Abstract
Sida cordifolia is a medicinal shrub that is conventionally used in the Indian system of medicine;however, the genes contributing to its medicinal properties have been minimally explored, thus limiting its application. High-throughputsequencing and Liquid Chromatography with tandem mass spectrometry(LC-MS/MS) technologies were applied to unravel the medicinally important bioactive compounds. As a result, transcriptomic sequencing generated more than 12 GB of clean data, and 187,215 transcripts were obtained by de novoassembly. These transcripts were broadly classified into 20 classes, based on the gene ontology classification, and 6551 unigenes were annotated using Kyoto Encyclopedia of Genes and Genomes (KEGG) database with more than 142 unigenes involved in the biosynthesis of secondary metabolites. LC-MS/MS analysis of three tissues of Sida cordifolia revealed that acacetin and procyanidin are some important metabolites identified thatcontribute to its medicinal value. Several key enzymes witha crucial role in phenylpropanoid and flavonoid biosynthetic pathways were identified, especially phenylalanine ammonia lyase, which might be an important rate-limiting enzyme. Real-Time Quantitative Reverse Transcription Polymerase chain reaction (qRT-PCR) analysis revealed enzymes, such as Phenylalanine ammonia lyase (PAL), Cinnamyl alcohol dehydrogenase 1 (CAD), Cinnamoyl-CoA reductase 1 (CF1) and Trans cinnamate 4-monooxygenase(TCM), which were predominantly expressed in root compared to leaf and stem tissue. The study provides a speculative insight for the screening of active metabolites and metabolic engineering in Sida cordifolia.
1. Introduction
Plants have been used as an immortal source of medicine forages. Globally, herbal medicines have played an important role in human health to treat chronic and acute conditions without any toxic effect. They are usually used as therapeutics to health conditions including diabetes mellitus, wounds, cancer, heart diseases, tuberculosis, hypertension, etc. These herbs are used in medicinal fields, as they are rich in bioactive phytocompounds, such as flavonoids, alkaloids, terpenoids, polyphenols, and tannins possessing various pharmacological properties [1]. A traditional sub shrub, Sida cordifolia, belonging to the family Malvaceae, is widely spread across countries like India, Brazil, and Africa and is extensively used in the ayurvedic system of medicine. It has laid the foundation to perform the transcriptomic approach to elucidate the various biosynthetic pathways responsible for its medicinal properties [2]. Some important constituents isolated from various extracts of Sida cordifolia are 1, 2, 3, 9-Terta hydropyrrolo[21-b]-quinazolin-3-yl-amine, ephedrine, vasicine, pseudo-ephedrine, vasicinone, hypaphorine, vasicinol, stigmasterol, and sterculic acid [3,4,5,6,7]. Pharmacological properties exhibited by different extracts of Sida cordifolia are antimicrobial, anti-inflammatory, analgesic, anti-ulcer, nephroprotective, anti-diabetic, hepatoprotective, anticancer, and central nervous system depressant activity [8]. The inevitable identification of genes responsible for the production of various secondary metabolites by S.cordifolia is achieved using the next-generation sequencing technology to unravel the novel transcripts from medicinal plants with respect to secondary metabolite biosynthesis and gene expression analysis [9]. The technology is employed to generate functional data for non-model plants and EST sequences for the annotation of genes and analysis of targeted discovery. The de novo sequencing prompts large volumes of information that can be used to identify various molecular markers, novel gene identification, or discovery and polymorphism [10,11,12,13]. Transcriptomics is the study of collection of all the transcripts available in the specific tissue andhas been used to study the structure and function of the gene at the molecular levels.The study of small molecular weight metabolites that exist in all cells to maintain their growth and function is referred to as metabolomics. The metabolite profiling reflects the overall biochemical and physiological conditions of the plant for the survival in that particular environment [14]. LC-MS analysis is preferable for evaluating large groups of secondary metabolites like phenolic compounds, flavonoids, and alkaloids [14]. Though the plant has been extensively used in herbal formulations, the biosynthetic pathways responsible for the synthesis of secondary metabolites that possess the pharmacological properties have yet to be unveiled. Thus, in this study, we aimed to annotate the transcripts, identify the putative transcripts that are involved in the major biosynthetic pathways attributing to the medicinal properties of the plant using transcriptomics, and analyze the different metabolites present in them by metabolomics.
2. Materials and Methods
2.1. Sample Collection and RNA Extraction
Root, stem, and leaf tissues were collected from a healthy plant near Potheri in Tamil Nadu, and identification was taxonomically done in SRMIST, IIISM. The RNA was extracted using TRizol®Reagent followed by phase-separation using Chloroform. After centrifugation at 12,000 rpm for 15 min at 4 °C. To the aqueous phase, 0.7 volume of isopropanol was added and centrifuged at 12,000 rpm for 10 min at 4 °C, followed by ethanol, which was to the pellet obtained from the previous step. Then, nuclease-free water was addedto the air-dried pellet. To remove the DNA contamination, the RNA was treated with DNase. A further purification of the total RNA was done using Qiagen RNeasy®MinElute clean-up kit. The integrity of RNA was analyzed using the Agilent Bioanalyzer with RIN value range of 7.4 to 9.2, a Thermo Scientific Nanodrop Lite spectrophotometer was used to check the quality of RNA, and the samples were visualized in the agarose gel electrophoresis [15,16].
2.2. RNA Library Preparation and Illumina Sequencing
Purified RNA was used for cDNA preparation. The poly A tail present in the mRNA was refined from the total RNA using magnetic beads attached to poly-T oligo, and a fragmentation buffer was used for fragmenting the mRNAs into smaller fragments. The first strand synthesis of these shorter fragments was carried out using Invitrogen’s Superscript II reverse transcriptase, and for the second strand synthesis, RNase H and DNA polymerase I were used. Single addition of dATP was done to the fragmented cDNA and then connected with adapters followed by a further selection of templates for PCR amplification. NextSeq 500 was used for sequencing, and the raw data in the FastQ format were obtained (Illumina Inc., San Diego, CA, USA) [15,16].
2.3. Assembly of Transcripts
The raw data obtained from sequencing were subjected to quality control and reads with (Phred score ≥ 30), were removed by the software FastQCv.0.11.9, and the adapter sequences were trimmed by Cutadapt software. TRINITY v2.14.0 de novo assembler was used for the re-construction of transcriptome data with three software modules: the Inchworm module assembles the transcripts by k-mer; the Chrysalis module clusters the contigs, thereby constructing a de Bruijn graph for the contigs; and the Butterfly module analyzes the contigs based on the graphs created and lists the isoforms. CD-HIT v4.8.1 program was used to reduce the sequence redundancy and increase the analysis performance [17,18].
2.4. Gene Ontology Classification and Functional Annotation
The de novo assembled transcripts of Sida cordifolia were collated against different databases including protein non-redundant database (nr) from NCBI with the BLASTX tool, and an E-value not greater than 1E-05 was considered as a significant match. Further Omics Box tools were used for gene ontology (GO) classification and annotation with enzyme codes (EC). The pathway mapping was done by KEGG Automated Annotation Server for retrieving the pathway maps of Kyoto Encyclopedia of Genes and Genomes (KEGG) [19].
2.5. SSR Identification
The microsatellites were identified using the Krait software, which offers an ultrafast and user-friendly graphic interface for investigating genome-wide microsatellites. The software also identifies VNTRS from a large genome size, locates the SSRs in the gene coding region, and statistically analyzes and plots the graphs [20]. Simple sequence repeats are short repeat DNA sequences with 1–6 bp in length, which have high polymorphism and can be used as a tool in genetic mapping, population genetics, and phylogenetic analysis [21].
2.6. Transcript Quantification
The SALMON quantification tool was used to quantify the transcript abundance from the sequenced data andcombines the dual-phase parallel interface algorithm. The tool quantifies the transcripts based on the GC content, quasi mapping was conducted for accuracy, and fast mapping of genes was conducted to study the expression levels [22].
2.7. Reverse Transcription PCR Validation
The validation assembled transcriptome data of Sida cordifolia from the root tissue by reverse transcription PCR was carried out. The full-length transcripts were found using the BLAST tool to find the complete transcript to design the primers. Then, the PRIMER BLAST tool from NCBI was used to design the primers, and in silico PCR was conducted, where Actin was considered as a housekeeping gene. The gene expression analysis was normalized using reverse transcription PCR, and gradient PCR was conductedto optimize the annealing temperatures from 52 °C to 60 °C. PCR amplification was carried out by the following parameters: 95 °C for 5 min, 95 °C for 30 s, 57–59 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min for 40 cycles.
2.8. Validation of Gene Expression Analysis
S.cordifolia transcriptome data were validated by quantitative real-time PCR with QuantStudio 5 (Thermo Scientific, Wilmington, DE, USA) PCR machine and the QuantiNovaSYBRGreen PCR (Qiagen Inc., GmbH, Germany) kit. Actin was used as an internal reference, and a negative control reaction was set up in all experiments where it was used. The relative gene expression levels were analyzed and calculated using the2-ΔΔCt method, which represents the cycle threshold of the target gene with the housekeeping gene Actin. qRT-PCR analysis was carried out with three replicates [23].
2.9. Extraction of Metabolites from Sida cordifolia
The leaf, root, and stem tissues of Sida cordifolia were collected, dried using a microwave oven, and pulverized using an electric blender. Then, 10 g of powdered plant tissues were transferred to a Schott bottle, and 100 mL of 99.9% methanol was added to it and macerated for three days. The solvent was evaporated using a rotary evaporator. The samples were centrifuged at 3000 rpm, and the supernatant was transferred into a fresh tube and stored at room temperature until further analysis.
2.10. LC-MS/MS Analysis
The secondary metabolites were analyzed using Shimadzu LC-MS/MS-8040 (QQQ) (liquid chromatography–mass spectrometry, Triple Quadrupole, North America) with a scan range of 50–1000 m/z in both positive and negative modes. Mobile phase A: 5 mM ammonium formateand 0.1% formic acid in water; phase B: 5 mM ammonium formateand 0.1% formic acid in methanol with a flow rate 0.6 ml/min an isocratic A—20% and B—80% were used. The column Union was used at a temperature 40 °C with a flow rate of 500 μL/min. The data acquisition was done by the software LabSolutions™ LCMS, and the compounds were identified from free-source online tools, such as METLIN, PUBCHEM, and KEGG databases, along with some previously published articles.
3. Results
3.1. RNA Sequencing Analysis of Raw and Processed Data
The root transcriptome of Sida cordifolia was obtained with 59,484,771 raw reads. From the raw reads, adapter sequences were trimmed to reduce the redundancy, and 59,484,597 clean reads were obtained further with a GC content of 40.52%. The raw reads were submitted to NCBI Sequence Read Achieve (SRA) and an accession number PRJNA841821 was obtained. Overall, 187,215 transcripts were assembled using a de novo assembler TRINITY.The average length of bases was 1035.19, and the median contig length was 717. The contig N50 value was found to be 1622 (Table 1).

Table 1.
Summary of Illumina paired-end sequencing and de novo assembly of Sida cordifolia.
3.2. Functional Annotation of Unigenes
A similarity search using BLASTX was conductedagainst the nr database from NCBI; with a total of 187,215 unigenes assembled, a total of 54,375 unigenes were non-annotated and 132,840 unigenes were annotated (Figure 1). Due to inadequate information of the Sida cordifolia genome, some annotated genes were classified into predicted, uncharacterized, or hypothetical proteins.The results from BLASTX were transferred to OMICS BOX to annotate further. The similarity search of assembled transcripts showed high similarity with Gossypium hirsutum, Gossypium raimondii, Gossypium arboretum, Duriozibethinus, Theobroma cacao, Herrania umbratica, and Quercus suber and the least similarity with Stipa magnifica and Stipa borysthenica (Supplementary Figure S1).
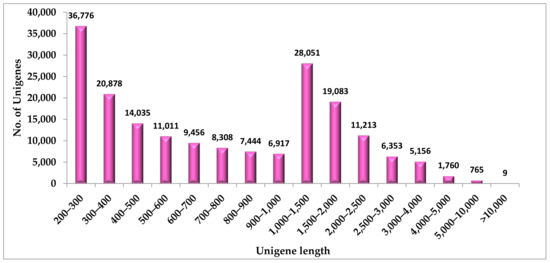
Figure 1.
Unigene length distribution of S.cordifolia root transcriptome.
3.3. Functional Classification of Unigenes
Using the OMICS BOX software, the assembled unigenes were annotated onto GO classification into three different categories viz. molecular function, cellular process, and biological process.The unigenes were categorized into 60 subcategories. In the cellular component, 9467 unigenes were classified into 20 classes. This category includes intracellular anatomical structure with high unigenes in the organelle, cytoplasm, and membrane and the leastunigenes in the nucleoplasm, supramolecular complex, and external encapsulating structure. In the molecular function, a total of 19,593 unigenes were classified into 20 different classes with high unigenes in organic cyclic compound binding, heterocyclic compound binding, and ion binding and low unigenesin isomerase activity, protein-containing complex binding, and carbohydrate binding. In the biological process, 25,315 unigenes were grouped into 20 classes with the maximum number of unigenes in organic substance metabolic process, cellular metabolic process, and primary metabolic process and the minimum number in signal transduction, response to chemicals, and vesicle-mediated transport (Supplementary Figure S2).
3.4. Biological Pathway Analysis
In total, 6551 unigenes were annotated using the KEGG database and assigned to 150 pathway maps. A total of 6409 unigenes were found to be annotated in metabolic pathways onto different categories, which include nucleotide metabolism (2539 unigenes), carbohydrate metabolism (347 unigenes), metabolism of co-factors and vitamins (1333 unigenes), energy metabolism (823 unigenes), biosynthesis of antibiotics (413 unigenes), xenobiotics biodegradation and metabolism (135 unigenes), lipid metabolism (267 unigenes), and amino acid metabolism (477 unigenes) (Supplementary Figure S3). Further, 142 unigenes were classified into biosynthesis of secondary metabolites, which was further subdivided into different categories: sesquiterpenoid and triterpenoid biosynthesis (57 unigenes), terpenoid backbone biosynthesis (22 unigenes), ubiquinone and other terpenoid-quinone biosynthesis (26 unigenes), phenylpropanoid biosynthesis (11 unigenes), isoquinoline alkaloid biosynthesis (8 unigenes), and flavonoid biosynthesis (6 unigenes) (Figure 2).
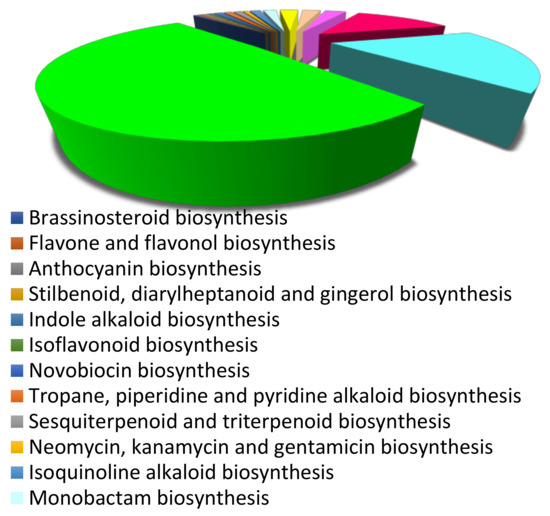
Figure 2.
KEGG pathway analysis based on secondary metabolite biosynthesis in root transcriptome of Sida cordifolia.
3.5. Untargeted Metabolic Profiling of Sida cordifolia
Transcriptomic analysis of metabolic pathways and the validation of key metabolites require further confirmation by the identification of metabolites. The metabolomic analysis of samples was completed by the untargeted metabolomic method. There were a total of 298 different metabolites in leaf, stem, and root tissues of S.cordifolia in both the positive and negative mode. Flavonoids were the major group of secondary metabolites found in the metabolome analysis of S.cordifolia where Naringin, Cinnamic acid, Cinnamaldehyde, Caffeic acid, Kaempferol derivatives, and Quercetin were predominantly present in root, stem, and leaf tissues. Some tissue-specific metabolites, such as Rosmarinic acid, Caffeic acid and its derivatives, Kaempferol derivatives, Rutin, Quercitrin, and Gallic acid were identified in root tissue. Apigenin, Gallagic acid, Quercetin and its derivates, Caffeic acid and its derivatives, and Kaempferol and its derivatives were found in leaf tissue. In stem tissue, Ferulic acid, Gallic acid, Quinic acid derivative, and Malic acid were found (Table 2). In some metabolic pathways, such as, flavonoid biosynthesis, isoflavonoid biosynthesis, flavones and flavonol biosynthesis, phenylalanine, tyrosine and tryptophan biosynthesis, and ubiquonone biosynthesis, intermediate metabolites could be identified in the mass spectra. The results revealed that the transcriptomic and metabolic analysis of the transcripts that catalyzes the enzymes is consistent with their metabolites that belong to the pathway.

Table 2.
Identification of various secondary metabolites from root, stem, and leaves tissue of Sida cordifolia by LC-MS/MS.
3.6. KEGG Enrichment Analysis of Transcripts Involved in Various Secondary Metabolite Biosynthesis
3.6.1. TranscriptsInvolvedin Phenylpropanoid Biosynthesis Pathway
The compounds derived from this pathway are initiated by the amino acid phenylalanine which serves as a precursor. Precursor phenylalanine, which is converted into Cinnamoyl-CoA, p-Coumaryl-CoA, p-Coumarylquinic acid, Caffeoylquinic acid, Caffeoyl-CoA, Feruloyl-CoA and Sinapoyl-CA. Eleven genes were intricate in the biosynthesis pathway. The root transcriptome analysis revealed the presence of the genes Phenylalanine ammonia lyase (EC: 4.3.1.24, EC: 4.3.1.25), 4-Coumarate CoA ligase (EC: 6.2.1.12), trans-cinnamate 4-monooxygenase (EC: 1.14.14.91), Cinnamoyl-CoA reductase 1 (EC: 1.2.1.44), and Cinnamyl alcohol dehydrogenase (EC: 1.1.1.195) (Supplementary Figure S4).
3.6.2. Transcripts Involved in Flavonoid Biosynthesis Pathway
The precursor for flavonoid biosynthesis is phenylalanine. The pathway is elucidated by the conversion of phenylalanine to p-Coumaryl-CoA, and the rate-limiting enzyme is Chalcone synthase. The genes identified from the transcriptome data are trans-cinnamate 4-monooxygenase (EC: 1.14.14.91), Chalcone-flavonone isomerase (EC: 5.5.1.6), and naringenin, 2-oxoglutarate 3-dioxygenase (EC: 1.14.11.9) (Supplementary Figure S5).
3.7. SSR Identification
A total of 187,215 transcripts were analyzed, and 36197 perfect SSRs were identified with a total length of 626,976 bp, and the average length of SSRs was 17.33. Relative abundance and density were found to be 186.77 and 3235.12, respectively. The most abundant repeat motifswere mono nucleotide (12,898;35.6%), followed by tri-nucleotide (12,732;35.2%), di-nucleotide (6868;19%), tetra-nucleotide (2526;6.98%), penta-nucleotide (653;1.8%), and hexa-nucleotide (520;1.44%). SSRs with 5tandem repeats were most common in Sida cordifolia, followed by 12 repeats and 7tandem repeats. Among mono-nucleotide repeat motifs, Awashighly abundant (35%).In tri-nucleotide repeats, the highest frequency was observed in AAG (11.51%) followed by AAT (4.36%) (Supplementary Figure S6)
3.8. Gene Expression Analysis of Sida cordifolia
The qRT-PCR analysis was done to validate the transcriptome date and the expression analysis of the selected genes. The selected transcripts included Phenylalanine ammonia lyase (EC: 4.3.1.24, 4.3.1.25), Trans-cinnamate4-monooxygenase (EC: 1.14.14.91), Chalcone-flavonone isomerase (EC: 5.5.1.6), and Cinnamyl alcohol dehydrogenase 1(EC: 1.1.1.219) (Table 3). The primer sequences used for the analysis are depicted in the Supplementary Table S1. The analysis revealed varied expression patterns of the selected genes, where all the genes were upregulated in the root tissue rather than the stem and leaf. Actin was used as a housekeeping gene. The results obtained showed significant agreement with the transcriptome data (Figure 3).

Table 3.
Major genes involved in phenylpropanoid and flavonoid biosynthesis pathways identified from Sida cordifolia root transcriptome.
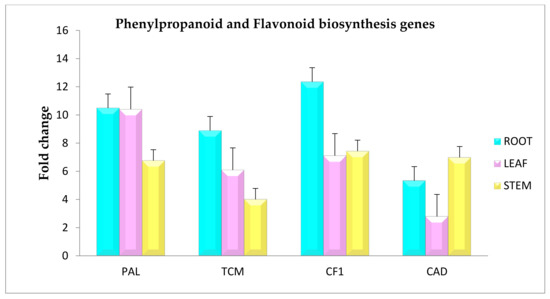
Figure 3.
Validation of gene expression of phenylpropanoid and flavonoid biosynthesis genes by real-time PCR from Sida cordifolia.
4. Discussion
4.1. De Novo Assembly, Functional Classification, and Annotation
RNA sequencing is a high-throughput sequencing method thathas been an integral part of metabolome research in non-model species with relatively high rapidity. This sequencing technology is effective to study the annotation and to elucidate various transcripts responsible for the biosynthetic pathways, gene expression analysis, and distribution of secondary metabolites in different tissues [52].The Illumina sequencing of the root tissue of Sida cordifolia revealed 59,484,771 reads, which were further processed to remove the adapter and redundant sequences. The de novo assembler TRINITY was used to assemble the transcripts, and a total of 187,215 numbers of transcripts were identified.
Functional annotation and classification comprise a vital step thatprovides us with the homology alignment. Gene ontology classification was done to annotate the transcripts into three major categories such as cellular function, molecular function, and biological process. In our study, out of 187,215 unigenes, a total of 54,375 unigenes were non-annotated, and 132,840 unigenes were annotated against different databases. Sida cordifolia showed high similarity with the genus Gossypium and the least similarity with Stipa genus. These results can be because of the scarcity of reference genomic resources for Sida genus.
4.2. KEGG Pathway Analysis and Identification of Candidate Genes Involved in Secondary Metabolite Biosynthesis
Biological pathway analysis against the KEGG database was conductedto identify the various pathways involved in the transcriptome data with a total of 150 pathway maps, out of which a higher number of transcripts were annotated into metabolism pathway than the secondary metabolite biosynthesis pathway maps. The phenylpropanoid biosynthesis pathway is the key biosynthetic pathway responsible for the production of various secondary metabolites. The pathway is initiated by catalyzing phenylalanine to cinnamate and ammonia by the enzyme Phenylalanine ammonialyase (PAL). PAL is the key regulatory enzyme involved in the pathway that is found ubiquitously in plants [53]. The enzyme serves as a precursor for the flavonoid and lignin biosynthetic pathways [54]. A total of 12 transcripts were identified that are responsible for synthesis of PAL gene. Trans-cinnamic acid is also a precursor for the flavonoid and biosynthetic pathways. The increased activity of the enzyme PAL in turn increases the production of phenylpropanoid products, which vary with different stress stimuli, developmental stage, and tissue and cell differentiation. The enzyme is delineated to be stimulated by infection, radiations, drastic change in temperature, or drought stress [55,56,57,58]. Cinnamoyl-CoA reductase 1 involved in the monolignol pathway engenders the conversion of p-coumaroyl-, feuloyl-, and sinapoyl-CoA to p-coumaraldehyde, coniferaldehyde, and sinapaldehyde [59,60].
Flavonoids are naturally occurring secondary metabolites that are rich in antioxidant, antibacterial, anti-inflammatory, antifungal, and antidiabetic activity and act as an anticancer agent. On the basis of a number of hydroxyl groups, flavonoids are grouped into flavones, flavonols, flavones, anthocyanidins, and isolflavones. Flavonoids are synthesized from the phenylpropanoid biosynthesis pathway with the key enzymes involved in them. The enzymes that are involved in the first step of flavonoids biosynthesis are 4-Coumaroyl-CoA ligase, 4-Coumarate-3-Hydroxylase, and Phenylalanine ammonialyase. The genes involved in flavonoid biosynthesis pathway are flavone synthase, Dihydroflavonol-4-Reductase, Chalcone synthase, and Chalcone isomerase [61], wherethe enzyme 4-coumarate-CoA ligase catalyzes the activation of 4-coumarate, which occurs in multiple isoenzymes forms and exhibits substrate affinity with the metabolic function. It has a pivotal role in the biosynthesis of secondary metabolites from general phenylpropanoid metabolism. Other secondary metabolite biosynthesis pathways identified are ubiquinone, triterpenoid, sesquiterpenoid and isoquinoline alkaloid biosynthesis pathways.
A flavonoid—quercetin—and its derivatives are extensively used due to their bioactive effects as they possess many pharmacological properties such as anti-arthritic, cardiovascular, anticancer, anti-Alzheimer’s, antimicrobial, and wound-healing effects[62,63,64]. Studies suggest that flavonoids are found to be potential inhibitors of coronaviruses against the SARS-CoV-2 infection by binding to the targets that promotes the replication and entry of viruses [65,66]. The KEGG pathway analysis revealed that the flavonoids and its derivatives are derived from the phenylpropanoid biosynthesis pathway from L-tyrosine with intermediate metabolites, such as Naringenin and Kaempferol. Phenolic compounds function by reducing the reactive oxygen species and stabbing the lipid peroxidation, thus acting as a potent antioxidant agent. They also play a vital role in prevention and treatment of chronic illnesses like neurodegenerative disorders. The integrated LC-MS/MS and transcriptomic analysis revealed that acacetin and procyanidin metabolites were some important secondary metabolites that could be responsible for the pharmacological properties exhibited by the plant, which is used to treat various inflammations and bronchodilator activity and has a cardiovascular protecting nature.
4.3. Simple Sequence Repeats Analysis
Molecular markers reveal the genetic relationship between the species as they are unaffected by the environment and have easy detection and high heritability [67,68]. These markers are widely used in plant improvement and genetic conservation [69]. A total of 36,197 SSRs were identified with highest number of mononucleotide motifs compared to di-, tri-, tetra-, penta-, and hexa-nucleotide motifs. SSRs with five tandem repeats were most common in Sida cordifolia with highly abundant A (35%) mono-nucleotide repeats and in tri-nucleotide repeats, and the highest frequency was observed in AAG (11.51%) followed by AAT (4.36%). The SSRs were discovered to help plant genetics for molecular reproduction as well as identification of candidate molecular markers for understanding the genetic variations in the Sida family.
5. Conclusions
To expedite molecular level studies in Sida cordifolia and the characterization of the root transcriptome to identify the unitranscripts responsible for the biosynthesis of secondary metabolites, since the root tissue of the plant contributesto several medicinal properties of the plant. The result suggests that flavonoids are the major secondary metabolites that are responsible for the medicinal properties exhibited by the plant. The assembled transcripts were validated using reverse transcription PCR method, and the expression levels of the key genes from the phenylpropanoid and flavonoid pathways wereobtainedusing qRT-PCR. The secondary metabolites identified from the metabolomic studies lead the way for understanding the molecular mechanism of pharmacological properties exhibited by S.cordifolia. This is the first study that integrates the molecular basis and metabolomics of this plant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13101909/s1, Figure S1: Top hit species distribution of Sida cordifolia. Figure S2: Gene ontology classification ofSida cordifolia. Figure S3: KEGG pathway analysis based on the metabolism pathways in Sida cordifolia. Figure S4: KEGG pathway analysis of phenylpropanoid biosynthesis. Figure S5: KEGG pathway analysis of flavonoid biosynthesis. Figure S6: SSR identification from root transcriptome ofSida cordifolia. Table S1: List of genes selected to validate gene expression and their primer sequences.
Author Contributions
Conceptualization, writing—review and editing, supervision, S.P.; software, P.N.; methodology, validation, writing—original draft preparation, D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used in the study was deposited in the SRA under the SRA accession number PRJNA841821.
Acknowledgments
All authors acknowledge the RNA-Seq support by SRM-DBT platform and High-Performance Computing Cluster Facility (Genome Server) at SRM Institute of Science and Technology (SRMIST).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prasathkumar, M.; Anisha, S.; Dhrisya, C.; Becky, R.; Sadhasivam, S. Therapeutic and Pharmacological Efficacy of Selective Indian Medicinal Plants—A Review. Phytomed. Plus 2021, 1, 100029. [Google Scholar] [CrossRef]
- Ahmed, H.; Juraimi, A.S.; Swamy, M.K.; Ahmad-Hamdani, M.S.; Omar, D.; Rafii, M.Y.; Sinniah, U.R.; Akhtar, M.S. Botany, Chemistry, and Pharmaceutical Significance of Sida cordifolia: A Traditional Medicinal Plant. In Anticancer Plants: Properties and Application; Springer: Singapore, 2018; Volume 1, pp. 517–537. [Google Scholar]
- Ghosal, S.; Chauhan, R.B.P.; Mehta, R. Alkaloids of Sida Cordifolia. Phytochemistry 1975, 14, 830–832. [Google Scholar] [CrossRef]
- Ghosh, S.; Dutta, A. Chemical examination of Sida cordifolia Linn. J. Indian Chem. Soc. 1930, 7, 825–829. [Google Scholar]
- Tamura, S.; Kaneko, M.; Shiomi, A.; Yang, G.M.; Yamaura, T.; Murakami, N. Unprecedented NES Non-Antagonistic Inhibitor for Nuclear Export of Rev from Sida Cordifolia. Bioorg. Med. Chem. Lett. 2010, 20, 1837–1839. [Google Scholar] [CrossRef]
- Aminah, N.S.; Laili, E.R.; Rafi, M.; Rochman, A.; Insanu, M.; Tun, K.N.W. Secondary metabolite compounds from Sida genus and their bioactivity. Heliyon 2021, 7, 2405–8440. [Google Scholar] [CrossRef]
- Momin, M.A.; Bellah, S.F.; Rahman, S.M.; Rahman, A.A.; Murshid, G.M.; Emran, T.B. Phytopharmacological evaluation of ethanol extract of Sida cordifolia L. roots. Asian Pac. J. Trop. Biomed. 2014, 4, 18–24. [Google Scholar] [CrossRef]
- Silva, R.L.; Melo, G.B.D.; Melo, V.A.D.; Antoniolli, Â.R.; Michellone, P.R.T.; Zucoloto, S.; Castro e Silva, O.D. Effect of the aqueous extract of Sida cordifolia on liver regeneration after partial hepatectomy. Acta Cir. Bras. 2006, 21, 37–39. [Google Scholar] [CrossRef]
- Köse, M.A.; Çetinsağ, N.; Gürcan, K. De Novo Transcriptome Assembly and SSR Marker Development in Apricot (Prunus Armeniaca). Turk. J. Agric. For. 2017, 41, 305–315. [Google Scholar] [CrossRef]
- Fan, J.; Lou, Y.; Shi, H.; Chen, L.; Cao, L. Transcriptomic Analysis of Dark-Induced Senescence in Bermudagrass (CynodonDactylon). Plants 2019, 8, 614. [Google Scholar] [CrossRef]
- Wu, T.; Luo, S.; Wang, R.; Zhong, Y.; Xu, X.; Lin, Y.; He, X.; Sun, B.; Huang, H. The First Illumina-Based de Novo Transcriptome Sequencing and Analysis of Pumpkin (Cucurbita MoschataDuch.) and SSR Marker Development. Mol. Breed 2014, 34, 1437–1447. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Yang, J.; Hu, H.; Wei, G.; Cui, J.; Xu, J. Transcriptome and Metabolome Analyses Reveal Differences in Terpenoid and Flavonoid Biosynthesis in Cryptomeria fortunei Needles Across Different Seasons. Front. Plant Sci. 2022, 13, 862746. [Google Scholar] [CrossRef]
- Fang, H.; Qi, X.; Li, Y.; Yu, X.; Xu, D.; Liang, C.; Li, W.; Liu, X. De Novo Transcriptomic Analysis of Light-Induced Flavonoid Pathway, Transcription Factors in the Flower Buds of. Trees 2020, 34, 267–283. [Google Scholar] [CrossRef]
- Khan, I.; Rahman, H.; Abd El-Salam, N.M.; Tawab, A.; Hussain, A.; Khan, T.A.; Khan, U.A.; Qasim, M.; Adnan, M.; Azizullah, A.; et al. Punica granatum peel extracts: HPLC fractionation and LC MS analysis to quest compounds having activity against multidrug resistant bacteria. BMC Complement Altern. Med. 2017, 17, 247. [Google Scholar] [CrossRef]
- Lateef, A.; Prabhudas, S.K.; Natarajan, P. RNA Sequencing and de Novo Assembly of Solanum Trilobatum Leaf Transcriptome to Identify Putative Transcripts for Major Metabolic Pathways. Sci. Rep. 2018, 8, 15375. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ files (Version 1.33) [Software]. 2011. Available online: https://github.com/najoshi/sickle (accessed on 23 September 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioin formatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.-H.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.-D. Prediction and Analysis of Essential Genes Using the Enrichments of Gene Ontology and KEGG Pathways. PLoS ONE 2017, 12, e0184129. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B.; Hancock, J. Krait: An Ultrafast Tool for Genome-Wide Survey of Microsatellites and Primer Design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Song, X.; Yang, Q.; Bai, Y.; Gong, K.; Wu, T.; Yu, T.; Pei, Q.; Duan, W.; Huang, Z.; Wang, Z.; et al. Comprehensive analysis of SSRs and database construction using all complete gene-coding sequences in major horticultural and representative plants. Hortic Res. 2021, 8, 122. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2 (– delta delta CT) methods for quantitative real-time poly merase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Zhang, J.; Chen, W.; Ju, W.; Liu, F.; Tan, H. Determination of plasma concentration of cinnamic acid by LC-MS-MS and study of pharmacokinetics in healthy volunteers after mailuoning injection. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J. Chin. Mater. Med. 2010, 35, 1887–1889. [Google Scholar]
- Romero, C.; Brenes, M.; García, P.; Garrido, A. Hydroxytyrosol 4-beta-D-glucoside, an important phenolic compound in olive fruits and derived products. J. Agric. Food Chem. 2002, 50, 3835–3839. [Google Scholar] [CrossRef]
- Al Kadhi., O.; Melchini, A.; Mithen, R.; Saha, S. Development of a LC-MS/MS Method for the Simultaneous Detection of Tricarboxylic Acid Cycle Intermediates in a Range of Biological Matrices. J. Anal. Methods Chem. 2017, 2017, 5391832. [Google Scholar] [CrossRef]
- Li, Y.; Guang, C.; Zhao, N.; Feng, X.; Qiu, F. LC–MS/MS Method for Simultaneous Determination of Linarin and Its Metabolites in Rat Plasma and Liver Tissue Samples: Application to Pharmacokinetic and Liver Tissue Distribution Study After Oral Administration of Linarin. Molecules 2019, 24, 3342. [Google Scholar] [CrossRef]
- Solomon, K.S.; Amoah.; Maique, W.; Biavatti, A. LC-MS validated method for quantification of rosmarinic acid and sesquiterpene lactones in Hedyosmum brasiliense. J. Chromatogr. Sci. 2018, 56, 812–818. [Google Scholar]
- Cheruvu, H.S.; Yadav, N.K.; Valicherla, G.R.; Arya, R.K.; Hussain, Z.; Sharma, C.; Arya, K.R.; Singh, R.K.; Datta, D.; Gayen, J.R. LC-MS/MS method for the simultaneous quantification of luteolin, wedelolactone and apigenin in mice plasma using hansen solubility parameters for liquid-liquid extraction: Application to pharmacokinetics of Eclipta alba chloroform fraction. J. Chromatogr. B Analyt. Technol. Biomed Life Sci. 2018, 1801, 76–86. [Google Scholar] [CrossRef]
- Espíndola, K.M.M.; Ferreira, R.G.; Narvaez, L.E.M.; Rosario, A.C.R.S.; de Silva, A.H.M.; Silva, A.G.B.; Vieira, A.P.O.; Monteiro, M.C. Chemical and Pharmacological Aspects of Caffeic Acid and Its Activity in Hepatocarcinoma. Front. Oncol. 2019, 9, 541. [Google Scholar] [CrossRef]
- Silva dos Santos, J.; Goncalves Cirino, J.P.; de Oliveira Carvalho, P.; Ortega, M.M. The Pharmacological Action of Kaempferol in Central Nervous System Diseases: A Review. Front. Pharmacol. 2021, 11, 565700. [Google Scholar] [CrossRef]
- Batiha, G.E.-S.; Beshbishy, A.M.; Ikram, M.; Mulla, Z.S.; El-Hack, M.E.A.; Taha, A.E.; Algammal, A.M.; Elewa, Y.H.A. The Pharmacological Activity, Biochemical Properties, and Pharmacokinetics of the Major Natural Polyphenolic Flavonoid: Quercetin. Foods 2020, 9, 374. [Google Scholar] [CrossRef]
- Rue, E.A.; Rush, M.D.; van Breemen, R.B. Procyanidins: A comprehensive review encompassing structure elucidation via mass spectrometry. Phytochem. Rev. 2018, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, A.M.; Basam, S.M.; Marzouk, H.S.; El-Hawary, S. LC–MS/MS and GC–MS profiling as well as the antimicrobial effect of leaves of selected Yucca species introduced to Egypt. Sci. Rep. 2020, 10, 17778. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.S.; Barroso, S.C.; Navoni, J.A.; Teles, M.M.R.S.; Barbosa-Filho, J.M.; Rocha, H.A.D.O.; do Amaral, V.S. Effect of Hecogenin on DNA Instability. Toxicol. Rep. 2016, 3, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Diantini, A.; Subarnas, A.; Lestari, K.; Halimah, E.; Susilawati, Y.; Supriyatna, S.; Julaeha, E.; Achmad, T.H.; Suradji, E.W.; Yamazaki, C.; et al. Kaempferol-3-O-Rhamnoside Isolated from the Leaves of SchimaWallichiiKorth. Inhibits MCF-7 Breast Cancer Cell Proliferation through Activation of the Caspase Cascade Pathway. Oncol. Lett. 2012, 3, 1069–1072. [Google Scholar] [CrossRef]
- Juang, Y.P.; Liang, P.H. Biological and Pharmacological Effects of Synthetic Saponins. Molecules 2020, 27, 4974. [Google Scholar] [CrossRef]
- Tatsimo, S.J.N.; Tamokou, J.D.D.; Havyarimana, L.; Csupor, D.; Forgo, P.; Hohmann, J.; Kuiate, J.-R.; Tane, P. Antimicrobial and antioxidant activity of kaempferol rhamnoside derivatives from Bryophyllumpinnatum. BMC Res. Notes 2012, 5, 158. [Google Scholar] [CrossRef]
- He, J.; Feng, Y.; Ouyang, H.Z.; Yu, B.; Chang, Y.X.; Pan, G.X.; Dong, G.Y.; Wang, T.; Gao, X.M. A sensitive LC-MS/MS method for simultaneous determination of six flavonoids in rat plasma: Application to a pharmacokinetic study of total flavonoids from mulberry leaves. J. Pharm. Biomed Anal. 2013, 84, 189–195. [Google Scholar]
- Sharma, U.K.; Sharma, A.K.; Gupta, A.; Kumar, R.; Pandey, A.; Pandey, A.K. Pharmacological activities of cinnamaldehyde and eugenol: Antioxidant, cytotoxic and anti-leishmanial studies. Cell Mol. Biol. 2017, 63, 73–78. [Google Scholar] [CrossRef]
- Grabarczyk, M.; Wińska, K.; Mączka, W.; Potaniec, B.; Anioł, M. Loliolide-the most ubiquitous lactone. Acta Univers. Lodz. Folia Biol.Oecol. 2015, 11, 1–8. [Google Scholar] [CrossRef]
- Bae, J.; Kim, N.; Shin, Y.; Kim, S.-Y.; Kim, Y.-J. Activity of catechins and their applications. Biomed. Dermatol. 2020, 4, 8. [Google Scholar]
- Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A.A.; Khan, G.J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; Fang, X.F.; Modarresi-Ghazani, F.; et al. Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother. 2018, 97, 67–74. [Google Scholar] [CrossRef]
- Serreli, G.; Sayec, M.L.; Thou, E.; Lacour, C.; Diotallevi, C.; Dhunna, M.A.; Deiana, M.; Spencer, J.P.E.; Corona, G. Ferulic Acid Derivatives and Avenanthramides Modulate Endothelial Function through Maintenance of Nitric Oxide Balance in HUVEC Cells. Nutrients 2021, 12, 2026. [Google Scholar] [CrossRef]
- Benali, T.; Bakrim, S.; Ghchime, R.; Benkhaira, N.; Omari, N.E.; Balahbib, A.; Taha, D.; Zengin, G.; Hasan, M.M.; Bibi, S.; et al. Pharmacological insights into the multifaceted biological properties of quinic acid. Biotechnol. Genet. Eng. Rev. 2022, 19, 1–30. [Google Scholar] [CrossRef]
- Semaming, Y.; Pannengpetch, P.; Chattipakorn, S.C.; Chattipakorn, N. Pharmacological properties of protocatechuic Acid and its potential roles as complementary medicine. Evid. Based Complement Altern. Med. Ecam 2015, 593902. [Google Scholar] [CrossRef]
- Hong, Y.; Wang, Z.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A.R. High-Throughput Screening and Characterization of Phenolic Compounds in Stone Fruits Waste by LC-ESI-QTOF-MS/MS and Their Potential Antioxidant Activities. Antioxidants. 2021, 10, 234. [Google Scholar] [CrossRef]
- Lee, D.E.; Lee, K.W.; Byun, S.; Jung, S.K.; Song, N.; Lim, S.H.; Heo, Y.S.; Kim, J.E.; Kang, N.J.; Kim, B.Y.; et al. 7,3′,4′-Trihydroxyisoflavone, a metabolite of the soy isoflavone daidzein, suppresses ultraviolet B-induced skin cancer by targeting Cot and MKK4. J. Biol. Chem. 2011, 22, 14246–14256. [Google Scholar] [CrossRef]
- Reddy, M.K.; Gupta, S.K.; Jacob, M.R.; Khan, S.I.; Ferreira, D. Antioxidant, antimalarial and antimicrobial activities of tannin-rich fractions, ellagitannins and phenolic acids from Punica granatum L. Planta Med. 2007, 73, 461–467. [Google Scholar] [CrossRef]
- Salehi, B.; Venditti, A.; Sharifi-Rad, M.; Kręgiel, D.; Sharifi-Rad, J.; Durazzo, A.; Lucarini, M.; Santini, A.; Souto, E.B.; Novellino, E.; et al. The Therapeutic Potential of Apigenin. Int. J. Mol. Sci. 2019, 15, 1305. [Google Scholar] [CrossRef]
- Kahkeshani, N.; Farzaei, F.; Fotouhi, M.; Alavi, S.S.; Bahramsoltani, R.; Naseri, R.; Momtaz, S.; Abbasabadi, Z.; Rahimi, R.; Farzaei, M.H.; et al. Pharmacological effects of gallic acid in health and diseases: A mechanistic review. Iran. J. Basic Med. Sci. 2019, 22, 225–237. [Google Scholar]
- Zhou, L.; Sun, J.; Zhang, T.; Tang, Y.; Liu, J.; Gao, C.; Zhai, Y.; Guo, Y.; Feng, L.; Zhang, X.; et al. Comparative Transcriptome Analyses of Different Rheum officinale Tissues Reveal Differentially Expressed Genes Associated with Anthraquinone, Catechin, and Gallic Acid Biosynthesis. Genes 2022, 13, 1592. [Google Scholar] [CrossRef]
- Kong, J.Q. Phenylalanine Ammonia-Lyase, a Key Component Used for Phenylpropanoids Production by Metabolic Engineering. RSC Adv. 2015, 5, 62587–62603. [Google Scholar] [CrossRef]
- Chun, H.J.; Bae, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef]
- Yamada, S.; Nabe, K.; Izuo, N.; Nakamichi, K.; Chibata, I. Production of l-Phenylalanine from trans-Cinnamic Acid with Rhodotorula glutinis Containing l-Phenylalanine Ammonia-Lyase Activity. Appl. Environ. Microbiol. 1981, 42, 773–778. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Hou, L.; Wang, L.; Wu, X.; Gao, W.; Zhang, J.X.; Huang, C.Y. Expression patterns of two pal genes of Pleurotus ostreatus across developmental stages and under heat stress. BMC Microbiol. 2019, 19, 231. [Google Scholar] [CrossRef]
- Bagal, U.R.; Leebens-Mack, J.H.; Lorenz, W.W.; Dean, J.F. The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC Genom. 2012, 13 (Suppl. 3), S1. [Google Scholar] [CrossRef]
- Umezawa, T. The cinnamate/monolignol pathway. Phytochem. Rev. 2010, 9, 1–17. [Google Scholar] [CrossRef]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef]
- Iwashina, T. The Structure and Distribution of the Flavonoids in Plants. J. Plant Res. 2000, 113, 287–299. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 28, 222. [Google Scholar] [CrossRef]
- Azeem, M.; Hanif, M.; Mahmood, K.; Ameer, N.; Chughtai, F.R.S.; Abid, U. An insight into anticancer, antioxidant, antimicrobial, antidiabetic and anti-inflammatory effects of quercetin: A review. Polym. Bull. 2022, 1–22. [Google Scholar] [CrossRef] [PubMed]
- David, A.V.A.; Arulmoli, R.; Parasuraman, S. Overviews of Biological Importance of Quercetin: A Bioactive Flavonoid. Pharmacogn. Rev. 2016, 10, 84–89. [Google Scholar]
- Saeedi-Boroujeni, A.; Mahmoudian-Sani, M.R. Anti-inflammatory potential of Quercetin in COVID-19 treatment. J. Inflamm. 2021, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Cherrak, S.A.; Merzouk, H.; Mokhtari-Soulimane, N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS ONE 2020, 15, e0240653. [Google Scholar] [CrossRef]
- Huang, C.J.; Chu, F.H.; Huang, Y.S.; Tu, Y.C.; Hung, Y.M.; Tseng, Y.H.; Pu, C.E.; Hsu, C.T.; Chao, C.H.; Chou, Y.S.; et al. SSR individual identification system construction and population genetics analysis for Chamaecyparis formosensis. Sci. Rep. 2022, 12, 4126. [Google Scholar]
- Yasodha, R.; Vasudeva, R.; Balakrishnan, S.; Sakthi, A.R.; Abel, N.; Binai, N.; Rajashekar, B.; Bachpai, V.K.W.; Pillai, C.; Dev, S.A. Draft Genome of a High Value Tropical Timber Tree, Teak (Tectona Grandis L. f): Insights into SSR Diversity, Phylogeny and Conservation. DNA Res. 2018, 25, 409–419. [Google Scholar] [CrossRef]
- Padmanabhan, D.; Lateef, A.; Natarajan, P.; Palanisamy, S. De novo transcriptome analysis of Justicia adhatoda reveals candidate genes involved in major biosynthetic pathway. Mol. Biol. Rep. 2022, 1–8. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).