PyunBBX18 Is Involved in the Regulation of Anthocyanins Biosynthesis under UV-B Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of BBX Gene Family Members in P. yunnanensis
2.2. Analysis of Basic Physicochemical and Subcellular Localization of PyunBBX Genes
2.3. Gene Structure and Motif Analysis of PyunBBX Genes
2.4. Cis-Acting Elements Analysis
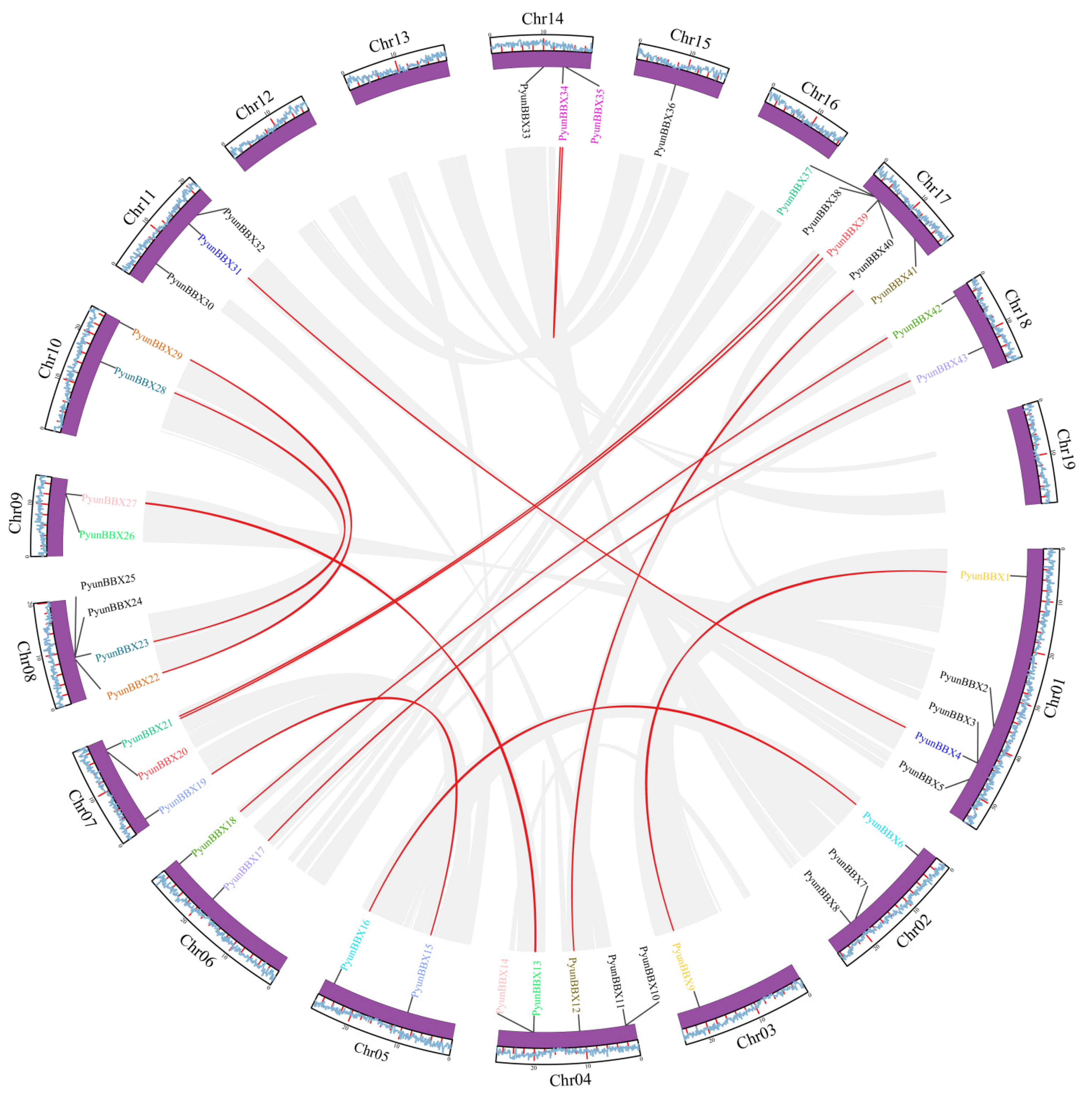
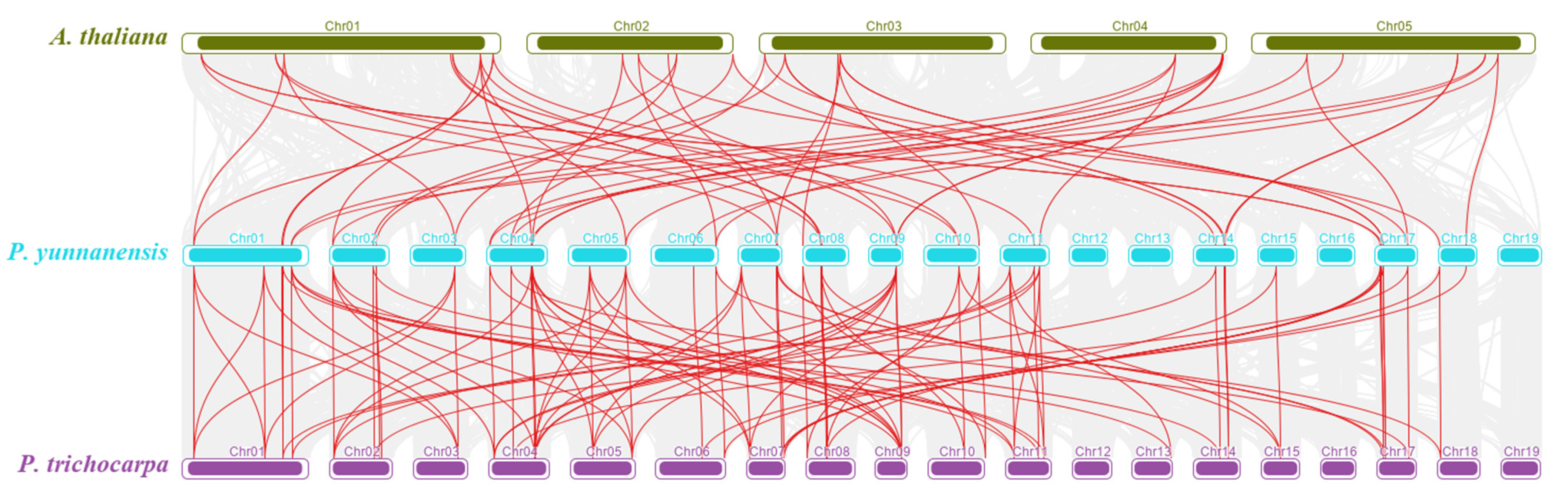
2.5. Chromosomal Location, Gene Duplication, and Synteny Analysis
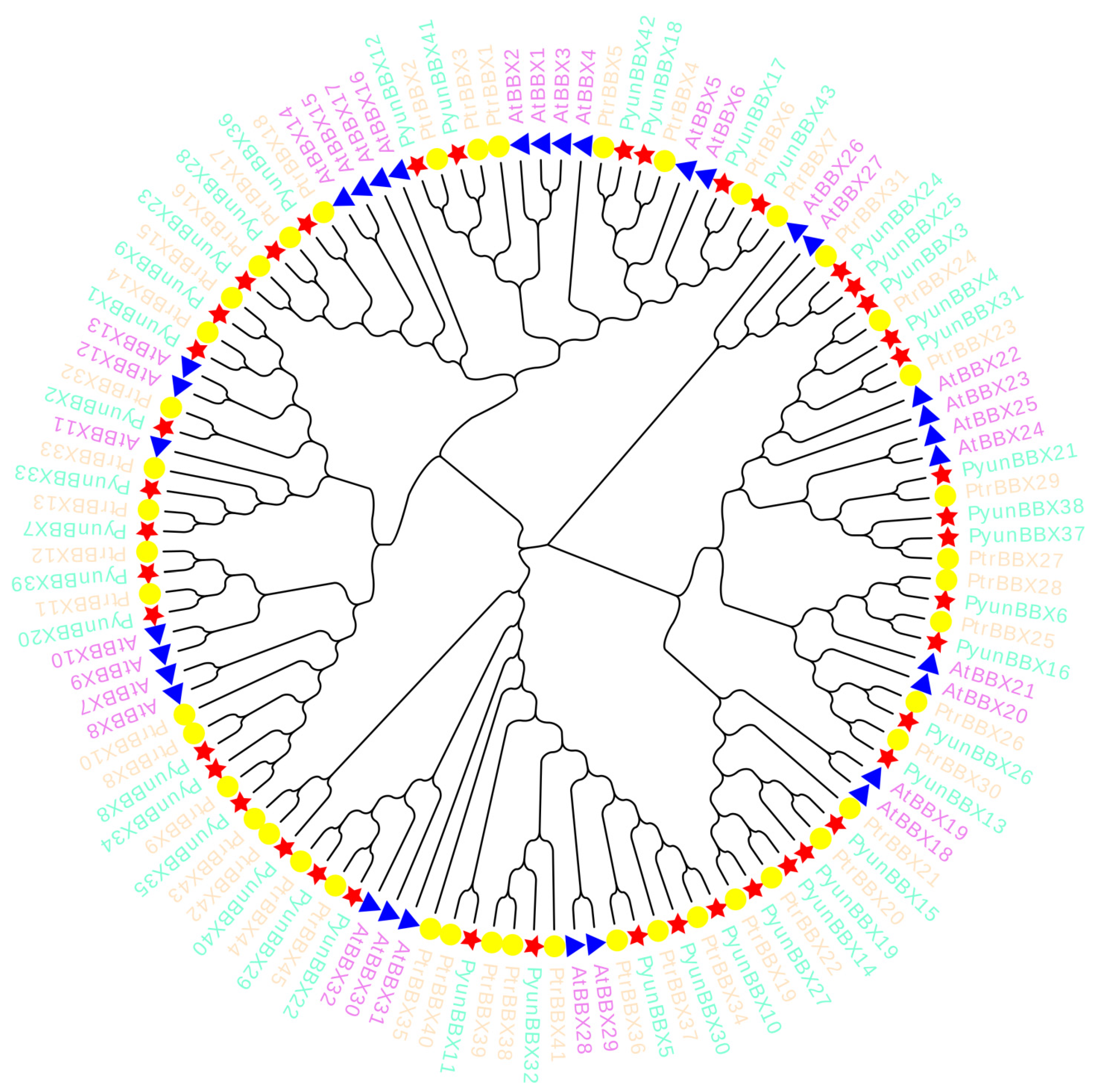
2.6. Evolutionary Analysis of BBX Gene Family Members
2.7. Plant Materials, Growth Conditions, and UV Treatment
2.8. Total RNA Isolation and cDNA Synthesis
2.9. Quantitative Real-Time PCR
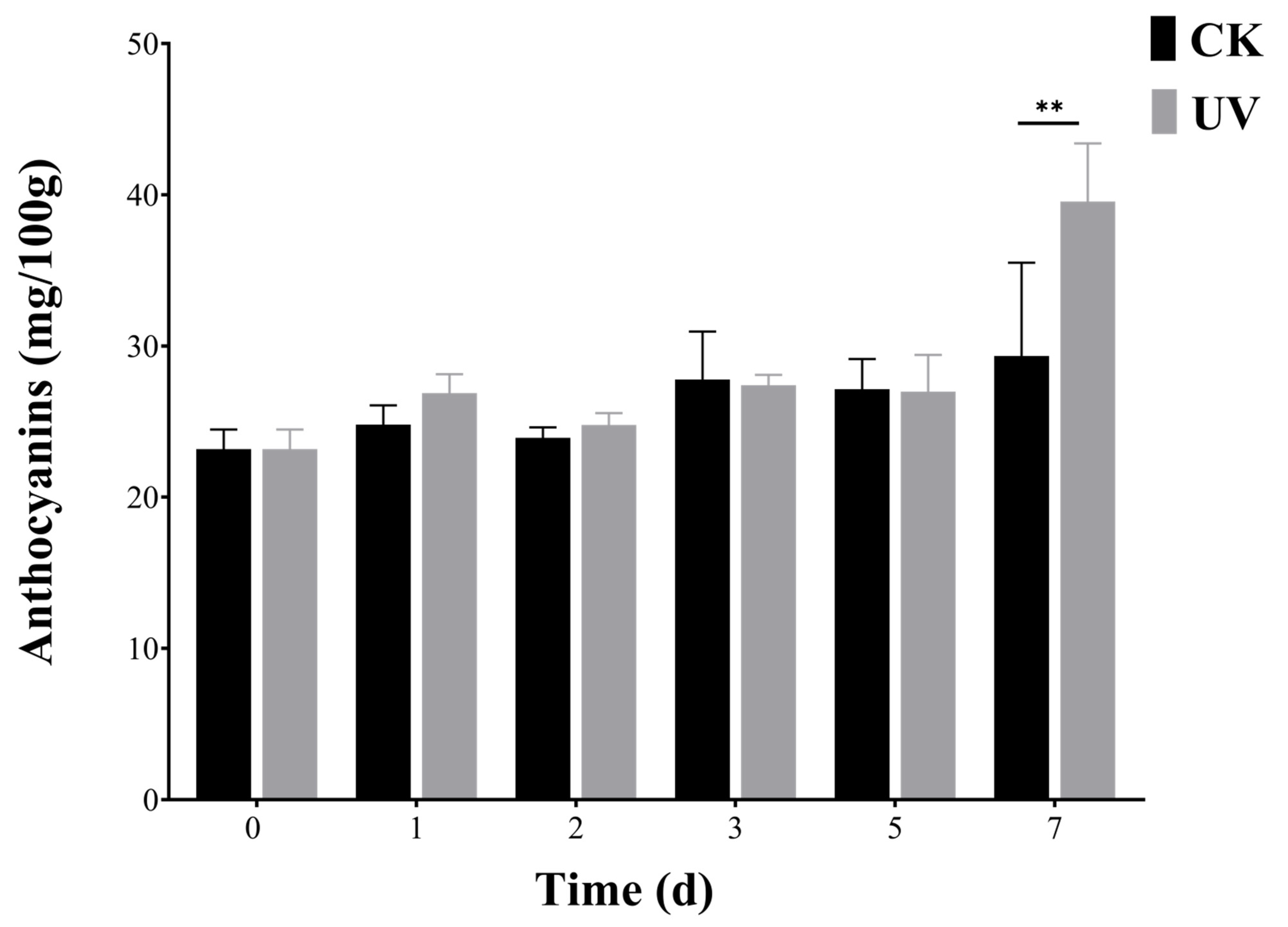
2.10. Extraction and Determination of Anthocyanins
2.11. Expression Patterns of BBX Family Members in P. yunnanensis by Rna-seq Data
2.12. Data Analysis
3. Results
3.1. Analysis of The Basic Characteristics of PyunBBX Genes
3.2. Gene Structure and Characteristics of Conserved Sequences of PyunBBX Gene Family Members
3.3. Collinearity Analysis
3.4. Cis-Acting Elements of PyunBBX Genes Analysis
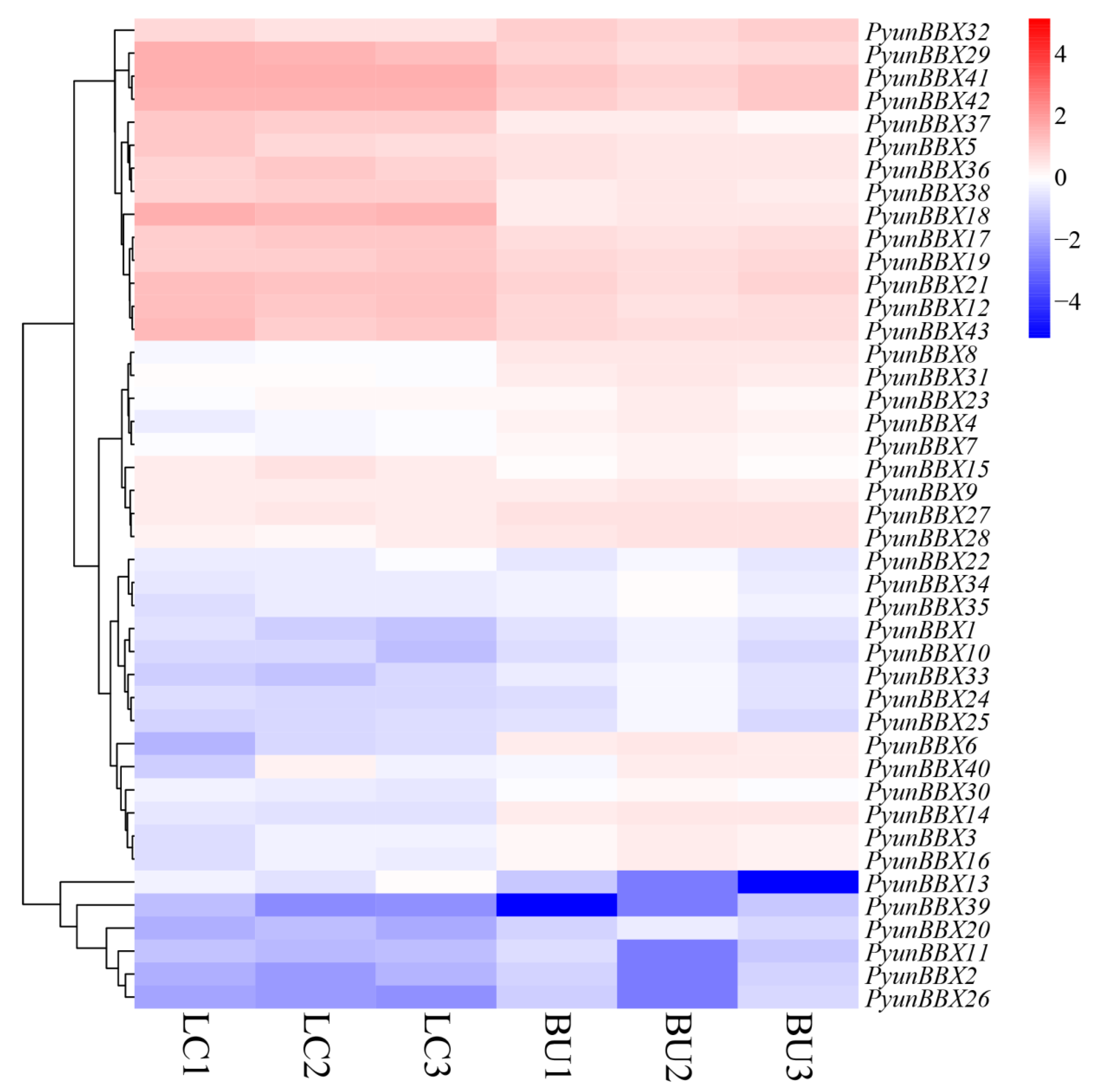
3.5. Expression Patterns of PyunBBX Genes in Leaf and Bark of P. yunnanensis by Rna-seq Data
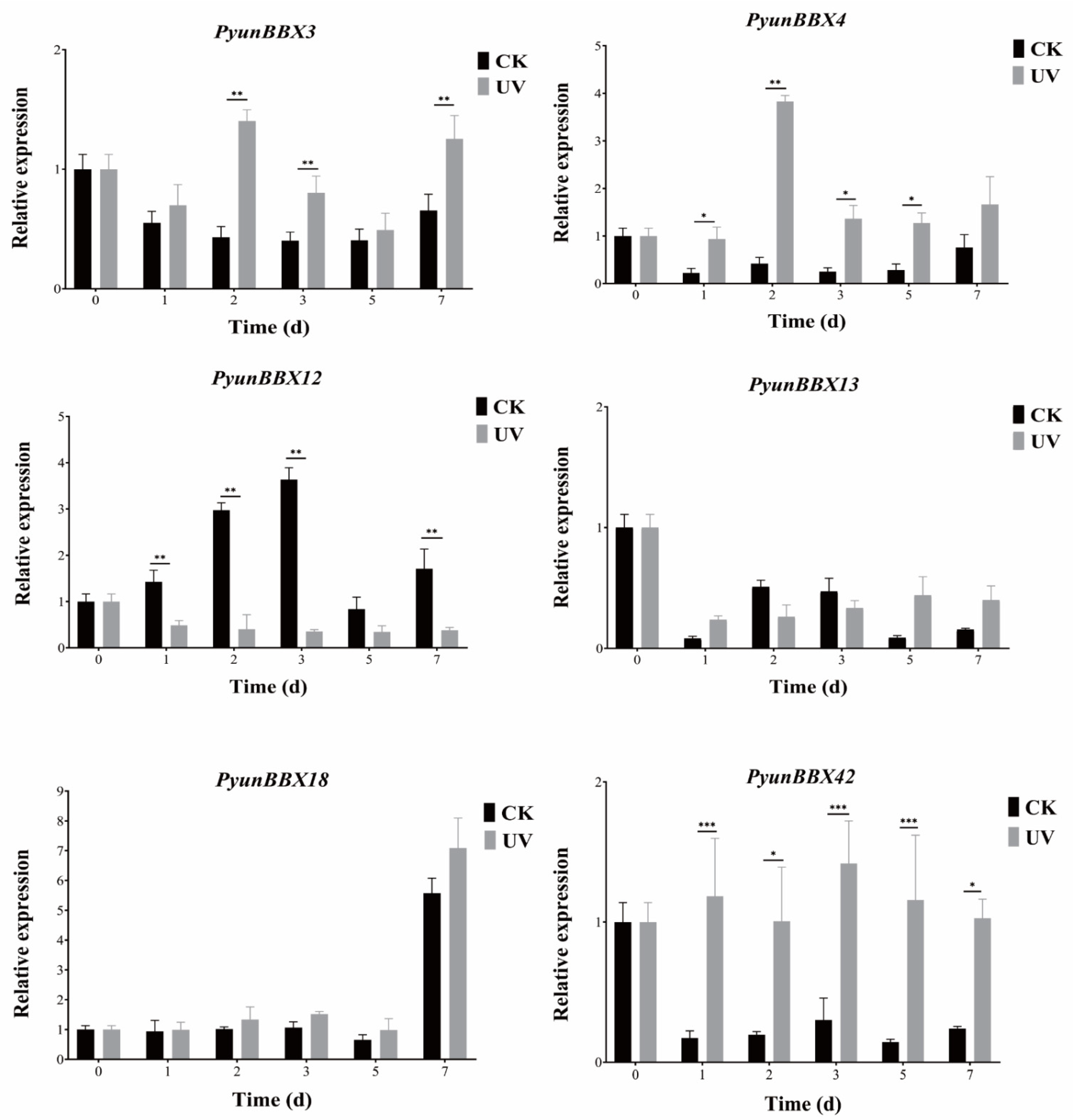
3.6. Expression Patterns of Six PyunBBXs under UV Treatment by RT-qPCR Analysis
4. Discussion
4.1. Identification and Analysis of PyunBBX Gene Family Members in P. yunnanensis
4.2. BBX Is Involved in Anthocyanins Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 7, 460–470. [Google Scholar] [CrossRef]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Holm, M.; Deng, X.W. The B-Box Domain Protein BBX21 Promotes Photomorphogenesis. Plant Physiol. 2018, 176, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-Box Proteins Regulate Photomorphogenesis by Oppositely Modulating HY5 through their Diverse C-Terminal Domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W. B-Box Containing Proteins BBX30 and BBX31, Acting Downstream of HY5, Negatively Regulate Photomorphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, C.; Dong, H.; Liu, X.; Guo, H.; Tong, B.; Fang, F.; Zhao, Y.; Yu, Y.; Liu, Y.; et al. Activation and negative feedback regulation of SlHY5 transcription by the SlBBX20/21–SlHY5 transcription factor module in UV-B signaling. Plant Cell 2022, 34, 2038–2055. [Google Scholar] [CrossRef]
- Li, C.; Pei, J.; Yan, X.; Cui, X.; Tsuruta, M.; Liu, Y.; Lian, C. A poplar B-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell Environ. 2021, 44, 3015–3033. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Wang, X.; Zhang, X.; Bi, S.; You, C.; Hao, Y. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Zhuang, W.B.; Liu, T.Y.; Shu, X.C.; Shen-Chun, Q.U.; Zhai, H.H.; Wang, T.; Zhang, F.J.; Wang, Z. The molecular regulation mechanism of anthocyanin biosynthesis and coloration in plants. Plant Physiol. J. 2018, 11, 1630–1644. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis Transcription Factor ANAC032 Represses Anthocyanin Biosynthesis in Response to High Sucrose and Oxidative and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Pradeep, S.; Oleg, F.; Dalia, M.; Noam, A. Improved Cold Tolerance of Mango Fruit with Enhanced Anthocyanin and Flavonoid Contents. Molecules 2018, 23, 1832. [Google Scholar] [CrossRef]
- Cui, Z.; Bi, W.; Hao, X.; Li, P.; Duan, Y.; Walker, M.A.; Xu, Y.; Wang, Q. Drought Stress Enhances Up-Regulation of Anthocyanin Biosynthesis in Grapevine leafroll-associated virus 3-Infected in vitro Grapevine (Vitis vinifera) Leaves. Plant Dis. 2017, 101, 1605–1615. [Google Scholar] [CrossRef]
- Bi, H.; Guo, M.; Wang, J.; Qu, Y.; Du, W.; Zhang, K. Transcriptome analysis reveals anthocyanin acts as a protectant in Begonia semperflorens under low temperature. Acta Physiol. Plant. 2018, 40, 10. [Google Scholar] [CrossRef]
- Roberts, J.A.; Evan, D.; Mcmanus, M.T.; Rose, J.K.C. Annual Plant Reviews. Functions of Flavonoid and Betalain Pigments in Abiotic Stress Tolerance in Plants. Annu. Plant Rev. Online 2018, 1, 1–41. [Google Scholar] [CrossRef]
- Zhang, T.; Chow, W.S.; Liu, X.; Zhang, P.; Liu, N.; Peng, C. A magic red coat on the surface of young leaves: Anthocyanins distributed in trichome layer protect Castanopsis fissa leaves from photoinhibition. Tree Physiol. 2016, 36, 1296–1306. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, X.; Sun, B.; Peng, C.; Chow, W.S. Over-expression of the CHS gene enhances resistance of Arabidopsis leaves to high light. Environ. Exp. Bot. 2018, 154, 33–43. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.; Jung, H.; Yang, T.; Hur, Y.; Nou, I. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 2014, 550, 46–55. [Google Scholar] [CrossRef]
- Hong, Y.; Wu, Y.; Song, X.; Li, M.; Dai, S. Molecular mechanism of light-induced anthocyanin biosynthesis in horticultural crops. Acta Hortic. Sin. 2021, 48, 1983–2000. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Sun, X.; Lin, H.; Chen, J.; Ren, J.; Hu, X.; Yang, Y. Comparative Physiological and Proteomic Analyses of Poplar (Populus yunnanensis) Plantlets Exposed to High Temperature and Drought. PLoS ONE 2014, 9, e107605. [Google Scholar] [CrossRef] [PubMed]
- Schmucki, D.A. Ultraviolet radiation in the Alps: The altitude effect. Proc. Spie 2002, 41, 3090–3095. [Google Scholar] [CrossRef]
- Costa, D.; Galvão, A.M.; Di Paolo, R.E.; Freitas, A.A.; Lima, J.C.; Quina, F.H.; Maçanita, A.L. Photochemistry of the hemiketal form of anthocyanins and its potential role in plant protection from UV-B radiation. Tetrahedron 2015, 71, 3157–3162. [Google Scholar] [CrossRef]
- Tsurunaga, Y.; Takahashi, T.; Katsube, T.; Kudo, A.; Kuramitsu, O.; Ishiwata, M.; Matsumoto, S. Effects of UV-B irradiation on the levels of anthocyanin, rutin and radical scavenging activity of buckwheat sprouts. Food Chem. 2013, 141, 552–556. [Google Scholar] [CrossRef]
- Kalidhasan, N.; Bhagavan, N.B.; Dhiraviam, K. Ultraviolet B (280-320 nm) enhanced radiation induced changes in secondary metabolites and photosystem-II of medicinal plant Withania somnifera Dunal. J. Med. Plant Res. 2013, 7, 3112–3120. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols; Link, A.J., Ed.; Humana Press: Totowa, NJ, USA, 1999; Volume 112. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Jingyuan, R.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica Rapa Shed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 2016, 8, 302–316. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2015, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Fan, G.; Han, Y.; Gu, Z.; Chen, D. Optimizing conditions for anthocyanins extraction from purple sweet potato using response surface methodology (RSM). LWT Food Sci. Technol. 2008, 41, 155–160. [Google Scholar] [CrossRef]
- Strayer, C. Cloning of the Arabidopsis Clock Gene TOC1, an Autoregulatory Response Regulator Homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef]
- Lau, K.; Podolec, R.; Chappuis, R.; Ulm, R.; Hothorn, M. Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 2019, 38, e102140. [Google Scholar] [CrossRef]
- Zhou, A.; Gan, P.; Zong, D.; Fei, X.; Zhong, Y.; Li, S.; Yu, J.; He, C. Bark tissue transcriptome analyses of inverted Populus yunnanensis cuttings reveal the crucial role of plant hormones in response to inversion. PeerJ 2019, 7, e7740. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Wagner, D.R. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; van der Woude, K.; Mol, J.N.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. Cell Mol. Biol. 1998, 13, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Spelt, C.; Quattrocchio, F.; Joseph, N.M.M.; Koes, R. anthocyanin1 of Petunia Encodes a Basic Helix-Loop-Helix Protein That Directly Activates Transcription of Structural Anthocyanin Genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Singh, R.K.; Navarre, D.A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013, 64, 5115–5131. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Yadav, A.; Ravindran, N.; Singh, D.; Rahul, P.V.; Datta, S. Role of Arabidopsis BBX proteins in light signaling. J. Plant Biochem. Biot. 2020, 29, 623–635. [Google Scholar] [CrossRef]
- Gourlay, G.; Ma, D.; Schmidt, A.; Constabel, C.P. MYB134-RNAi poplar plants show reduced tannin synthesis in leaves but not roots, and increased susceptibility to oxidative stress. J. Exp. Bot. 2020, 71, 6601–6611. [Google Scholar] [CrossRef]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef]





| Gene ID | Chromosomal Location | Amino Acid Length (aa) | Isoelectric Point (pI) | Molecular Weight (kDa) | Subcellular Localization |
|---|---|---|---|---|---|
| PyunBBX1 | Chr01: 5,540,646–5,543,961 | 513 | 5.75 | 56.51 | Nucleus |
| PyunBBX2 | Chr01: 36,111,780–36,114,290 | 4 30 | 5.54 | 48.42 | Nucleus |
| PyunBBX3 | Chr01: 43,995,595–43,999,866 | 297 | 5.68 | 31.88 | Nucleus |
| PyunBBX4 | Chr01: 44,107,170–44,111,390 | 297 | 5.99 | 31.88 | Nucleus |
| PyunBBX5 | Chr01: 47,801,948–47,803,497 | 271 | 4.31 | 29.56 | Nucleus |
| PyunBBX6 | Chr02: 1,909,069–1,910,516 | 311 | 6.05 | 34.36 | Nucleus |
| PyunBBX7 | Chr02: 19,567,193–19571319 | 362 | 5.7 | 40.72 | Nucleus |
| PyunBBX8 | Chr02: 20,923,421–20,934,900 | 416 | 5.68 | 45.58 | Nucleus |
| PyunBBX9 | Chr03: 20,261,596–20,265,145 | 541 | 6.19 | 59.46 | Nucleus |
| PyunBBX10 | Chr04: 1,800,513–1,801,098 | 184 | 4.15 | 20.32 | Nucleus |
| PyunBBX11 | Chr04: 1,805,089–1,811,255 | 236 | 4.67 | 25.65 | Nucleus |
| PyunBBX12 | Chr04: 11,046,033–11,048,035 | 369 | 5.92 | 40.86 | Nucleus |
| PyunBBX13 | Chr04: 20,203,185–20,203,893 | 206 | 5.72 | 22.9 | Nucleus |
| PyunBBX14 | Chr04: 20,382,296–20,384,272 | 203 | 5.98 | 22.75 | Nucleus |
| PyunBBX15 | Chr05: 9,739,681–9,741,476 | 193 | 7.56 | 21.42 | Nucleus |
| PyunBBX16 | Chr05: 25,453,505–25,455,163 | 311 | 6.2 | 34.34 | Nucleus |
| PyunBBX17 | Chr06: 18,887,979–18,889,903 | 384 | 6.23 | 42.36 | Nucleus |
| PyunBBX18 | Chr06: 28,508,368–28,511,098 | 346 | 6.09 | 37.58 | Nucleus |
| PyunBBX19 | Chr07: 1,484,594–1,486,875 | 192 | 6.49 | 21.18 | Nucleus |
| PyunBBX20 | Chr07: 16,742,283–16,744,039 | 433 | 5.26 | 47.03 | Nucleus |
| PyunBBX21 | Chr07: 17,319,158–17,322,042 | 235 | 4.8 | 26.05 | Nucleus |
| PyunBBX22 | Chr08: 292,896–293,927 | 250 | 8.66 | 27.54 | Nucleus |
| PyunBBX23 | Chr08: 8,052,050–8,054,730 | 444 | 5.58 | 49.27 | Nucleus |
| PyunBBX24 | Chr08: 8,604,637–8,611,266 | 500 | 5.81 | 56.13 | Nucleus |
| PyunBBX25 | Chr08: 8,669,884–8,676,275 | 500 | 5.78 | 56.09 | Nucleus |
| PyunBBX26 | Chr09: 12,288,823–12,289,565 | 217 | 6.24 | 24.06 | Nucleus |
| PyunBBX27 | Chr09: 12,435,516–12,440,014 | 238 | 7.05 | 26.38 | Nucleus |
| PyunBBX28 | Chr10: 156,680,74–15,670,342 | 447 | 5.1 | 48.93 | Nucleus |
| PyunBBX29 | Chr10: 24,419,560–24,421,322 | 263 | 9.26 | 28.89 | Nucleus |
| PyunBBX30 | Chr11: 4,714,212–4,714,775 | 187 | 4.12 | 20.59 | Nucleus |
| PyunBBX31 | Chr11: 15,010,674–15,015,143 | 298 | 5.85 | 32.2 | Nucleus |
| PyunBBX32 | Chr11: 17,403,770–17,405,035 | 336 | 4.34 | 37.13 | Nucleus |
| PyunBBX33 | Chr14: 9,996,595–10,004,305 | 315 | 5.3 | 35.29 | Nucleus |
| PyunBBX34 | Chr14: 13,912,158–13,917,939 | 416 | 5.09 | 45.2 | Nucleus |
| PyunBBX35 | Chr14: 14,454,914–14,460,689 | 416 | 5.04 | 45.34 | Nucleus |
| PyunBBX36 | Chr15: 8,537,097–8,539,444 | 426 | 5.12 | 48.43 | Nucleus |
| PyunBBX37 | Chr17: 2,781,131–2,783,909 | 238 | 4.77 | 26.02 | Nucleus |
| PyunBBX38 | Chr17: 3,054,640–3,057,429 | 238 | 4.77 | 25.99 | Nucleus |
| PyunBBX39 | Chr17: 4,089,911–4,091,481 | 421 | 5.85 | 45.62 | Nucleus |
| PyunBBX40 | Chr17: 4,094,941–4,096,953 | 108 | 8.61 | 12.11 | Nucleus |
| PyunBBX41 | Chr17: 14,748,917–14,751,095 | 380 | 5.25 | 41.61 | Nucleus |
| PyunBBX42 | Chr18: 966,086–968,260 | 350 | 5.6 | 37.89 | Nucleus |
| PyunBBX43 | Chr18: 12,620,866–12,622,557 | 382 | 5.96 | 41.94 | Nucleus |
| Syntenic Gene Pairs | Method | Ks | Ka | Ka/Ks | Duplicated Type | Divergence Time (Mya.) |
|---|---|---|---|---|---|---|
| PyunBBX22-PyunBBX29 | MA * | 0.117574 | 0.350705 | 0.335249 | Segmental | 3.92 |
| PyunBBX15-PyunBBX19 | MA | 0.0944959 | 0.335761 | 0.281438 | Segmental | 3.15 |
| PyunBBX13-PyunBBX26 | MA | 0.0922884 | 0.265914 | 0.347061 | Segmental | 3.08 |
| PyunBBX1-PyunBBX9 | MA | 0.0828391 | 0.301719 | 0.274557 | Segmental | 2.76 |
| PyunBBX23-PyunBBX28 | MA | 0.0735978 | 0.217724 | 0.338032 | Segmental | 2.45 |
| PyunBBX4-PyunBBX31 | MA | 0.0733725 | 0.226066 | 0.324562 | Segmental | 2.45 |
| PyunBBX14-PyunBBX27 | MA | 0.0726848 | 0.378882 | 0.19184 | Segmental | 2.42 |
| PyunBBX20-PyunBBX39 | MA | 0.0677018 | 0.192095 | 0.35244 | Segmental | 2.26 |
| PyunBBX17-PyunBBX43 | MA | 0.0672206 | 0.355935 | 0.188856 | Segmental | 2.24 |
| PyunBBX12-PyunBBX41 | MA | 0.0653516 | 0.3069 | 0.212941 | Segmental | 2.18 |
| PyunBBX6-PyunBBX16 | MA | 0.0542511 | 0.285174 | 0.190239 | Segmental | 1.81 |
| PyunBBX21-PyunBBX37 | MA | 0.0530882 | 0.237577 | 0.223457 | Segmental | 1.77 |
| PyunBBX18-PyunBBX42 | MA | 0.0426761 | 0.358764 | 0.118953 | Segmental | 1.42 |
| PyunBBX34-PyunBBX35 | MA | 0.00717976 | 0.0300601 | 0.238847 | Segmental | 0.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Ma, D.; Hu, Z.; Zong, D.; He, C. PyunBBX18 Is Involved in the Regulation of Anthocyanins Biosynthesis under UV-B Stress. Genes 2022, 13, 1811. https://doi.org/10.3390/genes13101811
Zhang Q, Ma D, Hu Z, Zong D, He C. PyunBBX18 Is Involved in the Regulation of Anthocyanins Biosynthesis under UV-B Stress. Genes. 2022; 13(10):1811. https://doi.org/10.3390/genes13101811
Chicago/Turabian StyleZhang, Qin, Dongxiao Ma, Zhixu Hu, Dan Zong, and Chengzhong He. 2022. "PyunBBX18 Is Involved in the Regulation of Anthocyanins Biosynthesis under UV-B Stress" Genes 13, no. 10: 1811. https://doi.org/10.3390/genes13101811
APA StyleZhang, Q., Ma, D., Hu, Z., Zong, D., & He, C. (2022). PyunBBX18 Is Involved in the Regulation of Anthocyanins Biosynthesis under UV-B Stress. Genes, 13(10), 1811. https://doi.org/10.3390/genes13101811






