne, FL, USA, 1989. [Google Scholar]
- Yoon, S.H.; Kim, W. Phylogenetic Relationships Among Six Vetigastropod Subgroups (Mollusca, Gastropoda) Based on 18S rDNA Sequences. Mol. Cells 2005, 19, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Tauana, J.C.; James, D.R.; Gonzalo, G. Investigating Sources of Conflict in Deep Phylogenomics of Vetigastropod Snails. Syst. Biol. 2022, 71, 1009–1022. [Google Scholar] [CrossRef]
- Stoger, I.; Schrodl, M. Mitogenomics does not resolve deep molluscan relationships (yet?). Mol. Phylogenet. Evol. 2013, 69, 376–392. [Google Scholar] [CrossRef]
- Rawlings, T.; MacInnis, M.; Bieler, R.; Boore, J.; Collins, T. Sessile snails, dynamic genomes: Gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BMC Genom. 2010, 11, 440. [Google Scholar] [CrossRef] [PubMed]
- Schrodl, M.; Stoger, I. A review on deep molluscan phylogeny: Old markers, integrative approaches, persistent problems. J. Nat. Hist. 2014, 48, 2773–2804. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).