Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Collection
2.2. Mitochondrial Genome Sequencing, Assembly, and Analysis
2.2.1. PCR Amplification and Sequencing
2.2.2. Sequence Splicing and Gene Annotations
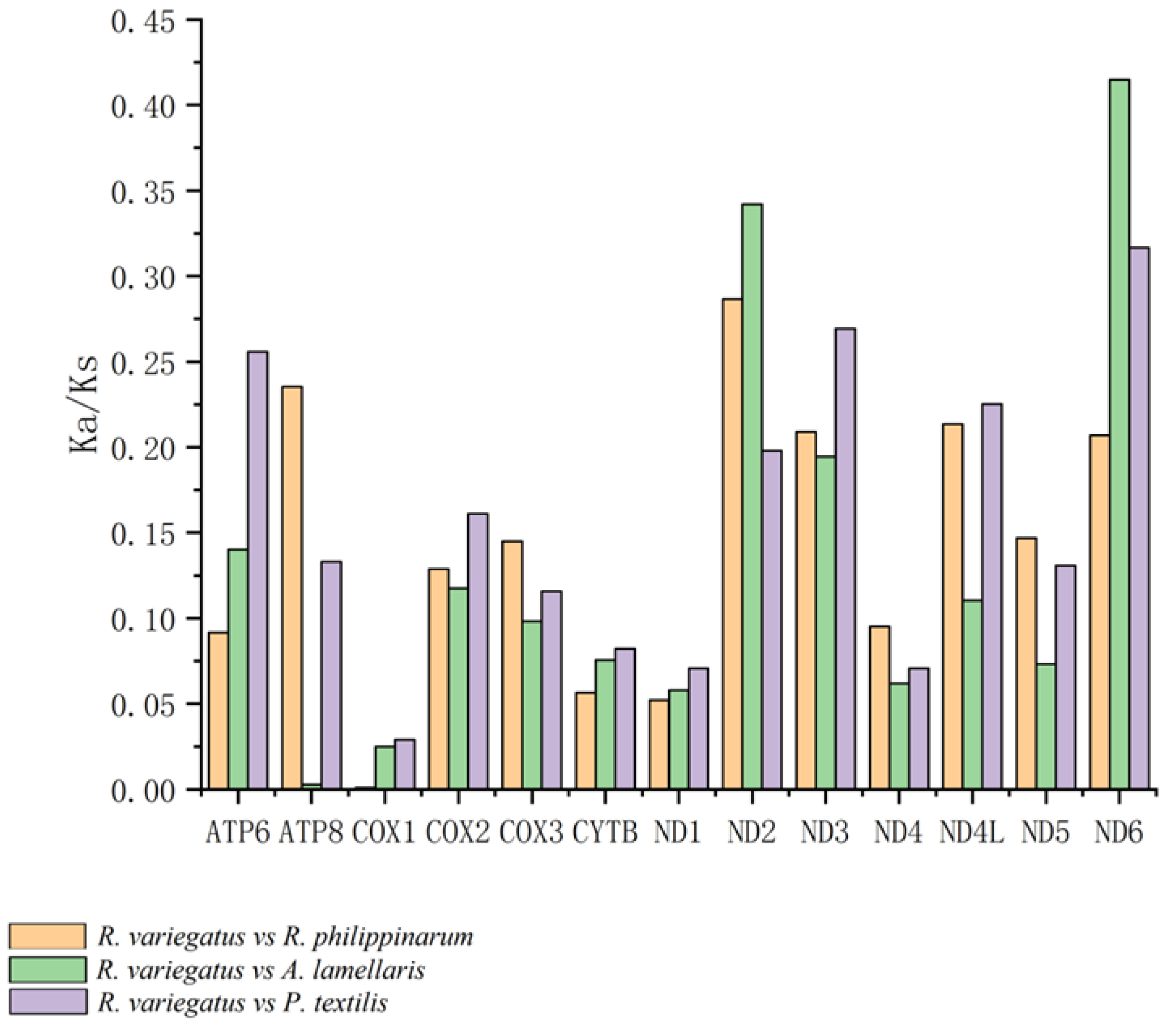
2.2.3. Analysis of Selective Pressure on the PCGs
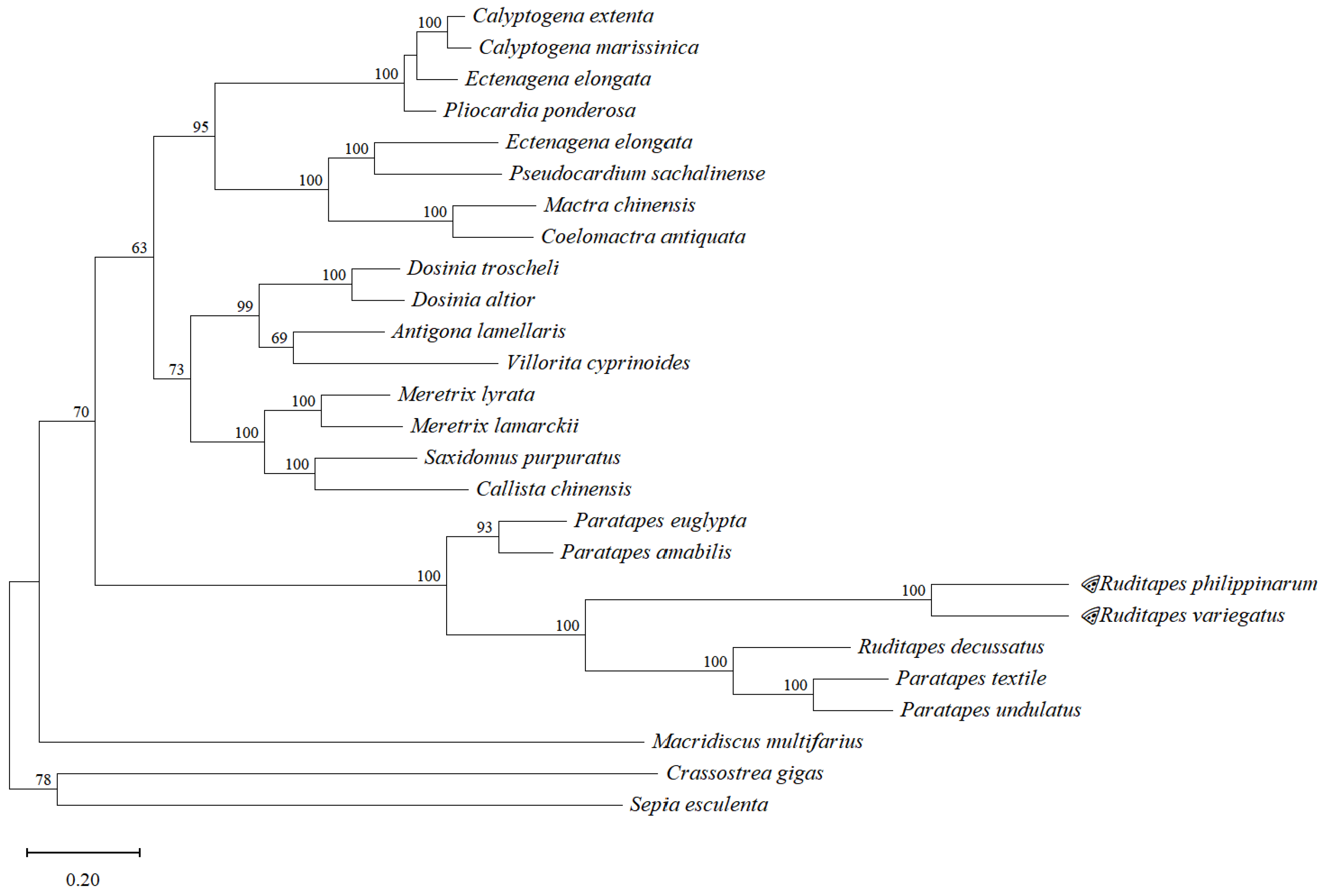
2.2.4. Phylogenetic Analysis
3. Results
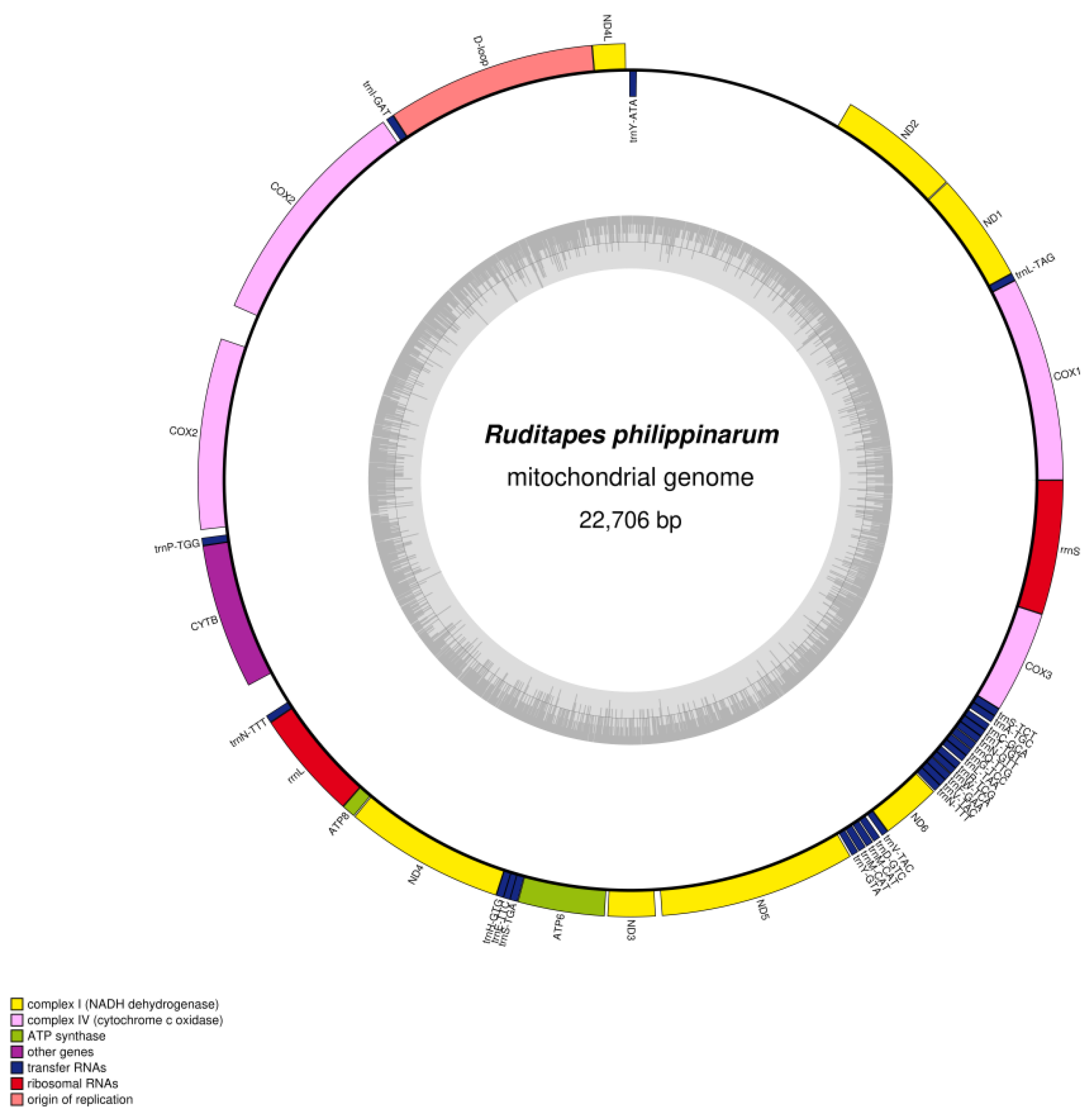
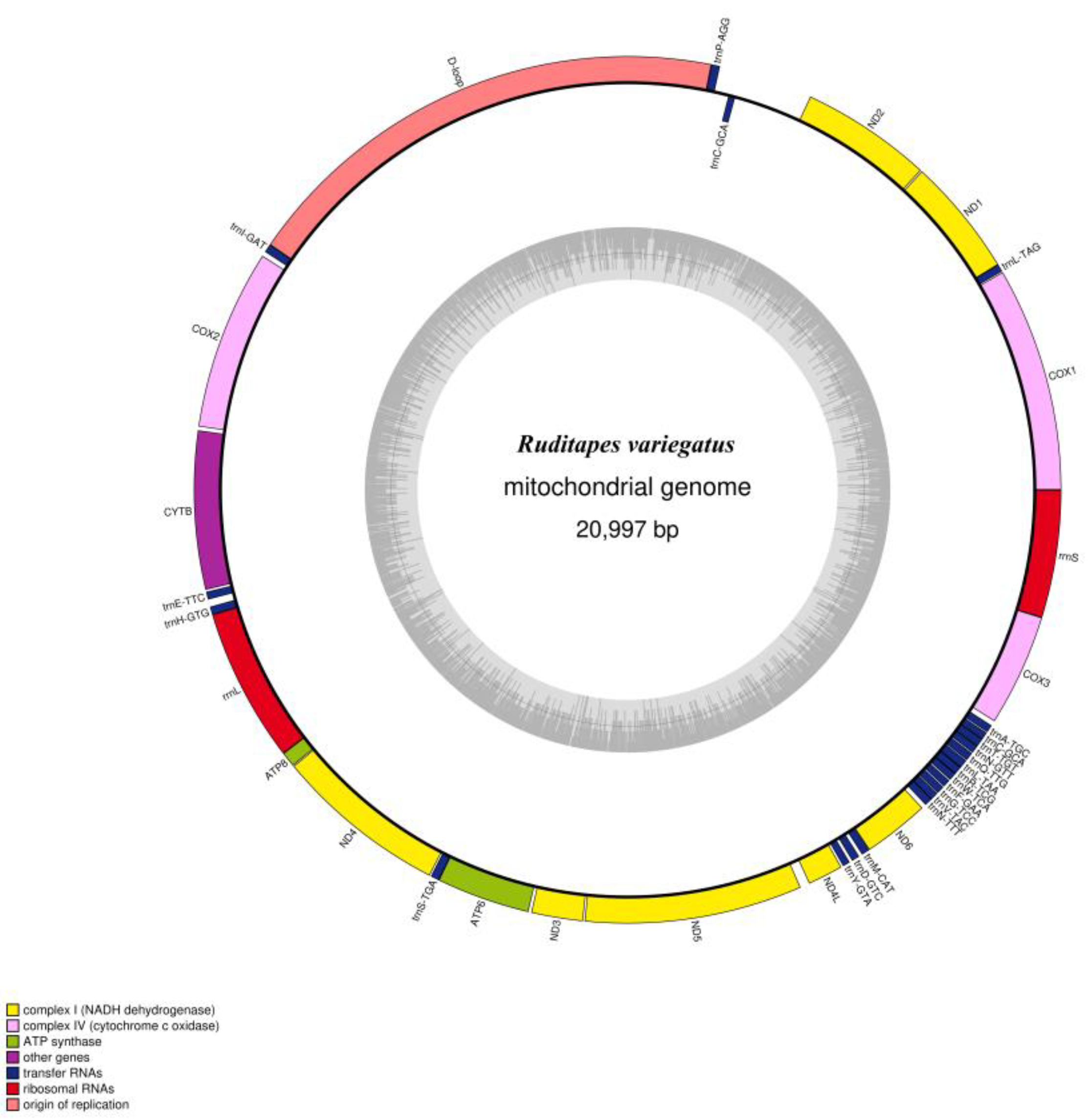
3.1. Mitochondrial Genome Features of R. variegatus and R. philippinarum
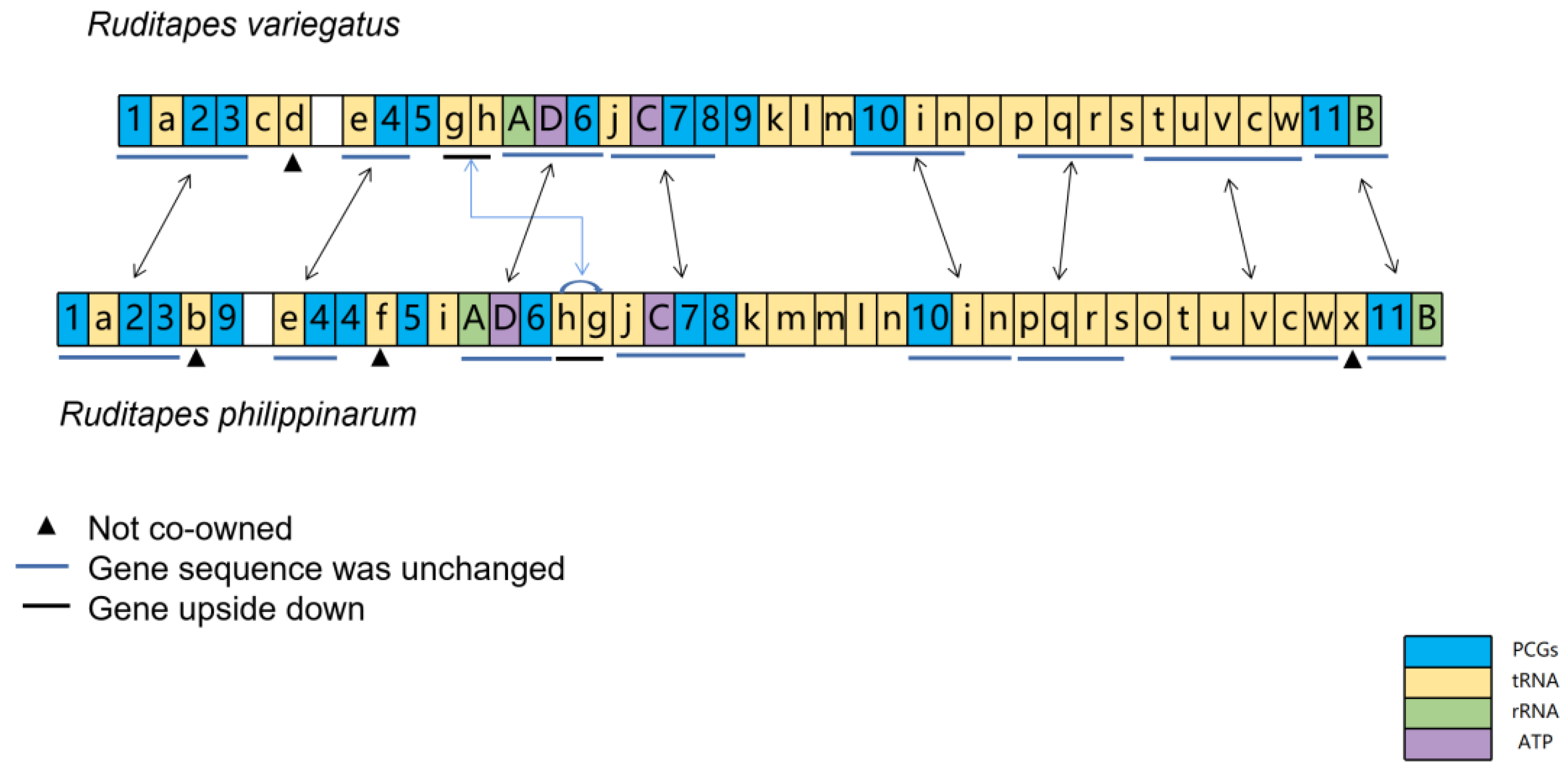
3.2. Comparison of Gene Order in R. philippinarum and R. variegatus
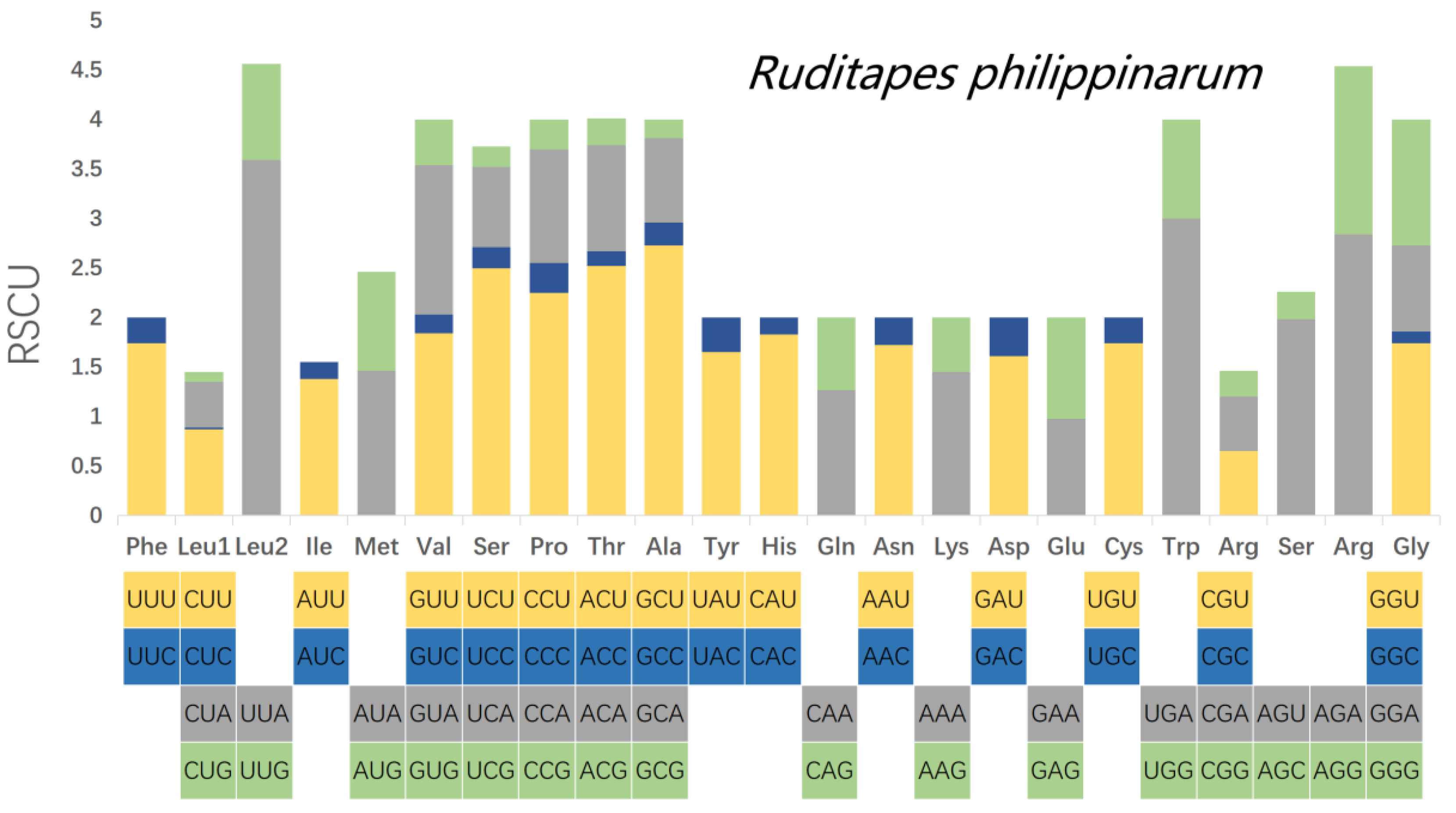
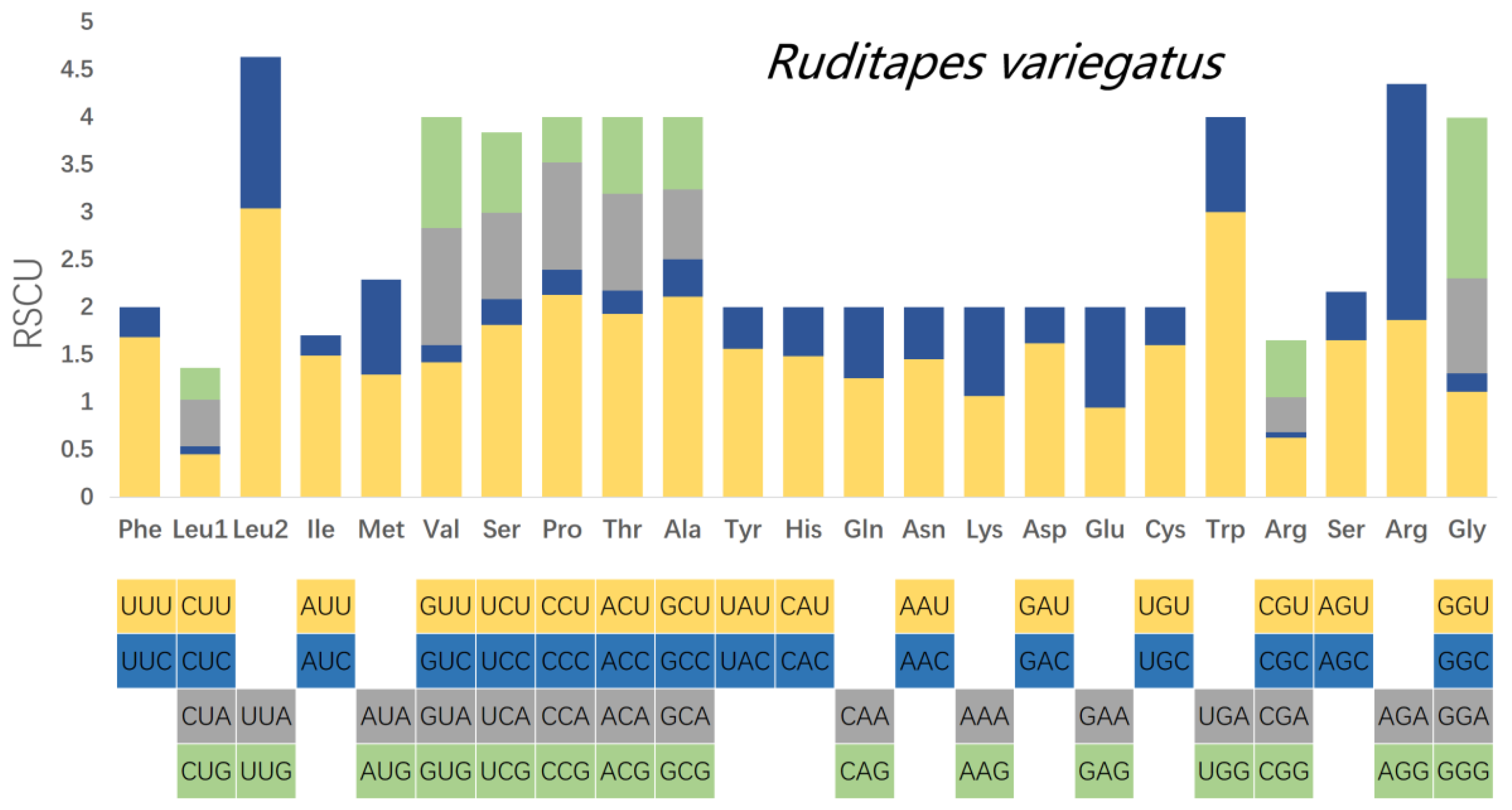
3.3. Analysis of Codon Usage Patterns of the PCGs
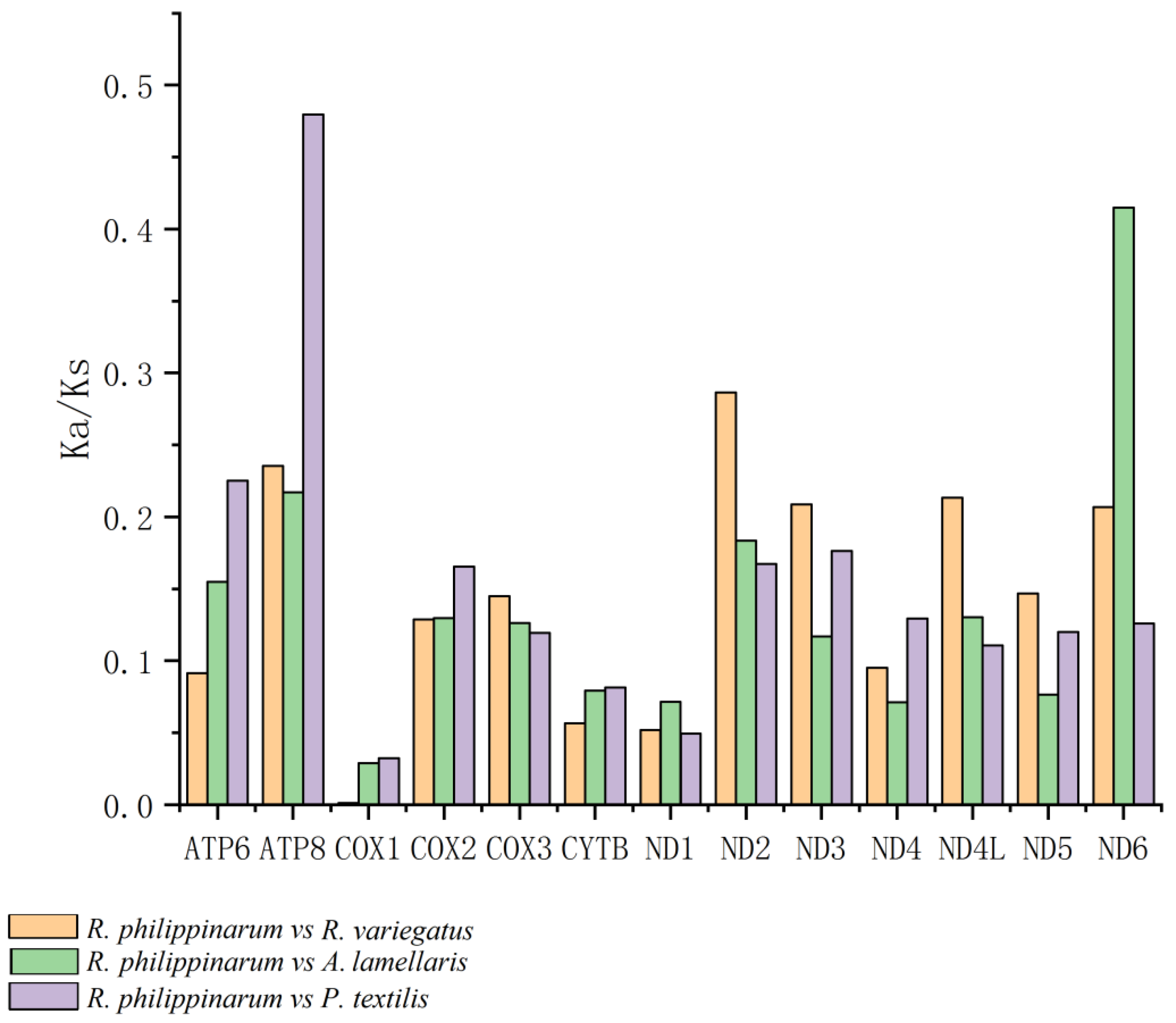
3.4. Codon-Based Analysis of Selective Pressure
4. Discussion
4.1. Expression of PCGs under Selective Pressure
4.2. Comparison of Gene Order
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, X. The Culture Biology and Technology and Selective Breeding in Manila Clam, Ruditapes philippinarum; Institute of Oceanology, Chinese Academy of Science: Qingdao, China, 2005. (In Chinese) [Google Scholar]
- Zheng, K.; Yang, M.; Yan, C.; Tang, M.; Song, Z. Mitochondrial Dynamics and Apoptosis. Chin. J. Cell Biol. 2019, 41, 1467–1476. (In Chinese) [Google Scholar]
- Passamonti, M. De Novo Assembly of the Manila Clam Ruditapes philippinarum Transcriptome Provides New Insights into Expression Bias, Mitochondrial Doubly Uniparental Inheritance and Sex Determination. Mol. Biol. Evol. 2012, 29, 771–786. [Google Scholar] [CrossRef]
- Milani, L.; Ghiselli, F.; Iannello, M.; Passamonti, M. Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr. Genet. 2014, 60, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bai, Y.; Nie, H.; Xu, Q.; Yin, Z.; Zhang, Y.; Yin, X.; Yan, X. Molecular mechanisms of wound healing and regeneration of siphon in the Manila clam Ruditapes philippinarum revealed by transcriptomic analysis. Genomics 2021, 113, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.-D.; Tan, Y.; Wang, J.; Xie, Q.; Huo, Z.; Fang, L.; Yan, X. Effect of estradiol stimulation on Dmrt gene expression in Manila clam Ruditapes philippinarum. J. Dalian Ocean. Univ. 2019, 34, 362–369. [Google Scholar] [CrossRef]
- Guerra, D.; Ghiselli, F.; Milani, L.; Breton, S.; Passamonti, M. Early replication dynamics of sex-linked mitochondrial DNAs in the doubly uniparental inheritance species Ruditapes philippinarum (Bivalvia, Veneridae). Heredity 2016, 116, 324–332. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; You, L.; Yu, J.; Zhao, J.; Li, L.; Wang, Q.; Li, F.; Li, C.; Liu, D.; et al. Differential toxicological effects induced by mercury in gills from three pedigrees of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Ecotoxicology 2011, 20, 177–186. [Google Scholar] [CrossRef]
- Delaporte, M.; Soudant, P.; Moal, J.; Lambert, C.; Quéré, C.; Miner, P.; Choquet, G.; Paillard, C.; Samain, J. Effect of a mono-specific algal diet on immune functions in two bivalve species—Crassostrea gigas and Ruditapes philippinarum. J. Exp. Biol. 2003, 206, 3053–3064. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Wei, Y.-H. Mitochondrial role in life and death of the cell. J. Biomed. Sci. 2000, 7, 2–15. [Google Scholar] [CrossRef]
- Suzuki, A.; Tsutomi, Y.; Yamamoto, N.; Shibutani, T.; Akahane, K. Mitochondrial regulation of cell death: Mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol. Cell. Biol. 1999, 19, 3842–3847. [Google Scholar] [CrossRef]
- Tang, B.-P.; Liu, Y.; Xin, Z.-Z.; Zhang, D.-Z.; Wang, Z.-F.; Zhu, X.-Y.; Wang, Y.; Zhang, H.-B.; Zhou, C.-L.; Chai, X.-Y.; et al. Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: Varunidae) and comparison with other Brachyuran crabs. Genomics 2018, 110, 221–230. [Google Scholar] [CrossRef]
- Iannelli, F.; Pesole, G.; Sordino, P.; Gissi, C. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 2007, 23, 419–422. [Google Scholar] [CrossRef]
- Shen, C.; Sun, S.-C. Mitochondrial genome of Micrura bella (Nemertea: Heteronemertea), the largest mitochondrial genome known to phylum Nemertea. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 2899–2900. [Google Scholar] [CrossRef]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, J.; Li, Y.; Cui, Y.; Xie, Q.; Bu, W.; Hillis, D.M. A Mitochondrial Genome of Rhyparochromidae (Hemiptera: Heteroptera) and a Comparative Analysis of Related Mitochondrial Genomes. Sci. Rep. 2016, 6, 35175. [Google Scholar] [CrossRef] [PubMed]
- Herd, K.E.; Barker, S.C.; Shao, R. The mitochondrial genome of the chimpanzee louse, Pediculus schaeffi: Insights into the process of mitochondrial genome fragmentation in the blood-sucking lice of great apes. BMC Genom. 2015, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zheng, Y.; Miura, I.; Wong, P.B.; Murphy, R.W.; Zeng, X. The evolution of mitochondrial genomes in modern frogs (Neobatrachia): Nonadaptive evolution of mitochondrial genome reorganization. BioMed Cent. 2014, 15, 691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, G.; Cao, D.; Li, S.; Su, A.; Geng, J.; Grover, C.E.; Hu, S.; Hua, J. The Complete Mitochondrial Genome of Gossypium hirsutum and Evolutionary Analysis of Higher Plant Mitochondrial Genomes. PLoS ONE 2013, 8, e69476. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.-D.; Shao, R.; Yuan, M.-L.; Dou, W.; Barker, S.C.; Wang, J.-J. The Multipartite Mitochondrial Genome of Liposcelis bostrychophila: Insights into the Evolution of Mitochondrial Genomes in Bilateral Animals. PLoS ONE 2012, 7, e33973. [Google Scholar] [CrossRef]
- Brockman, S.A.; McFadden, C.S. The Mitochondrial Genome of Paraminabea aldersladei (Cnidaria: Anthozoa: Octocorallia) Supports Intramolecular Recombination as the Primary Mechanism of Gene Rearrangement in Octocoral Mitochondrial Genomes. Genome Biol. Evol. 2012, 4, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Yang, T.; Du, T.; Huang, Y.; Chen, J.; Yan, J.; He, J.; Guan, R. Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC Genom. 2011, 12, 497. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Lu, C.; Deng, J.; Huang, X. The First Complete Mitochondrial Genome of Lachninae Species and Comparative Genomics Provide New Insights into the Evolution of Gene Rearrangement and the Repeat Region. Insects 2021, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, S. Characterization and phylogenetic analysis of the mitochondrial genome of Macdunnoughia hybrida (Lepidoptera: Noctuidae: Plusiinae). Mitochondrial DNA Part B 2021, 6, 2294–2296. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, Y.; Guo, X.; Wang, G.; Li, W.; Murányi, D. Two Complete Mitochondrial Genomes from Leuctridae (Plecoptera: Nemouroidea): Implications for the Phylogenetic Relationships among Stoneflies. J. Insect Sci. 2021, 21, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-T.; Gao, X.-G.; Li, Y.-F.; Liu, W.-D.; Liu, Y.; He, C.-B. Comparison of mitochondrial genomes of bivalves. Hereditas 2009, 31, 1127–1134. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Li, Q.; Kong, L.; Yu, H. Complete mitochondrial genomes of Trisidos kiyoni and Potiarca pilula: Varied mitochondrial genome size and highly rearranged gene order in Arcidae. Sci. Rep. 2016, 6, 33794. [Google Scholar] [CrossRef] [PubMed]
- Teimori, A.; Esmaeili, M.R.; Motamedi, M. Scanning electron microscopy and morphological analysis reveal size-dependent changes in the scale surface ornamentation of tooth-carp Aphaniops hormuzensis (Teleostei; Aphaniidae). Microsc. Res. Tech. 2021, 84, 1710–1720. [Google Scholar] [CrossRef]
- Nie, H.; Jiang, L.; Huo, Z.; Liu, L.; Yang, F.; Yan, X. Transcriptomic responses to low temperature stress in the Manila clam, Ruditapes philippinarum. Fish Shellfish. Immunol. 2016, 55, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985, 2, 13–34. [Google Scholar]
- Śmietanka, B.; Burzyński, A.; Wenne, R. Comparative Genomics of Marine Mussels (Mytilus spp.) Gender Associated mtDNA: Rapidly Evolving atp8. J. Mol. Evol. 2010, 71, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yan, P.; Liang, C.; Guo, X. Bioinformatics analysis of the ATP6 and ATP8 genes in yak. <The Sixth China Cattle Industry Development Conference> proceedings. Association. 2011, pp. 346–349. (In Chinese). Available online: https://xueshu.baidu.com/usercenter/paper/show?paperid=0560b0332c2d4c2713115353af28f783 (accessed on 18 October 2022).
- Uddin, A.; Mazumder, T.H.; Barbhuiya, P.A.; Chakraborty, S. Similarities and dissimilarities of codon usage in mitochondrial ATP genes among fishes, aves, and mammals. IUBMB Life 2020, 72, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Akimoto, S.-I. Development of new barcoding loci in gall-forming aphids (Eriosomatinae: Eriosomatini): Comparing three mitochondrial genes, ATP6, ATP8, and COI. J. Asia-Pac. Èntomol. 2015, 18, 267–275. [Google Scholar] [CrossRef]
- Bernhard, E.; Lutz, B.; Bastian, F. Atp8 is in the ground pattern of flatworm mitochondrial genomes. BMC Genom. 2017, 18, 414. [Google Scholar]
- Yi, L.; Ai, Y.; Ming, L.; Hai, L.; He, J.; Guo, F.-C.; Qiao, X.-Y.; Ji, R. Molecular diversity and phylogenetic analysis of domestic and wild Bactrian camel populations based on the mitochondrial ATP8 and ATP6 genes. Livest. Sci. 2017, 199, 95–100. [Google Scholar] [CrossRef]
- Zhang, W.; Hou, L.; Wang, T.; Lu, W.; Tao, Y.; Chen, W.; Du, X.; Huang, Y. The expression characteristics of mt-ND2 gene in chicken. Mitochondrial DNA Part A 2015, 27, 3787–3792. [Google Scholar] [CrossRef]
- Shi, Y.; Hu, Y.; Wang, J.; Elzo, M.A.; Yang, X.; Lai, S. Genetic diversities of MT-ND1 and MT-ND2 genes are associated with high-altitude adaptation in yak. Mitochondrial DNA Part A 2017, 29, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.P.; Lanyon, S.M. Molecular Systematics of the Grackles and Allies, and the Effect of Additional Sequence (Cyt B and ND2). Ornithology 1999, 116, 759–768. [Google Scholar] [CrossRef]
- Jones, D.A.; Gibbs, H.L. Intra- and Interspecific Sequence Variation in a Portion of the Mitochondrial ND6 Gene in Cuckoos. Ornithol. Appl. 1997, 99, 815–818. [Google Scholar] [CrossRef]
- Bi, C.; Lu, N.; Xu, Y.; He, C.; Lu, Z. Characterization and Analysis of the Mitochondrial Genome of Common Bean (Phaseolus vulgaris) by Comparative Genomic Approaches. Int. J. Mol. Sci. 2020, 21, 3778. [Google Scholar] [CrossRef]
- Moritz, C.; Brown, W.M. Tandem Duplications in Animal Mitochondrial DNAs: Variation in Incidence and Gene Content among Lizards. Proc. Natl. Acad. Sci. USA 1987, 84, 7183–7187. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.K.-S. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 2006, 259–263. [Google Scholar] [CrossRef]
- Arndt, A.; Smith, M.J. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol. Biol. Evol. 1998, 15, 1009–1016. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Lang, B.F. Poriferan mt DNA and animal phylogeny based on mitochondrial gene arrangements. Syst. Biol. 2005, 54, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Du, S. Molecular Evolution and Phylogenetic Study of the Mitochondrial Genome of Freshwater Crabs in Gecarcinucidae Rathbun. Master’s Thesis, Nanjing Normal University, Nanjing, China, 2021. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?doi=10.27245/d.cnki.gnjsu.2021.000959&dbcode=CMFD (accessed on 18 October 2022). (In Chinese).
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Poulton, J.; Deadman, M.E.; Bindoff, L.; Morten, K.; Land, J.; Brown, G. Families of mtDNA rearrangements can be detected in patients with mtDNA deletions: Duplications may be a transient intermediate form. Hum. Mol. Genet. 1993, 223–230. [Google Scholar] [CrossRef]

| Material | Shell | Shell | Shell | Tissue Dry | Meat |
|---|---|---|---|---|---|
| Length | Width | Height | Weight | Condition | |
| R. variegatus | 32.66 ± 2.07 | 14.37 ± 1.56 | 23.67 ± 1.57 | 0.27 ± 0.05 | 6.11 ± 1.85 |
| R. philippinarum | 34.96 ± 1.33 | 15.28 ± 0.69 | 23.56 ± 1.03 | 0.28 ± 0.06 | 7.30 ± 1.40 |
| Species | Family | bp Length | GenBank Accession No. |
|---|---|---|---|
| Calyptogena marissinica | Vesicomyidae | 17,374 | NC_044766.1 |
| Calyptogena extenta | Vesicomyidae | 16,106 | MF981085.1 |
| Ectenagena elongata | Vesicomyidae | 16,827 | NC_051454.1 |
| Pliocardia ponderosa | Vesicomyidae | 16,275 | MF981084.1 |
| Pseudocardium sachalinense | Mactridae | 17,978 | MG431821.1 |
| Mactra antiquata | Mactridae | 17,199 | JQ423460.1 |
| Mactra chinensis | Mactridae | 17,285 | KJ754823.1 |
| Lutraria maxima | Mactridae | 17,082 | MF784266.1 |
| Paphia amabilis | Veneridae | 19,629 | JF969276.1 |
| Paratapes textilis | Veneridae | 18,561 | JF969277.1 |
| Paratapes undulatus | Veneridae | 18,154 | JF969278.1 |
| Paphia euglypta | Veneridae | 18,643 | GU269271.1 |
| Macridiscus multifarius | Veneridae | 20,171 | NC_045888.1 |
| Ruditapes decussatus | Veneridae | 18,995 | KP089983.1 |
| Dosinia altior | Veneridae | 17,536 | NC_037916.1 |
| Dosinia troscheli | Veneridae | 17,229 | NC_037917.1 |
| Saxidomus purpurata | Veneridae | 19,637 | KP419933.1 |
| Callista chinensis | Veneridae | 19,703 | MT742541.1 |
| Meretrix lamarckii | Veneridae | 21,209 | GU071281.1 |
| Villorita cyprinoides | Veneridae | 15,880 | NC_050989.1 |
| Meretrix lyrata | Veneridae | 21,625 | KC832317.1 |
| Antigona lamellaris | Veneridae | 17,532 | NC_051506.1 |
| Sepia esculenta | Sepiidae | 16,199 | NC_009690.1 |
| Crassostrea gigas | Ostreidae | 18,224 | MZ497416.1 |
| Group of Genes | Gene Name |
|---|---|
| complex Ⅰ (NADH dehydrogenase) | ND1, ND2, ND3, ND4, ND5, ND6, ND4L |
| complex Ⅳ (cytochrome c oxidase) | COX1, COX2 (2 copies), COX3 |
| ATP synthase | ATP6, ATP8 |
| other genes | CYTB |
| transfer RNAs | trnY-ATA, trnY-GTA, trnl-GAT, trnP-TGG, trnN-TTT, trnN-GTT (2 copies), trnH-GTG, trnE-TTC, trnS-TGA, trnS-TCT, trnM-CAT (2 copies), trnM-CAT, trnD-GTC, trnV-TAC (2 copies), trnF-GAA, trnW-TCA, trnR-TCG, trnG-TCC, trnQ-TTG, trnT-GCA, trnA-TGC, trnL-TAG, trnL-TAA |
| ribosomal RNAs | RrnL, rrnS |
| origin of replication | D-loop |
| Ruditapes philippinarum | A% | C% | G% | T% | GC% | Size (bp) | Proportion in Genome (%) |
|---|---|---|---|---|---|---|---|
| Whole genome | 30.2 | 9.80 | 20.5 | 39.6 | 30.30 | 22,706 | 100 |
| Protein-coding genes | 27.4 | 9.9 | 21.7 | 41.1 | 31.57 | 14,454 | 63.66 |
| tRNA genes | 32.8 | 11.8 | 18.2 | 37.3 | 29.97 | 1685 | 7.42 |
| rRNA genes | 35.4 | 10.4 | 18.5 | 35.7 | 28.87 | 2054 | 9.05 |
| D-loop | 34.7 | 12.0 | 21.6 | 31.7 | 33.65 | 1777 | 7.83 |
| Group of Genes | Gene Name |
|---|---|
| complex Ⅰ (NADH dehydrogenase) | ND4, ND3, ND5, ND4L, ND6, ND1, ND2 |
| complex Ⅳ (cytochrome c oxidase) | COX2, COX3, COX1 |
| ATP synthase | ATP8, ATP6 |
| other genes | CYTB |
| transfer RNAs | trnC-GCA (2 copies), trnP-AGG, trnl-GAT, trnE-TTC, trnH-GTG, trnS-TGA, trnY-GAT, trnD-GTC, trnM-CAT, trnN-TTT, trnN-GTT, trnV-TAC, trnG-TCC, trnF-GAA, trnW-TCA, trnR-TCG, trnL-TAA, trnL-TAG, trnQ-TTG, trnT-TGT, trnA-TGC |
| ribosomal RNAs | rrnL, rrnS |
| origin of replication | D-loop |
| Ruditapes variegatus | A% | C% | G% | T% | GC% | Size (bp) | Proportion in Genome (%) |
|---|---|---|---|---|---|---|---|
| Whole genome | 28.1 | 10.7 | 25.7 | 35.5 | 36.4 | 20,997 | 100 |
| Protein-coding genes | 24.7 | 10.8 | 26.2 | 38.3 | 37.0 | 12,378 | 59.0 |
| tRNA genes | 30.9 | 12.7 | 22.7 | 33.7 | 35.4 | 1414 | 6.7 |
| rRNA genes | 33.5 | 11.3 | 22.1 | 33.1 | 33.4 | 2207 | 10.5 |
| D-loop | 33.4 | 9.4 | 27.1 | 30.1 | 36.5 | 3892 | 18.5 |
| Ruditapes philippinarum | Ruditapes variegatus | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Length | Start Codon | Stop Codon | Direction | Gene Name | Length | Start Codon | Stop Codon | Direction |
| COX1 | 1719 | ATA | TAG | + | COX1 | 1758 | GTG | TAG | + |
| ND1 | 930 | ATA | TAA | + | ND1 | 933 | ATG | TAG | + |
| ND2 | 1035 | ATT | TAG | + | ND2 | 1029 | ATG | TAG | + |
| ND4L | 282 | ATT | TAA | + | COX2 | 1416 | TTG | TAG | + |
| COX2 | 1995 | ATC | TAA | + | CYTB | 1239 | ATT | TAA | + |
| COX2 | 1608 | ATA | TAA | + | ATP8 | 120 | TTG | TAA | + |
| CYTB | 1224 | ATT | TAA | + | ND4 | 1356 | ATG | TAA | + |
| ATP8 | 120 | ATT | TAG | + | ATP6 | 735 | ATG | TAG | + |
| ND4 | 1359 | ATG | TAA | + | ND3 | 405 | ATG | TAA | + |
| ATP6 | 738 | ATA | TAG | + | ND5 | 1704 | GTG | TAA | + |
| ND3 | 405 | GTG | TAA | + | ND4L | 279 | ATG | TAG | + |
| ND5 | 1665 | TTG | TAA | + | ND6 | 522 | ATG | TAG | + |
| ND6 | 510 | TTG | TAA | + | COX3 | 882 | ATT | TAA | + |
| COX3 | 864 | ATT | TAA | + | |||||
| Ruditapes philippinarum | Ruditapes variegatus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU |
| UUU(F) | 430 | 1.74 | GCG(A) | 9 | 0.19 | UUU(F) | 324 | 1.68 | GCG(A) | 37 | 0.76 |
| UUC(F) | 63 | 0.26 | UAU(Y) | 171 | 1.65 | UUC(F) | 61 | 0.32 | UAU(Y) | 140 | 1.56 |
| UUA(L) | 430 | 3.59 | UAC(Y) | 36 | 0.35 | UUA(L) | 265 | 3.04 | UAC(Y) | 40 | 0.44 |
| UUG(L) | 116 | 0.97 | CAU(H) | 73 | 1.83 | UUG(L) | 139 | 1.59 | CAU(H) | 46 | 1.48 |
| CUU(L) | 104 | 0.87 | CAC(H) | 7 | 0.17 | CUU(L) | 39 | 0.45 | CAC(H) | 16 | 0.52 |
| CUC(L) | 2 | 0.02 | CAA(Q) | 38 | 1.27 | CUC(L) | 7 | 0.08 | CAA(Q) | 38 | 1.25 |
| CUA(L) | 55 | 0.46 | CAG(Q) | 22 | 0.73 | CUA(L) | 43 | 0.49 | CAG(Q) | 23 | 0.75 |
| CUG(L) | 12 | 0.1 | AAU(N) | 153 | 1.72 | CUG(L) | 30 | 0.34 | AAU(N) | 72 | 1.45 |
| AUU(I) | 239 | 1.38 | AAC(N) | 25 | 0.28 | AUU(I) | 174 | 1.49 | AAC(N) | 27 | 0.55 |
| AUC(I) | 29 | 0.17 | AAA(K) | 138 | 1.45 | AUC(I) | 25 | 0.21 | AAA(K) | 71 | 1.06 |
| AUA(M) | 253 | 1.46 | AAG(K) | 53 | 0.55 | AUA(M) | 151 | 1.29 | AAG(K) | 63 | 0.94 |
| AUG(M) | 79 | 1 | GAU(D) | 99 | 1.61 | AUG(M) | 98 | 1 | GAU(D) | 77 | 1.62 |
| GUU(V) | 238 | 1.84 | GAC(D) | 24 | 0.39 | GUU(V) | 154 | 1.42 | GAC(D) | 18 | 0.38 |
| GUC(V) | 25 | 0.19 | GAA(E) | 91 | 0.98 | GUC(V) | 20 | 0.18 | GAA(E) | 66 | 0.94 |
| GUA(V) | 195 | 1.51 | GAG(E) | 95 | 1.02 | GUA(V) | 134 | 1.23 | GAG(E) | 74 | 1.06 |
| GUG(V) | 59 | 0.46 | UGU(C) | 105 | 1.74 | GUG(V) | 127 | 1.17 | UGU(C) | 88 | 1.6 |
| UCU(S) | 141 | 2.5 | UGC(C) | 16 | 0.26 | UCU(S) | 68 | 1.81 | UGC(C) | 22 | 0.4 |
| UCC(S) | 12 | 0.21 | UGA(W) | 65 | 3 | UCC(S) | 10 | 0.27 | UGA(W) | 39 | 3 |
| UCA(S) | 46 | 0.81 | UGG(W) | 72 | 1 | UCA(S) | 34 | 0.91 | UGG(W) | 65 | 1 |
| UCG(S) | 12 | 0.21 | CGU(R) | 35 | 0.65 | UCG(S) | 32 | 0.85 | CGU(R) | 30 | 0.62 |
| CCU(P) | 82 | 2.25 | CGC(R) | 0 | 0 | CCU(P) | 66 | 2.13 | CGC(R) | 3 | 0.06 |
| CCC(P) | 11 | 0.3 | CGA(R) | 30 | 0.55 | CCC(P) | 8 | 0.26 | CGA(R) | 18 | 0.37 |
| CCA(P) | 42 | 1.15 | CGG(R) | 14 | 0.26 | CCA(P) | 35 | 1.13 | CGG(R) | 29 | 0.6 |
| CCG(P) | 11 | 0.3 | AGU(S) | 112 | 1.98 | CCG(P) | 15 | 0.48 | AGU(S) | 62 | 1.65 |
| ACU(T) | 104 | 2.52 | AGC(S) | 16 | 0.28 | ACU(T) | 57 | 1.93 | AGC(S) | 19 | 0.51 |
| ACC(T) | 6 | 0.15 | AGA(R) | 154 | 2.84 | ACC(T) | 7 | 0.24 | AGA(R) | 90 | 1.86 |
| ACA(T) | 44 | 1.07 | AGG(R) | 92 | 1.7 | ACA(T) | 30 | 1.02 | AGG(R) | 121 | 2.49 |
| ACG(T) | 11 | 0.27 | GGU(G) | 180 | 1.74 | ACG(T) | 24 | 0.81 | GGU(G) | 98 | 1.11 |
| GCU(A) | 129 | 2.73 | GGC(G) | 12 | 0.12 | GCU(A) | 103 | 2.11 | GGC(G) | 17 | 0.19 |
| GCC(A) | 11 | 0.23 | GGA(G) | 90 | 0.87 | GCC(A) | 19 | 0.39 | GGA(G) | 88 | 1 |
| GCA(A) | 40 | 0.85 | GGG(G) | 131 | 1.27 | GCA(A) | 36 | 0.74 | GGG(G) | 149 | 1.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Huo, Z.; Liu, Y.; Wang, Y.; Zuo, L.; Fang, L.; Zhao, W.; Tan, Y.; Yan, X. Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus. Genes 2022, 13, 2157. https://doi.org/10.3390/genes13112157
Tang Y, Huo Z, Liu Y, Wang Y, Zuo L, Fang L, Zhao W, Tan Y, Yan X. Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus. Genes. 2022; 13(11):2157. https://doi.org/10.3390/genes13112157
Chicago/Turabian StyleTang, Yumei, Zhongming Huo, Yang Liu, Yuhang Wang, Luya Zuo, Lei Fang, Wen Zhao, Yue Tan, and Xiwu Yan. 2022. "Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus" Genes 13, no. 11: 2157. https://doi.org/10.3390/genes13112157
APA StyleTang, Y., Huo, Z., Liu, Y., Wang, Y., Zuo, L., Fang, L., Zhao, W., Tan, Y., & Yan, X. (2022). Full Mitochondrial Genomes Reveal Species Differences between the Venerid Clams Ruditapes philippinarum and R. variegatus. Genes, 13(11), 2157. https://doi.org/10.3390/genes13112157





