Selection Signature and CRISPR/Cas9-Mediated Gene Knockout Analyses Reveal ZC3H10 Involved in Cold Adaptation in Chinese Native Cattle
Abstract
1. Background
2. Material and methods
2.1. FLK and hapFLK Genomic Scans and Local Tree Building of ZC3H10
2.2. RT-PCR
2.3. Construction and Transfection of ZC3H10 Gene Knockout Vector
2.4. Cell Culture and Cold-Stress Treatment
2.5. Library Construction and Sequencing
2.6. Reads Filtering and Differential Expression Analysis
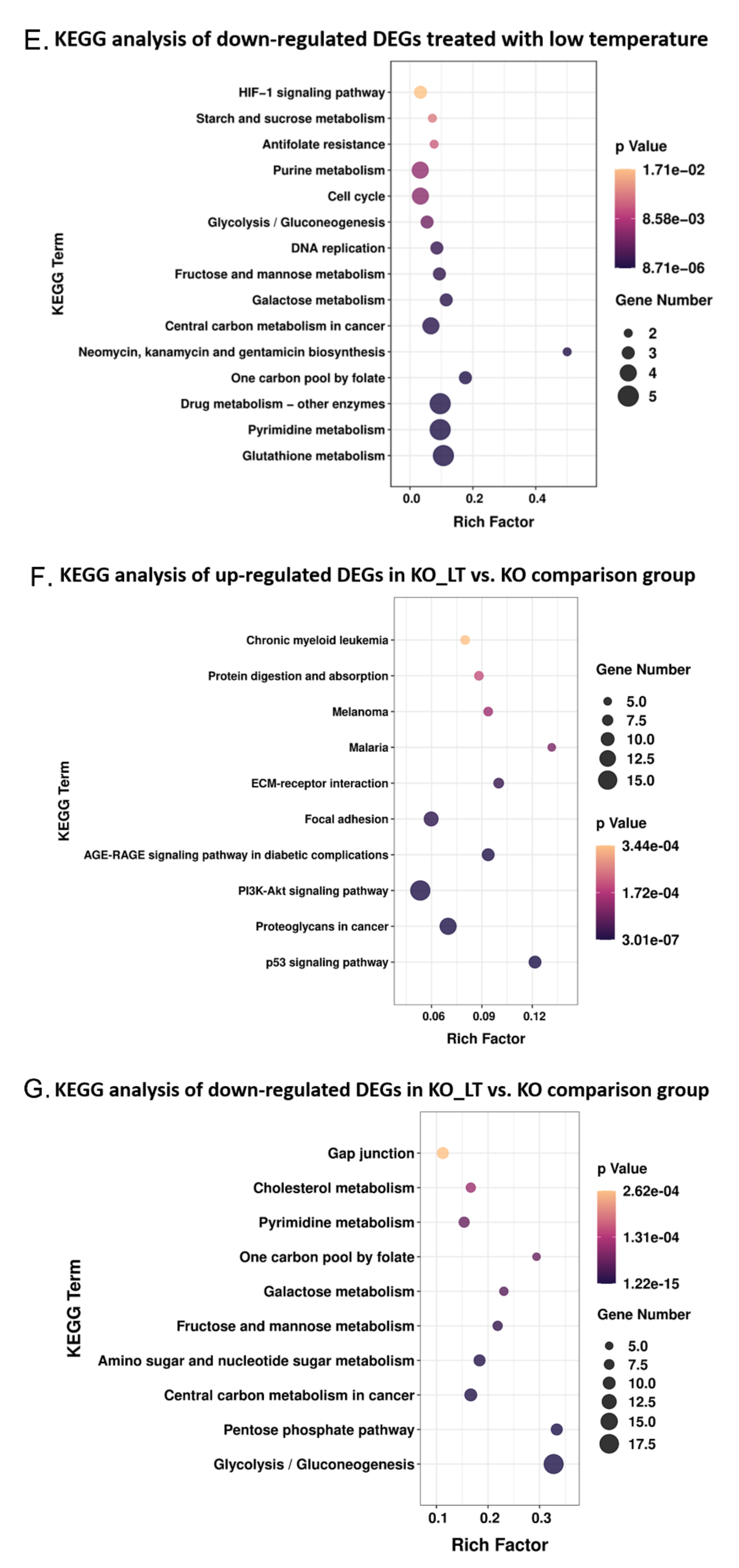
2.7. Enrichment Analysis of GO and KEGG
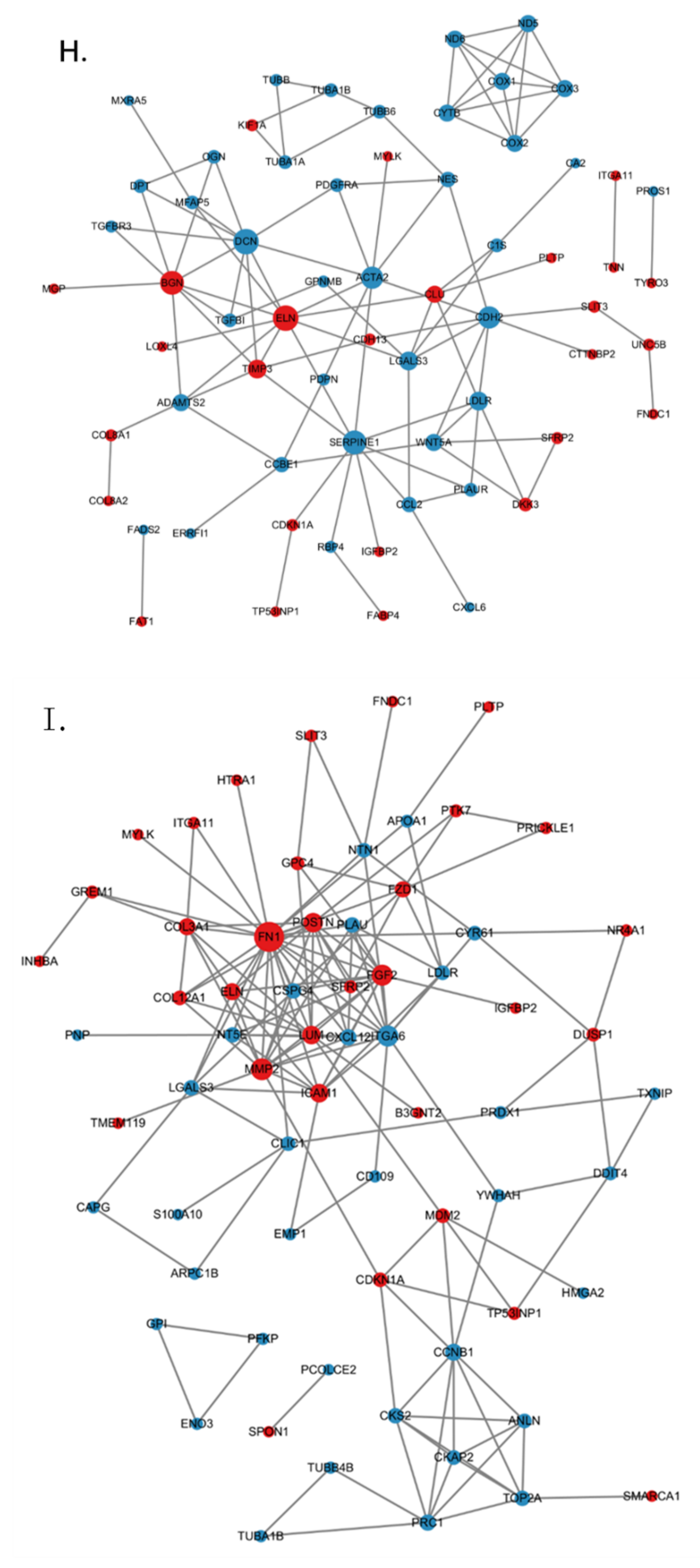
2.8. Establishment of a PPI Network
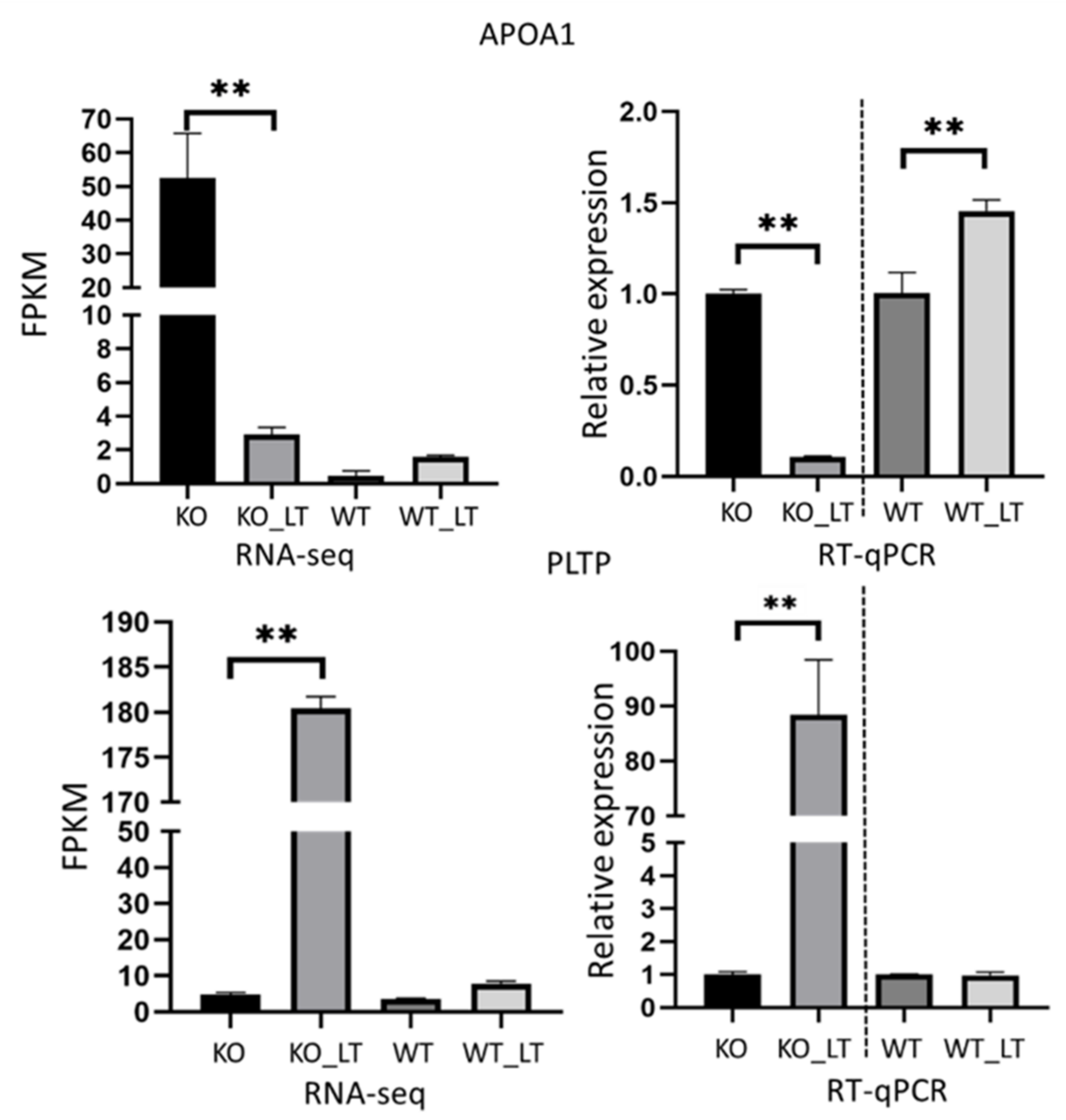
2.9. Validation of Gene Expression Using RT-qPCR
3. Results
3.1. Signatures of Selection in the Genomes of Cattle Breed in Cold and Heat Areas
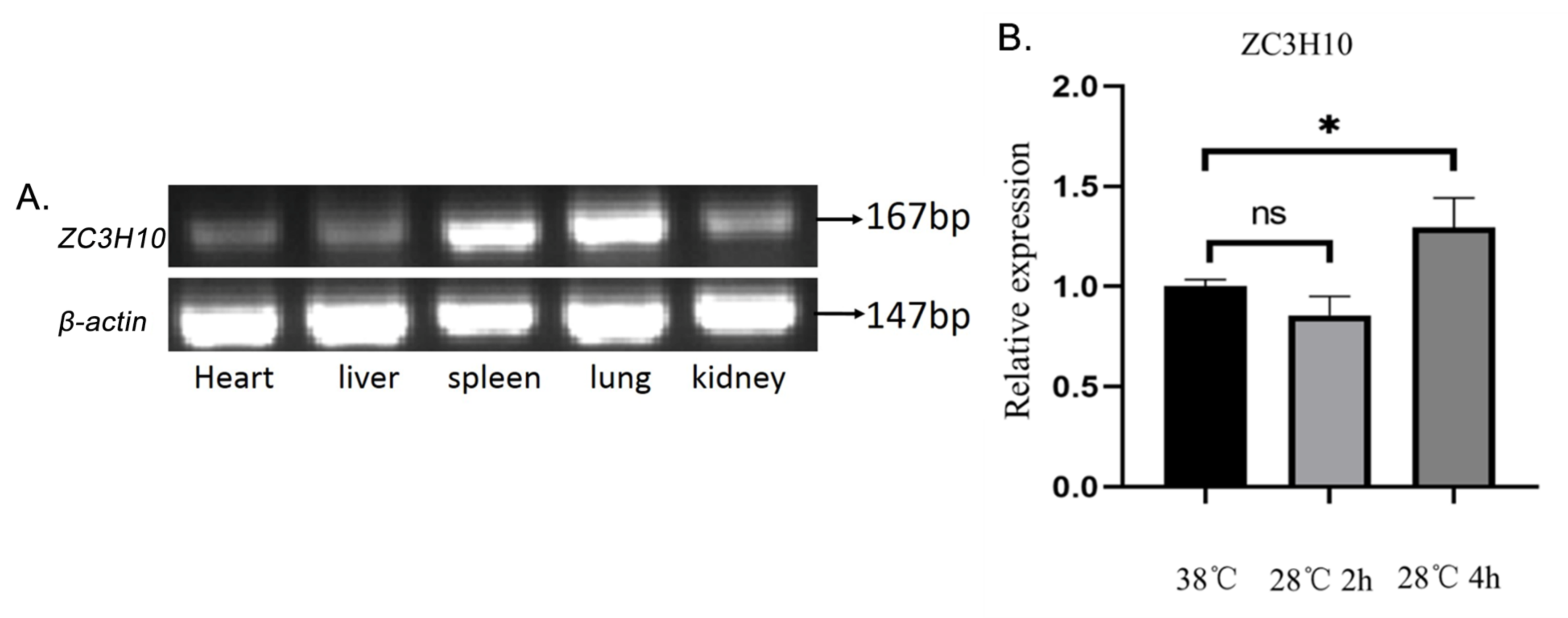
3.2. Expression of ZC3H10 mRNA in Bovine Different Tissues and in Cold-Treated Cells
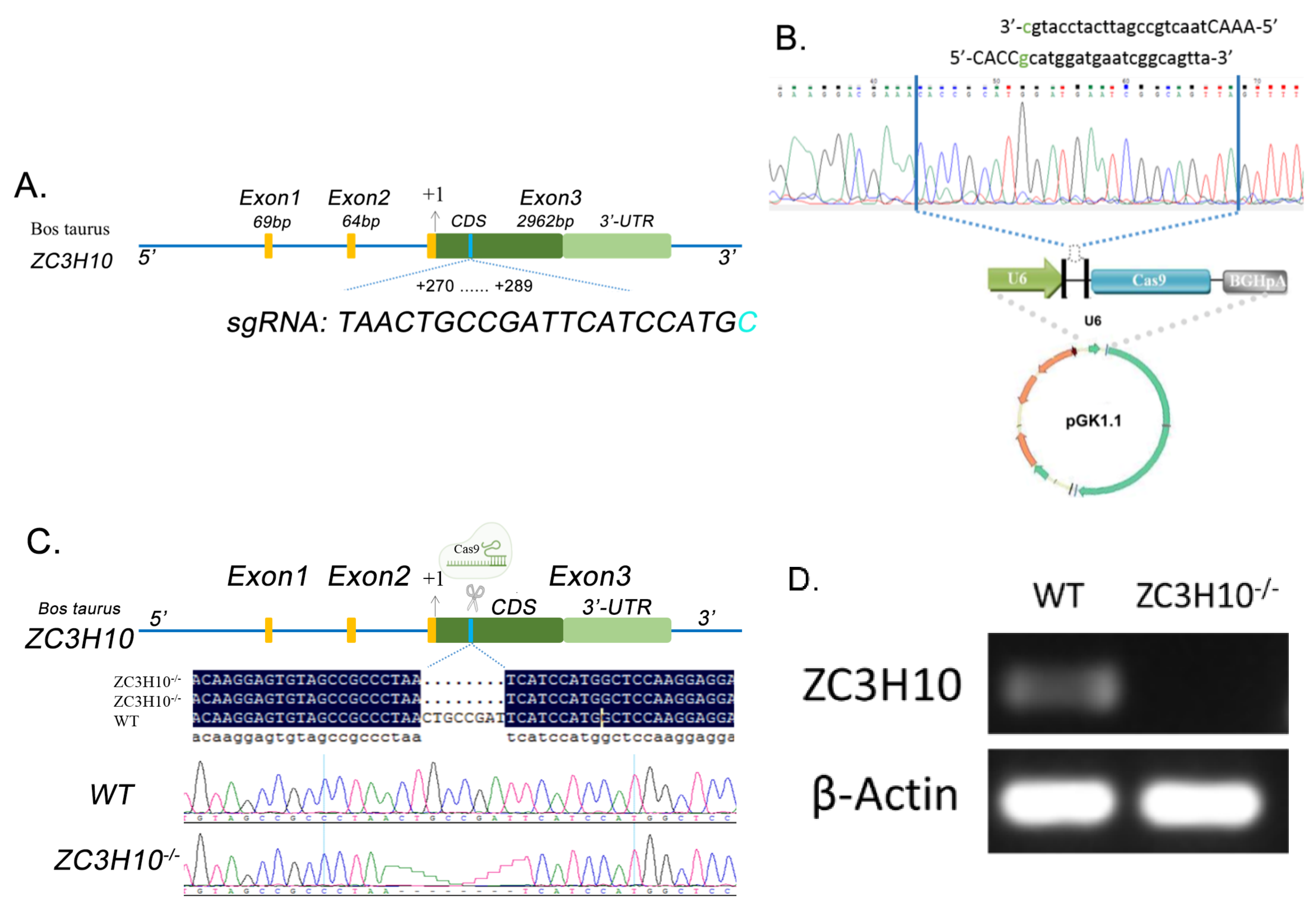
3.3. Production and Screening of ZC3H10 Knockout Cell Line
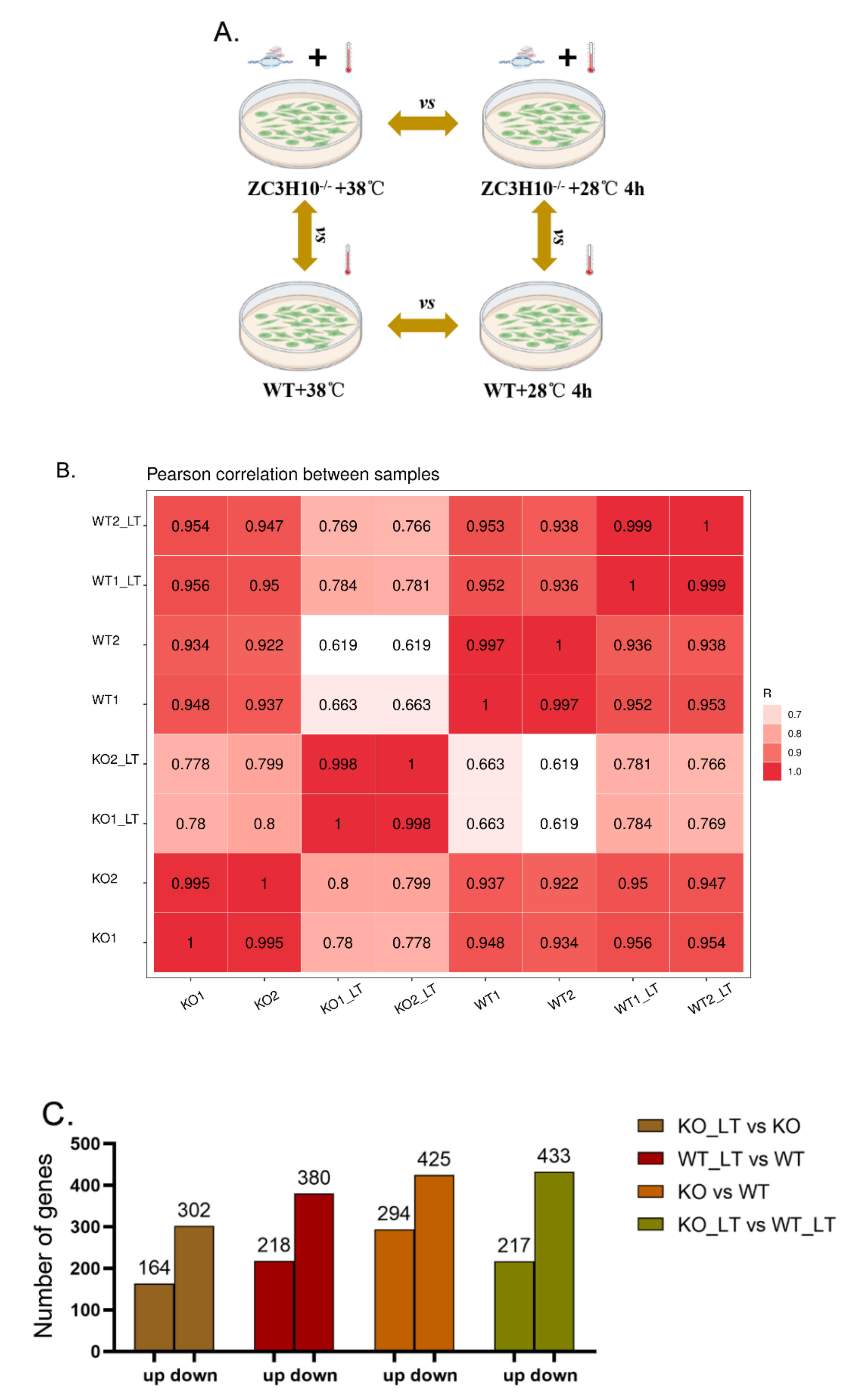
3.4. Overview of RNA Sequencing Data
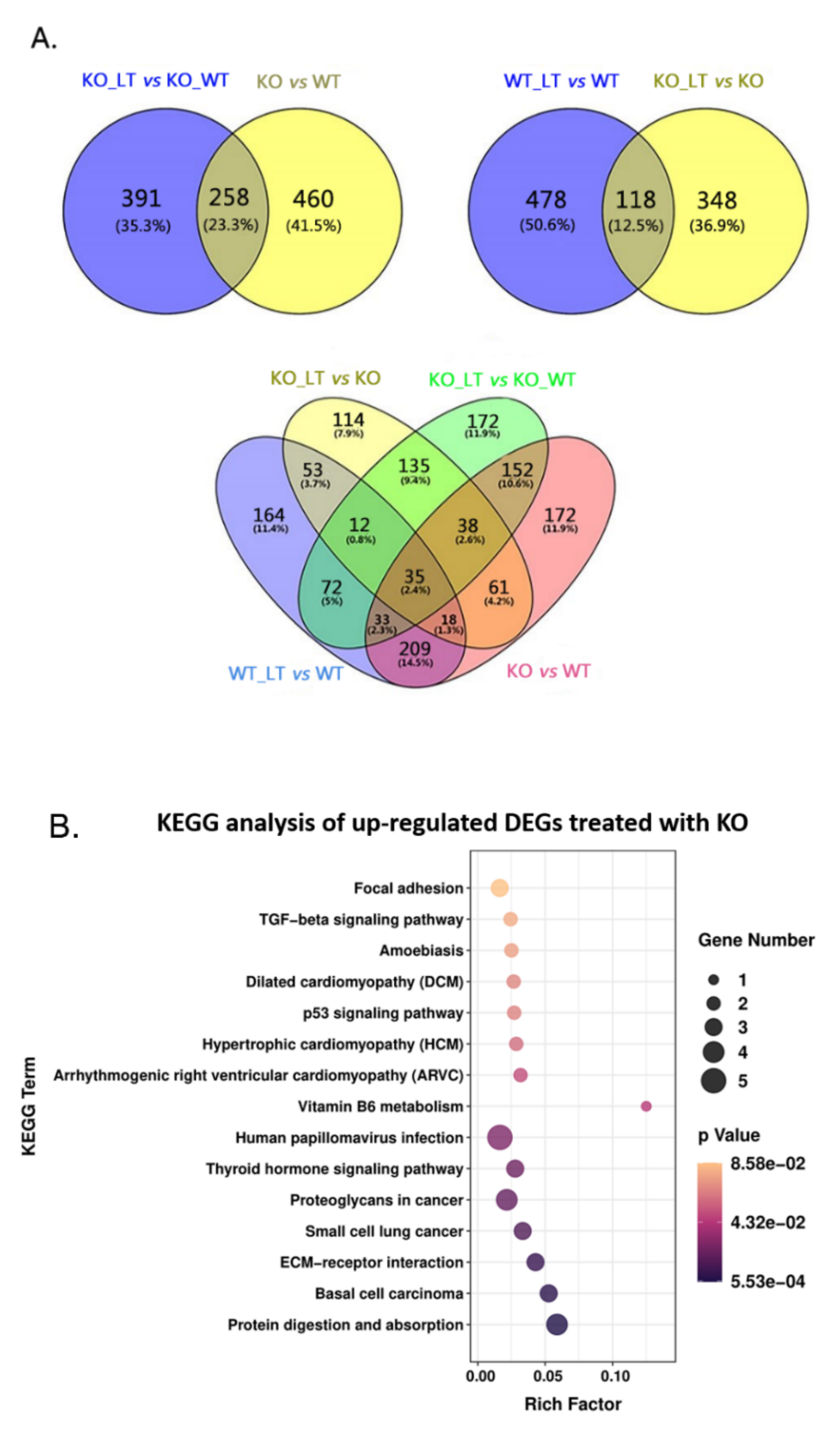
3.5. ZC3H10 Knockout in BFFs Dysregulated Pathways Associated with Thermogenesis and Immunity
3.6. Low Temperature-Activated Pathways Associated with Energy Metabolism and Thermogenesis Regulation
3.7. Low Temperature and Deletion of ZC3H10 Altered the Gene Expression of PPAR Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chebel, R.C.; Braga, F.A.; Dalton, J.C. Factors affecting reproductive performance of Holstein heifers. Anim. Reprod. Sci. 2007, 101, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Qanbari, S.; Pausch, H.; Jansen, S.; Somel, M.; Strom, T.M.; Fries, R.; Nielsen, R.; Simianer, H. Classic selective sweeps revealed by massive sequencing in cattle. PLoS Genet. 2014, 10, e1004148. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Fariello, M.I.; Boitard, S.; Naya, H.; SanCristobal, M.; Servin, B. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 2013, 193, 929–941. [Google Scholar] [CrossRef]
- Wang, X.; Ju, Z.; Jiang, Q.; Zhong, J.; Liu, C.; Wang, J.; Hoff, J.L.; Schnabel, R.D.; Zhao, H.; Gao, Y. Introgression, admixture, and selection facilitate genetic adaptation to high-altitude environments in cattle. Genomics 2021, 113, 1491–1503. [Google Scholar] [CrossRef]
- Audano, M.; Pedretti, S.; Ligorio, S.; Gualdrini, F.; Polletti, S.; Russo, M.; Ghisletti, S.; Bean, C.; Crestani, M.; Caruso, D. Zc3h10 regulates adipogenesis by controlling translation and F-actin/mitochondria interaction. J. Cell Biol. 2021, 220, e202003173. [Google Scholar] [CrossRef]
- Yi, D.; Dempersmier, J.M.; Nguyen, H.P.; Viscarra, J.A.; Dinh, J.; Tabuchi, C.; Wang, Y.; Sul, H.S. Zc3h10 acts as a transcription factor and is phosphorylated to activate the thermogenic program. Cell Rep. 2019, 29, 2621-2633.e4. [Google Scholar] [CrossRef]
- Audano, M.; Pedretti, S.; Cermenati, G.; Brioschi, E.; Diaferia, G.R.; Ghisletti, S.; Cuomo, A.; Bonaldi, T.; Salerno, F.; Mora, M. Zc3h10 is a novel mitochondrial regulator. EMBO Rep. 2018, 19, e45531. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, H.; Zheng, P.; Feng, R.; Wang, X.; Huang, F.; Ma, M.; Tian, Y.; Zhang, G. The alleviative effect of thyroid hormone on cold stress-induced apotosis via HSP70 and mitochondrial apoptosis signal pathway in bovine Sertoli cells. Cryobiology 2022, 105, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, C.; Sul, H.S. Signaling pathways regulating thermogenesis. Front. Endocrinol. 2021, 12, 595020. [Google Scholar] [CrossRef] [PubMed]
- Mariman, E.; Wang, P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010, 67, 1277–1292. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. P53 and the immune response: 40 years of exploration—A plan for the future. Int. J. Mol. Sci. 2020, 21, 541. [Google Scholar] [CrossRef]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M.F. Tumour necrosis factor signalling in health and disease. F1000Research 2019, 8, 111. [Google Scholar] [CrossRef]
- Paluch, E.K.; Aspalter, I.M.; Sixt, M. Focal Adhesion-Independent Cell Migration. Annu. Rev. Cell Dev. Biol. 2016, 32, 469–490. [Google Scholar] [CrossRef]
- Maekubo, H.; Moriya, K.; Hiroshige, T. Role of ketone bodies in nonshivering thermogenesis in cold-acclimated rats. J. Appl. Physiol. 1977, 42, 159–165. [Google Scholar] [CrossRef]
- Behrouz, V.; Yari, Z. A review on differential effects of dietary fatty acids on weight, appetite and energy expenditure. Crit. Rev. Food Sci. Nutr. 2022, 62, 2235–2249. [Google Scholar] [CrossRef]
- Mäkelä, A.M.; Hohtola, E.; Miinalainen, I.J.; Autio, J.A.; Schmitz, W.; Niemi, K.J.; Hiltunen, J.K.; Autio, K.J. Mitochondrial 2, 4-dienoyl-CoA reductase (Decr) deficiency and impairment of thermogenesis in mouse brown adipose tissue. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARs in health and disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, Y.; Yasuda, O.; Soejima, H.; Miyata, K.; Yamamoto, E.; Izumiya, Y.; Maeda, N.; Ohishi, M.; Rakugi, H.; Oike, Y. Tissue inhibitor of metalloproteinase-3 knockout mice exhibit enhanced energy expenditure through thermogenesis. PLoS ONE 2014, 9, e94930. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Z.; Jiang, S.; Zhang, L.; Yu, Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling. Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Michurina, S.; Stafeev, I.; Menshikov, M.; Parfyonova, Y.V. Mitochondrial dynamics keep balance of nutrient combustion in thermogenic adipocytes. Mitochondrion 2021, 59, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Yeung, Y.T.; Aziz, F.; Guerrero-Castilla, A.; Arguelles, S. Signaling pathways in inflammation and anti-inflammatory therapies. Curr. Pharm. Des. 2018, 24, 1449–1484. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Lian, S.; Guo, J.-R.; Wang, J.-F.; Zhang, L.-P.; Li, S.-Z.; Yang, H.-M. Activation of the MAPK signaling pathway induces upregulation of pro-apoptotic proteins in the hippocampi of cold stressed adolescent mice. Neurosci. Lett. 2019, 699, 97–102. [Google Scholar] [CrossRef]
- Lee, S.; Dong, H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017, 233, R67. [Google Scholar] [CrossRef]
- Ren, J.; Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Characterization of biological pathways regulating acute cold resistance of zebrafish. Int. J. Mol. Sci. 2021, 22, 3028. [Google Scholar] [CrossRef]
- Lee, J.-W.; Bae, S.-H.; Jeong, J.-W.; Kim, S.-H.; Kim, K.-W. Hypoxia-inducible factor (HIF-1) α: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12. [Google Scholar] [CrossRef]
- Rieder, C.L.; Cole, R.W. Cold-shock and the Mammalian cell cycle. Cell Cycle 2002, 1, 168–174. [Google Scholar] [CrossRef]
- Li, S.-T.; Dai, Q.; Zhang, S.-X.; Liu, Y.-J.; Yu, Q.-Q.; Tan, F.; Lu, S.-H.; Wang, Q.; Chen, J.-W.; Huang, H.-Q. Ulinastatin attenuates LPS-induced inflammation in mouse macrophage RAW264. 7 cells by inhibiting the JNK/NF-κB signaling pathway and activating the PI3K/Akt/Nrf2 pathway. Acta Pharmacol. Sin. 2018, 39, 1294–1304. [Google Scholar] [CrossRef]
- Liu, F.; Chu, T.; Wang, M.; Zhan, W.; Xie, Q.; Lou, B. Transcriptome analyses provide the first insight into the molecular basis of cold tolerance in Larimichthys polyactis. J. Comp. Physiol. B 2020, 190, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.S.; Pal, U.; Chowdhury, N.; Maiti, N.C.; Bagchi, A.; Swarnakar, S. Omeprazole prevents stress induced gastric ulcer by direct inhibition of MMP-2/TIMP-3 interactions. Free Radic. Biol. Med. 2022, 181, 221–234. [Google Scholar] [CrossRef]
- Kay, A.M.; Simpson, C.L.; Stewart, J.A. The role of AGE/RAGE signaling in diabetes-mediated vascular calcification. J. Diabetes Res. 2016, 2016, 6809703. [Google Scholar] [CrossRef] [PubMed]
- Guasch-Ferré, M.; Santos, J.L.; Martínez-González, M.A.; Clish, C.B.; Razquin, C.; Wang, D.; Liang, L.; Li, J.; Dennis, C.; Corella, D. Glycolysis/gluconeogenesis-and tricarboxylic acid cycle–related metabolites, Mediterranean diet, and type 2 diabetes. Am. J. Clin. Nutr. 2020, 111, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar]
- Li, H.; Zhang, X.; Huang, C.; Liu, H.; Zhang, Q.; Sun, Q.; Jia, Y.; Liu, S.; Dong, M.; Hou, M. FGF2 disruption enhances thermogenesis in brown and beige fat to protect against adiposity and hepatic steatosis. Mol. Metab. 2021, 54, 101358. [Google Scholar] [CrossRef] [PubMed]
- Graja, A.; Garcia-Carrizo, F.; Jank, A.M.; Gohlke, S.; Ambrosi, T.H.; Jonas, W.; Ussar, S.; Kern, M.; Schürmann, A.; Aleksandrova, K. Loss of periostin occurs in aging adipose tissue of mice and its genetic ablation impairs adipose tissue lipid metabolism. Aging Cell 2018, 17, e12810. [Google Scholar] [CrossRef] [PubMed]
- Koerner, C.M.; Roberts, B.S.; Neher, S.B. Endoplasmic reticulum quality control in lipoprotein metabolism. Mol. Cell. Endocrinol. 2019, 498, 110547. [Google Scholar] [CrossRef]
- Ngai, Y.F.; Quong, W.L.; Glier, M.B.; Glavas, M.M.; Babich, S.L.; Innis, S.M.; Kieffer, T.J.; Gibson, W.T. Ldlr−/− mice display decreased susceptibility to Western-type diet-induced obesity due to increased thermogenesis. Endocrinology 2010, 151, 5226–5236. [Google Scholar] [CrossRef]
- Liao, X.; Bao, H.; Meng, Y.; Plastow, G.; Moore, S.; Stothard, P. Sequence, structural and expression divergence of duplicate genes in the bovine genome. PLoS ONE 2014, 9, e102868. [Google Scholar] [CrossRef]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019, 4, eaau6085. [Google Scholar] [CrossRef]
- Xue, G.; Yin, J.; Zhao, N.; Liu, Y.; Fu, Y.; Zhang, R.; Bao, J.; Li, J. Intermittent mild cold stimulation improves the immunity and cold resistance of spleens in broilers. Poult. Sci. 2021, 100, 101492. [Google Scholar] [CrossRef]
- Jia, Z.; Chen, A.; Wang, C.; He, M.; Xu, J.; Fu, H.; Zhang, X.; Lv, W.; Guo, Z. Amelioration effects of Kaempferol on immune response following chronic intermittent cold-stress. Res. Vet. Sci. 2019, 125, 390–396. [Google Scholar] [CrossRef]
- Spinu, M.; Degen, A. Effect of cold stress on performance and immune responses of Bedouin and White Leghorn hens. Br. Poult. Sci. 1993, 34, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.C. Complement and humoral immunity. Vaccine 2008, 26, I28–I33. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Casolaro, V.; Bermúdez-Humarán, L.G.; Keyvani, H.; Taghinezhad-S, S. Modulation of the PI3K/Akt/mTOR signaling pathway by probiotics as a fruitful target for orchestrating the immune response. Gut Microbes 2021, 13, 1886844. [Google Scholar] [CrossRef]
- Wang, C.; Zeng, X.; Zhou, Z.; Zhao, J.; Pei, G. β-arrestin-1 contributes to brown fat function and directly interacts with PPARα and PPARγ. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Karavia, E.A.; Zvintzou, E.; Petropoulou, P.-I.; Xepapadaki, E.; Constantinou, C.; Kypreos, K.E. HDL quality and functionality: What can proteins and genes predict? Expert Rev. Cardiovasc. Ther. 2014, 12, 521–532. [Google Scholar] [CrossRef]
- Xepapadaki, E.; Nikdima, I.; Zvintzou, E.; Karavia, E.A.; Kypreos, K.E. Tissue-specific functional interaction between apolipoproteins A1 and E in cold-induced adipose organ mitochondrial energy metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158859. [Google Scholar] [CrossRef]
- Rao, R.; Albers, J.J.; Wolfbauer, G.; Pownall, H.J. Molecular and macromolecular specificity of human plasma phospholipid transfer protein. Biochemistry 1997, 36, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Sponton, C.H.; Hosono, T.; Taura, J.; Jedrychowski, M.P.; Yoneshiro, T.; Wang, Q.; Takahashi, M.; Matsui, Y.; Ikeda, K.; Oguri, Y. The regulation of glucose and lipid homeostasis via PLTP as a mediator of BAT–liver communication. EMBO Rep. 2020, 21, e49828. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Gao, Y.; Wang, J.; Huang, N.; Jiang, Q.; Ju, Z.; Yang, C.; Wei, X.; Xiao, Y.; Zhang, Y.; et al. Selection Signature and CRISPR/Cas9-Mediated Gene Knockout Analyses Reveal ZC3H10 Involved in Cold Adaptation in Chinese Native Cattle. Genes 2022, 13, 1910. https://doi.org/10.3390/genes13101910
Wang L, Gao Y, Wang J, Huang N, Jiang Q, Ju Z, Yang C, Wei X, Xiao Y, Zhang Y, et al. Selection Signature and CRISPR/Cas9-Mediated Gene Knockout Analyses Reveal ZC3H10 Involved in Cold Adaptation in Chinese Native Cattle. Genes. 2022; 13(10):1910. https://doi.org/10.3390/genes13101910
Chicago/Turabian StyleWang, Luyu, Yaping Gao, Jinpeng Wang, Ning Huang, Qiang Jiang, Zhihua Ju, Chunhong Yang, Xiaochao Wei, Yao Xiao, Yaran Zhang, and et al. 2022. "Selection Signature and CRISPR/Cas9-Mediated Gene Knockout Analyses Reveal ZC3H10 Involved in Cold Adaptation in Chinese Native Cattle" Genes 13, no. 10: 1910. https://doi.org/10.3390/genes13101910
APA StyleWang, L., Gao, Y., Wang, J., Huang, N., Jiang, Q., Ju, Z., Yang, C., Wei, X., Xiao, Y., Zhang, Y., Yang, L., & Huang, J. (2022). Selection Signature and CRISPR/Cas9-Mediated Gene Knockout Analyses Reveal ZC3H10 Involved in Cold Adaptation in Chinese Native Cattle. Genes, 13(10), 1910. https://doi.org/10.3390/genes13101910




