Basement-Membrane-Related Gene Signature Predicts Prognosis in WHO Grade II/III Gliomas
Abstract
1. Introduction
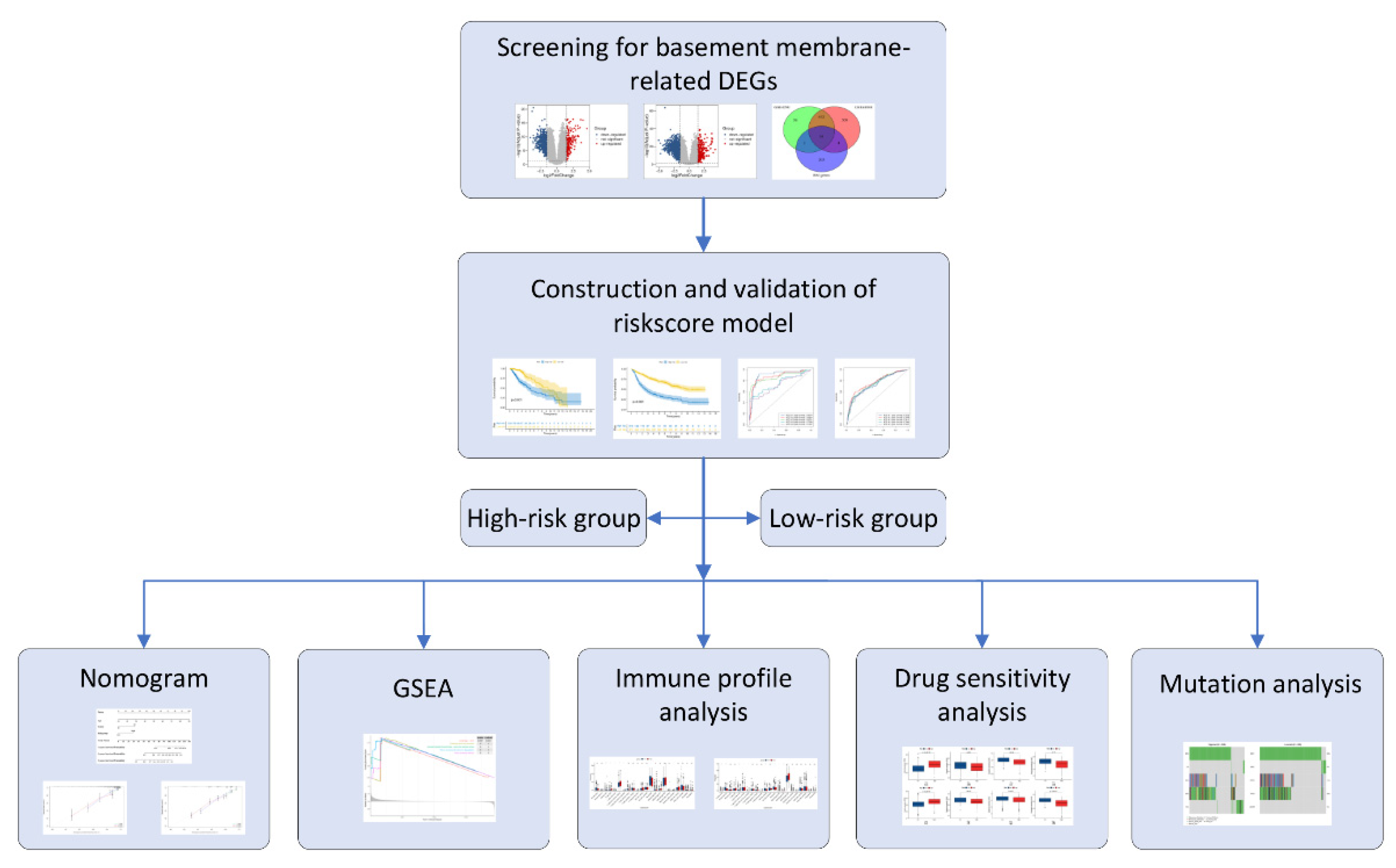
2. Materials and Methods
2.1. Data Extraction
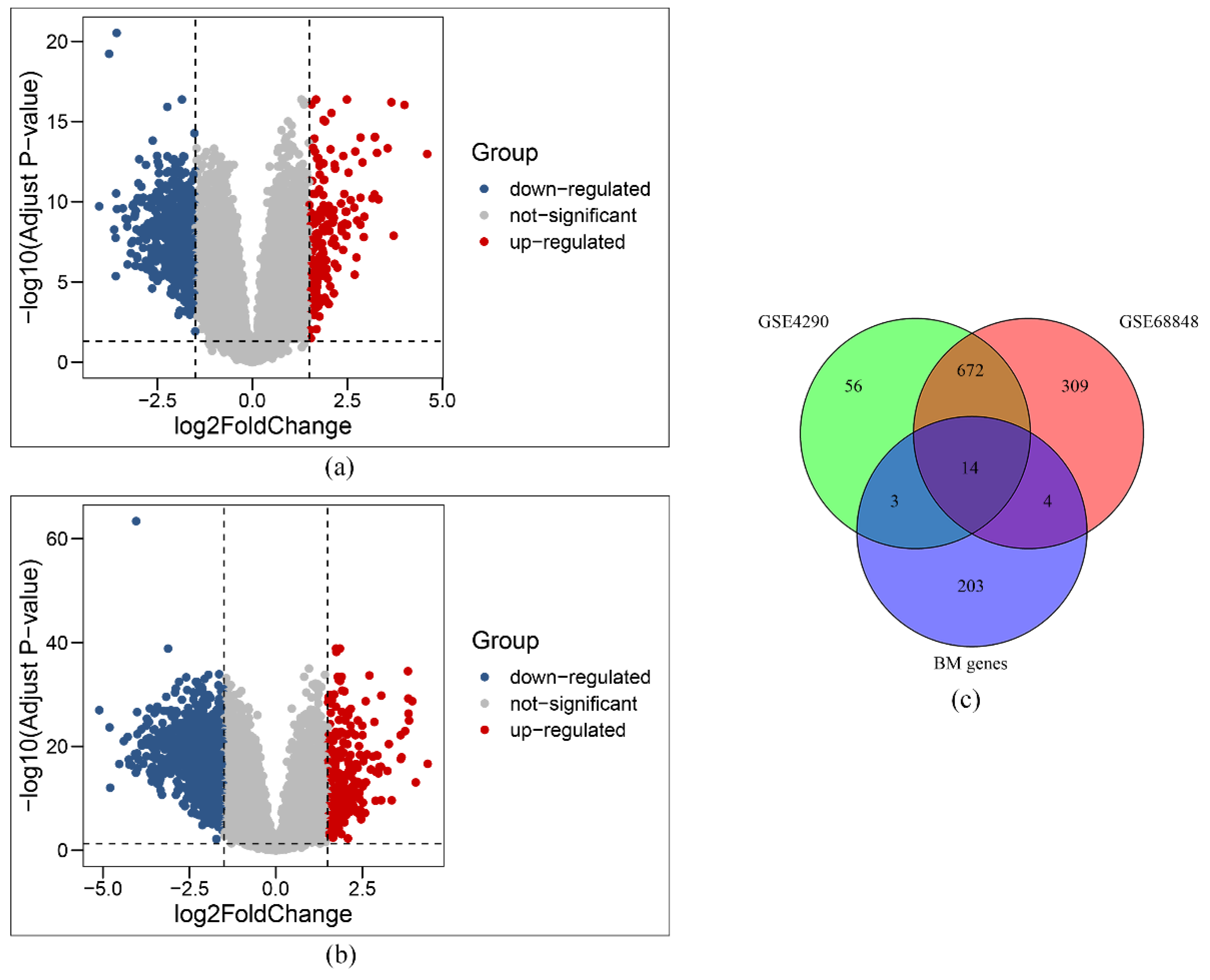
2.2. Initial Screening of BM-Related Genes
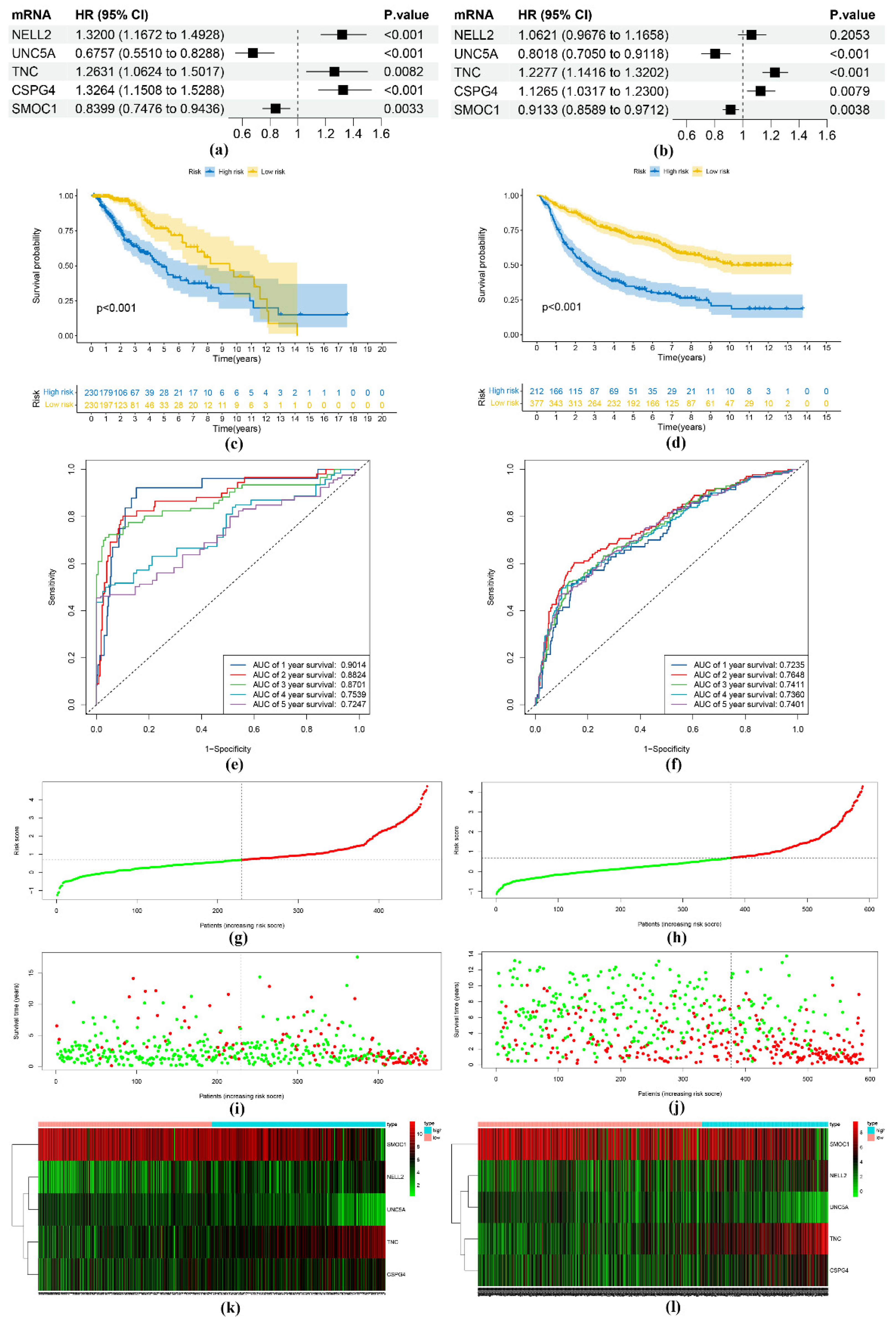
2.3. Development of the Risk Score Model
2.4. Establishment and Verification of a Predictive Nomogram
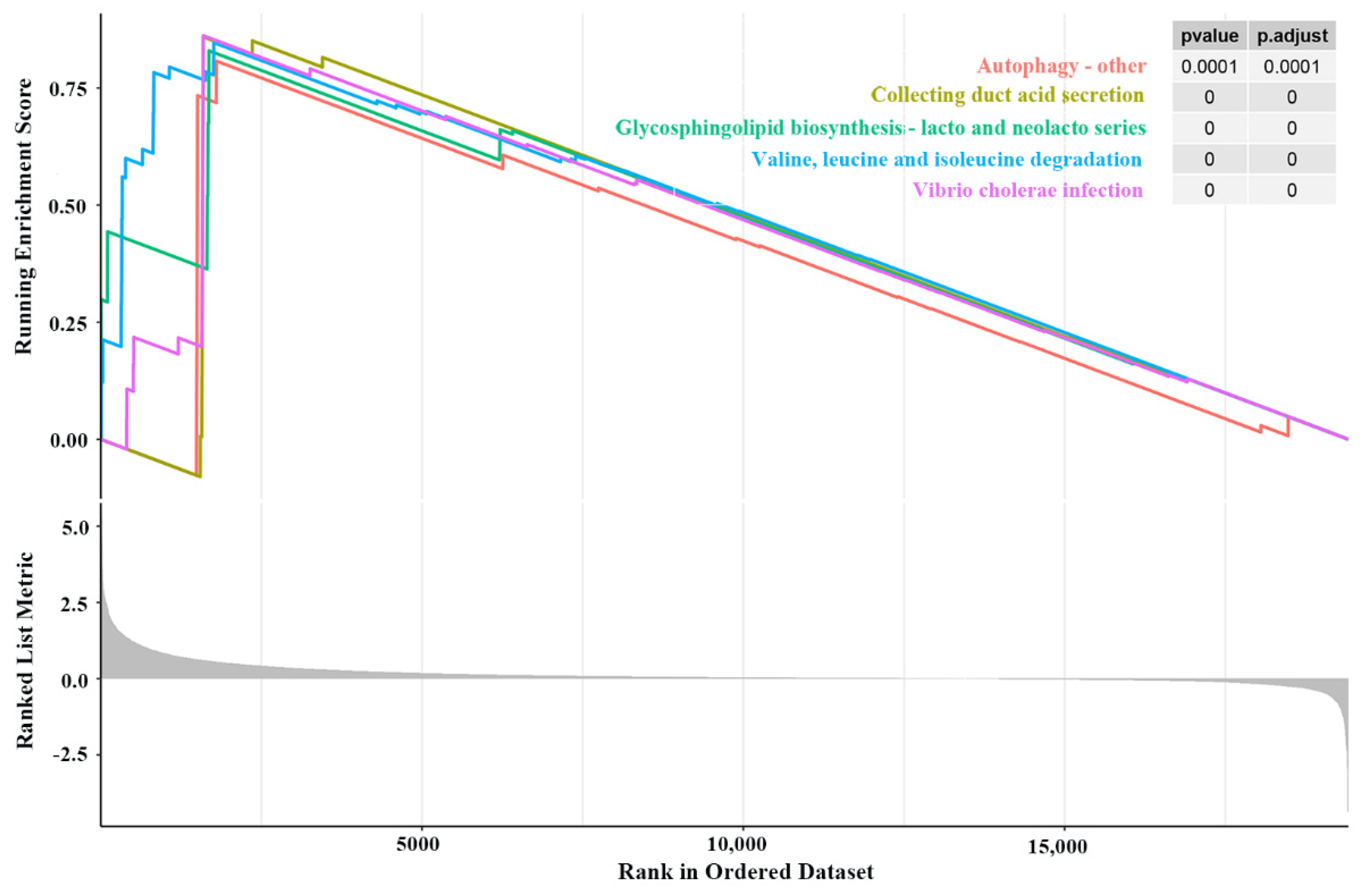
2.5. Gene Set Enrichment Analysis (GSEA)
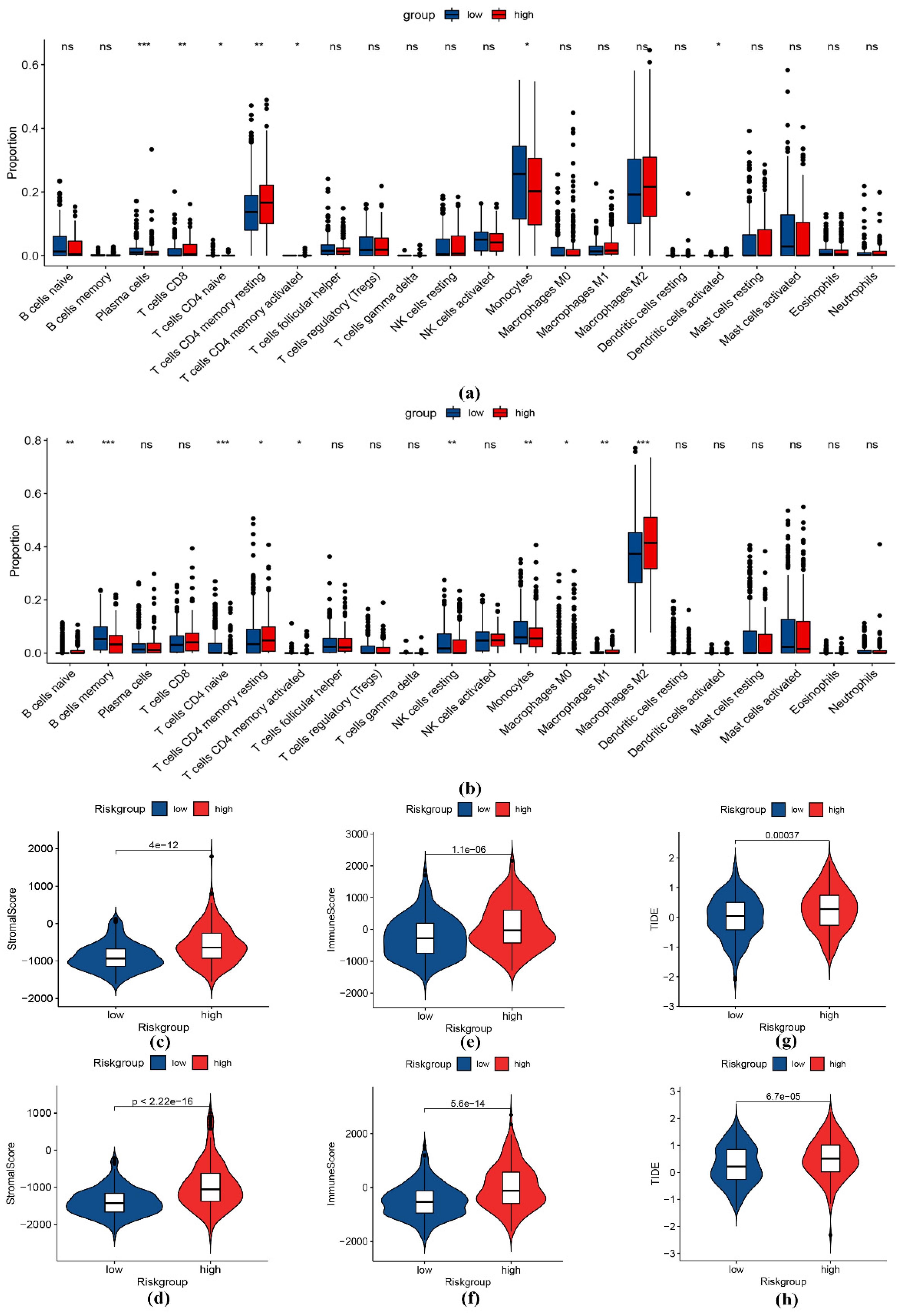
2.6. Immune Infiltration Analysis
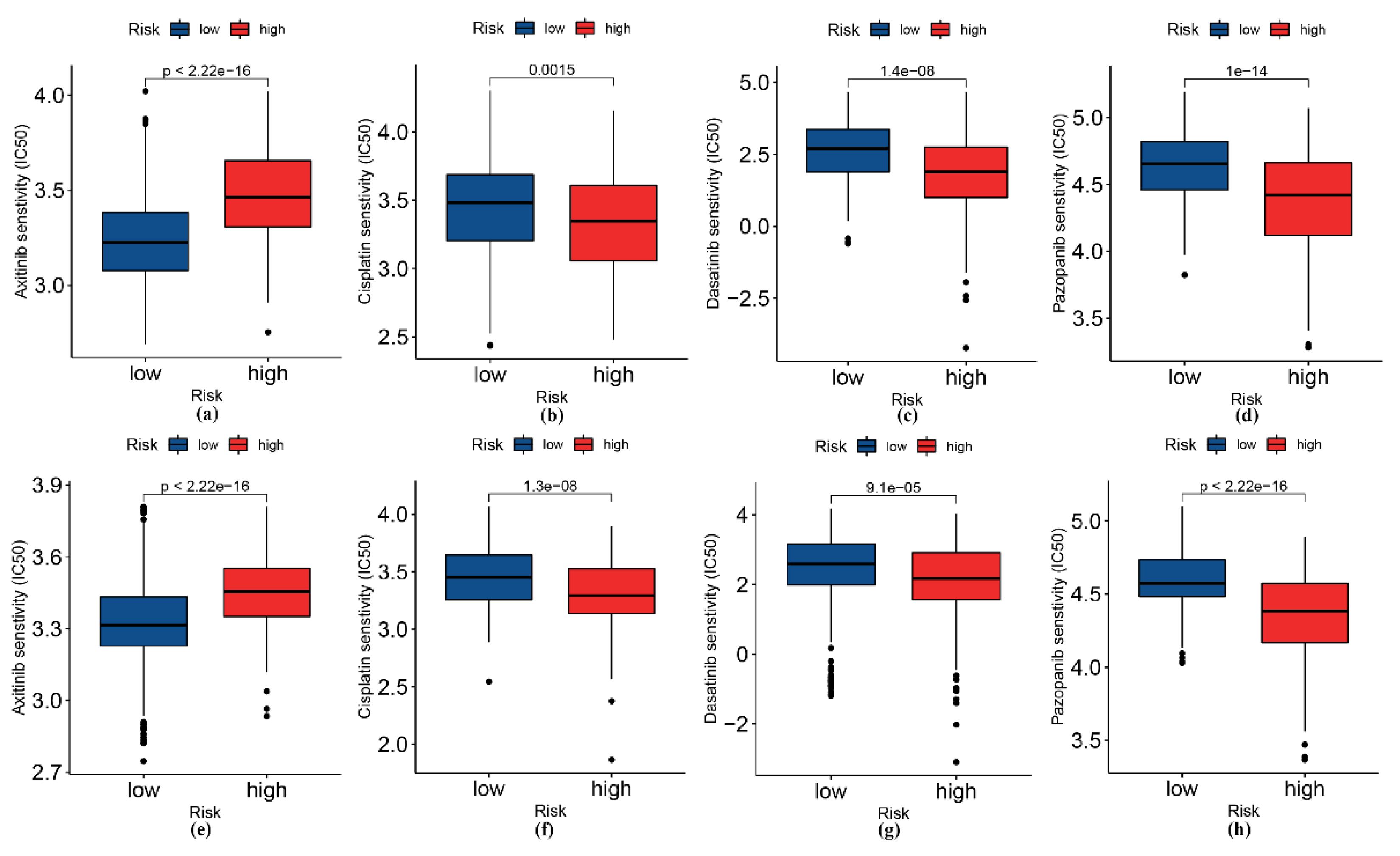
2.7. Drug Sensitivity Analysis
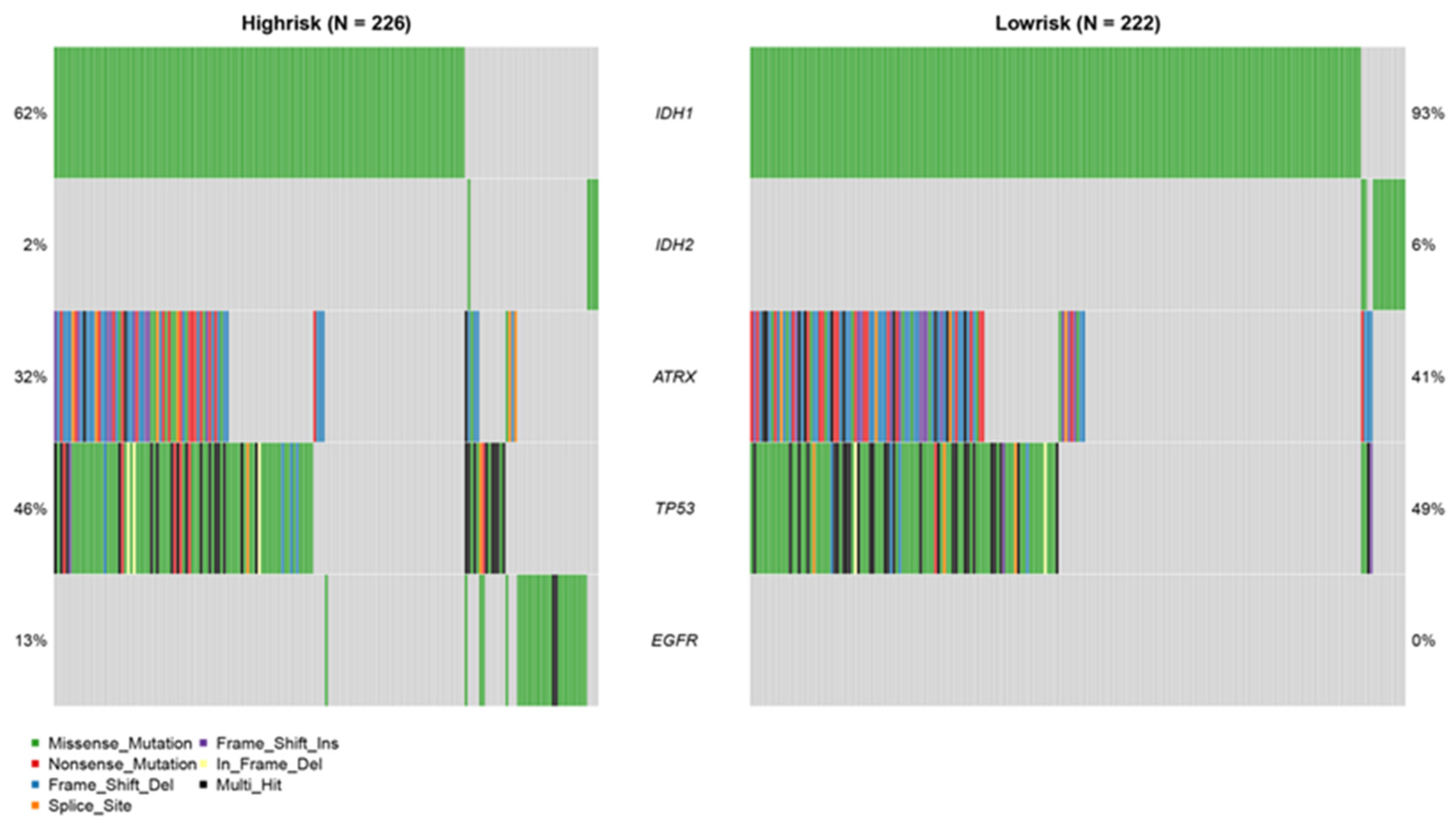
2.8. Mutation Analysis
2.9. Statistical Analysis
3. Results
3.1. Initial Selection of BM-Related Genes
3.2. Construction and Verification of the Risk Score Model
3.3. Establishment and Verification of a Nomogram
3.4. GSEA Findings
3.5. Immune Infiltration Analysis
3.6. Analysis of Patients’ Sensitivity to Selected Drugs
3.7. Mutation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Mair, M.J.; Geurts, M.; van den Bent, M.J.; Berghoff, A.S. A basic review on systemic treatment options in WHO grade II-III gliomas. Cancer Treat. Rev. 2021, 92, 102124. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Ren, X.; Chen, X.; Zhu, C.; Wu, A. Multiomics Profiling and Clustering of Low-Grade Gliomas Based on the Integrated Stress Status. Biomed. Res. Int. 2021, 2021, 5554436. [Google Scholar] [CrossRef]
- Jayadev, R.; Morais, M.; Ellingford, J.M.; Srinivasan, S.; Naylor, R.W.; Lawless, C.; Li, A.S.; Ingham, J.F.; Hastie, E.; Chi, Q.; et al. A basement membrane discovery pipeline uncovers network complexity, regulators, and human disease associations. Sci. Adv. 2022, 8, eabn2265. [Google Scholar] [CrossRef]
- Pozzi, A.; Yurchenco, P.D.; Iozzo, R.V. The nature and biology of basement membranes. Matrix Biol. 2017, 57–58, 1–11. [Google Scholar] [CrossRef]
- Sekiguchi, R.; Yamada, K.M. Basement Membranes in Development and Disease. Curr. Top. Dev. Biol. 2018, 130, 143–191. [Google Scholar] [CrossRef]
- Jayadev, R.; Sherwood, D.R. Basement membranes. Curr. Biol. 2017, 27, R207–R211. [Google Scholar] [CrossRef]
- Leclech, C.; Natale, C.F.; Barakat, A.I. The basement membrane as a structured surface - role in vascular health and disease. J. Cell Sci. 2020, 133, jcs239889. [Google Scholar] [CrossRef]
- Sherwood, D.R. Basement membrane remodeling guides cell migration and cell morphogenesis during development. Curr. Opin. Cell Biol. 2021, 72, 19–27. [Google Scholar] [CrossRef]
- Glentis, A.; Gurchenkov, V.; Matic Vignjevic, D. Assembly, heterogeneity, and breaching of the basement membranes. Cell Adh. Migr. 2014, 8, 236–245. [Google Scholar] [CrossRef]
- Kohrman, A.Q.; Matus, D.Q. Divide or Conquer: Cell Cycle Regulation of Invasive Behavior. Trends Cell Biol. 2017, 27, 12–25. [Google Scholar] [CrossRef]
- Matus, D.Q.; Li, X.Y.; Durbin, S.; Agarwal, D.; Chi, Q.; Weiss, S.J.; Sherwood, D.R. In vivo identification of regulators of cell invasion across basement membranes. Sci. Signal. 2010, 3, ra35. [Google Scholar] [CrossRef]
- Kalluri, R. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer 2003, 3, 422–433. [Google Scholar] [CrossRef]
- Qu, M.; Qiu, B.O.; Xiong, W.; Chen, D.; Wu, A. Expression of a-disintegrin and metalloproteinase 10 correlates with grade of malignancy in human glioma. Oncol. Lett. 2015, 9, 2157–2162. [Google Scholar] [CrossRef]
- Zheng, X.; Jiang, F.; Katakowski, M.; Lu, Y.; Chopp, M. ADAM17 promotes glioma cell malignant phenotype. Mol. Carcinog. 2012, 51, 150–164. [Google Scholar] [CrossRef]
- Yu, H.; Ding, J.; Zhu, H.; Jing, Y.; Zhou, H.; Tian, H.; Tang, K.; Wang, G.; Wang, X. LOXL1 confers antiapoptosis and promotes gliomagenesis through stabilizing BAG2. Cell Death Differ. 2020, 27, 3021–3036. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, A.; Han, X.; Li, Y.; Zhang, Z.; Song, L.; Wang, W.; Lou, M. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas. Cancer Immunol. Immunother. 2022, 71, 645–660. [Google Scholar] [CrossRef]
- Cai, H.; Xue, Y.; Liu, W.; Li, Z.; Hu, Y.; Li, Z.; Shang, X.; Liu, Y. Overexpression of Roundabout4 predicts poor prognosis of primary glioma patients via correlating with microvessel density. J. Neurooncol. 2015, 123, 161–169. [Google Scholar] [CrossRef]
- Yamanaka, R.; Arao, T.; Yajima, N.; Tsuchiya, N.; Homma, J.; Tanaka, R.; Sano, M.; Oide, A.; Sekijima, M.; Nishio, K. Identification of expressed genes characterizing long-term survival in malignant glioma patients. Oncogene 2006, 25, 5994–6002. [Google Scholar] [CrossRef]
- Sun, L.; Hui, A.M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Zhao, Z.; Meng, F.; Wang, W.; Wang, Z.; Zhang, C.; Jiang, T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data 2017, 4, 170024. [Google Scholar] [CrossRef]
- Bao, Z.S.; Chen, H.M.; Yang, M.Y.; Zhang, C.B.; Yu, K.; Ye, W.L.; Hu, B.Q.; Yan, W.; Zhang, W.; Akers, J.; et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014, 24, 1765–1773. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, T.; You, G.; Peng, X.; Chen, C.; You, Y.; Yao, K.; Wu, C.; Ma, J.; Sha, Z.; et al. Localizing seizure-susceptible brain regions associated with low-grade gliomas using voxel-based lesion-symptom mapping. Neuro Oncol. 2015, 17, 282–288. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Qian, Z.; Sun, Z.; Xu, K.; Wang, K.; Liu, S.; Fan, X.; Li, S.; Zhang, Z.; et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin. 2018, 20, 1070–1077. [Google Scholar] [CrossRef]
- Nelson, B.R.; Claes, K.; Todd, V.; Chaverra, M.; Lefcort, F. NELL2 promotes motor and sensory neuron differentiation and stimulates mitogenesis in DRG in vivo. Dev. Biol. 2004, 270, 322–335. [Google Scholar] [CrossRef]
- Williams, M.E.; Lu, X.; McKenna, W.L.; Washington, R.; Boyette, A.; Strickland, P.; Dillon, A.; Kaprielian, Z.; Tessier-Lavigne, M.; Hinck, L. UNC5A promotes neuronal apoptosis during spinal cord development independent of netrin-1. Nat. Neurosci. 2006, 9, 996–998. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Futamura, M.; Kitamura, N.; Nakamura, Y.; Baba, H.; Arakawa, H. Identification of UNC5A as a novel transcriptional target of tumor suppressor p53 and a regulator of apoptosis. Int. J. Oncol. 2010, 36, 1253–1260. [Google Scholar] [CrossRef]
- Midwood, K.S.; Chiquet, M.; Tucker, R.P.; Orend, G. Tenascin-C at a glance. J. Cell Sci. 2016, 129, 4321–4327. [Google Scholar] [CrossRef]
- Ampofo, E.; Schmitt, B.M.; Menger, M.D.; Laschke, M.W. The regulatory mechanisms of NG2/CSPG4 expression. Cell. Mol. Biol. Lett. 2017, 22, 4. [Google Scholar] [CrossRef]
- Benassi, M.S.; Pazzaglia, L.; Chiechi, A.; Alberghini, M.; Conti, A.; Cattaruzza, S.; Wassermann, B.; Picci, P.; Perris, R. NG2 expression predicts the metastasis formation in soft-tissue sarcoma patients. J. Orthop. Res. 2009, 27, 135–140. [Google Scholar] [CrossRef]
- Vannahme, C.; Smyth, N.; Miosge, N.; Gosling, S.; Frie, C.; Paulsson, M.; Maurer, P.; Hartmann, U. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J. Biol. Chem. 2002, 277, 37977–37986. [Google Scholar] [CrossRef]
- Brellier, F.; Ruggiero, S.; Zwolanek, D.; Martina, E.; Hess, D.; Brown-Luedi, M.; Hartmann, U.; Koch, M.; Merlo, A.; Lino, M.; et al. SMOC1 is a tenascin-C interacting protein over-expressed in brain tumors. Matrix Biol. 2011, 30, 225–233. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Movahedi, K.; Laoui, D.; Gysemans, C.; Baeten, M.; Stange, G.; Van den Bossche, J.; Mack, M.; Pipeleers, D.; In’t Veld, P.; De Baetselier, P.; et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010, 70, 5728–5739. [Google Scholar] [CrossRef]
- Griffith, T.S.; Wiley, S.R.; Kubin, M.Z.; Sedger, L.M.; Maliszewski, C.R.; Fanger, N.A. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J. Exp. Med. 1999, 189, 1343–1354. [Google Scholar] [CrossRef]
- Luo, C.; Liu, Z.; Ye, W.; Liu, F. Immune Infiltration-Related Signature Predicts Risk Stratification and Immunotherapy Efficacy in Grade II and III Gliomas. Front. Cell Dev. Biol. 2021, 9, 756005. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, Y.B.; Wu, W.; Zhang, L.; Wang, Z.; Dai, Z.; Feng, S.; Cao, H.; Cheng, Q.; Liu, Z. The molecular feature of macrophages in tumor immune microenvironment of glioma patients. Comput. Struct. Biotechnol. J. 2021, 19, 4603–4618. [Google Scholar] [CrossRef]
- Fan, C.; Du, J.; Liu, N. Identification of a Transcription Factor Signature That Can Predict Breast Cancer Survival. Comput. Math. Methods Med. 2021, 2021, 2649123. [Google Scholar] [CrossRef]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef]
- You, F.; Zhang, C.; Liu, X.; Ji, D.; Zhang, T.; Yu, R.; Gao, S. Drug repositioning: Using psychotropic drugs for the treatment of glioma. Cancer Lett. 2022, 527, 140–149. [Google Scholar] [CrossRef]
- Matteoni, S.; Matarrese, P.; Ascione, B.; Buccarelli, M.; Ricci-Vitiani, L.; Pallini, R.; Villani, V.; Pace, A.; Paggi, M.G.; Abbruzzese, C. Anticancer Properties of the Antipsychotic Drug Chlorpromazine and Its Synergism With Temozolomide in Restraining Human Glioblastoma Proliferation In Vitro. Front. Oncol. 2021, 11, 635472. [Google Scholar] [CrossRef]
- Hosseinimehr, S.J.; Najafi, S.H.; Shafiee, F.; Hassanzadeh, S.; Farzipour, S.; Ghasemi, A.; Asgarian-Omran, H. Fluoxetine as an antidepressant medicine improves the effects of ionizing radiation for the treatment of glioma. J. Bioenerg. Biomembr. 2020, 52, 165–174. [Google Scholar] [CrossRef]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 345. [Google Scholar] [CrossRef]
- Sun, H.; Yin, L.; Li, S.; Han, S.; Song, G.; Liu, N.; Yan, C. Prognostic significance of IDH mutation in adult low-grade gliomas: A meta-analysis. J. Neurooncol. 2013, 113, 277–284. [Google Scholar] [CrossRef]
- Haase, S.; Garcia-Fabiani, M.B.; Carney, S.; Altshuler, D.; Nunez, F.J.; Mendez, F.M.; Nunez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, Y.; Yang, C.; Zhang, X.; Xu, C.; Qiao, X.; Xu, J.; Tian, S.; Fang, C.; Kang, C. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol. Med. 2019, 16, 784–796. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Han, B.; Cai, J.; Gao, W.; Meng, X.; Gao, F.; Wu, P.; Duan, C.; Wang, R.; Dinislam, M.; Lin, L.; et al. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett. 2018, 419, 280–290. [Google Scholar] [CrossRef]
- Hao, Z.; Guo, D. EGFR mutation: Novel prognostic factor associated with immune infiltration in lower-grade glioma; an exploratory study. BMC Cancer 2019, 19, 1184. [Google Scholar] [CrossRef]
- Binder, Z.A.; Thorne, A.H.; Bakas, S.; Wileyto, E.P.; Bilello, M.; Akbari, H.; Rathore, S.; Ha, S.M.; Zhang, L.; Ferguson, C.J.; et al. Epidermal Growth Factor Receptor Extracellular Domain Mutations in Glioblastoma Present Opportunities for Clinical Imaging and Therapeutic Development. Cancer Cell 2018, 34, 163–177.e167. [Google Scholar] [CrossRef]
- Xu, L.; Nirwane, A.; Yao, Y. Basement membrane and blood-brain barrier. Stroke Vasc. Neurol. 2019, 4, 78–82. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef]
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Chintala, S.K.; Gokaslan, Z.L.; Go, Y.; Sawaya, R.; Nicolson, G.L.; Rao, J.S. Role of extracellular matrix proteins in regulation of human glioma cell invasion in vitro. Clin. Exp. Metastasis 1996, 14, 358–366. [Google Scholar] [CrossRef]
- Xu, X.; Ban, Y.; Zhao, Z.; Pan, Q.; Zou, J. MicroRNA-1298-3p inhibits proliferation and invasion of glioma cells by downregulating Nidogen-1. Aging 2020, 12, 7761–7773. [Google Scholar] [CrossRef]
- Howe, M.D.; Furr, J.W.; Munshi, Y.; Roy-O’Reilly, M.A.; Maniskas, M.E.; Koellhoffer, E.C.; d’Aigle, J.; Sansing, L.H.; McCullough, L.D.; Urayama, A. Transforming growth factor-beta promotes basement membrane fibrosis, alters perivascular cerebrospinal fluid distribution, and worsens neurological recovery in the aged brain after stroke. Geroscience 2019, 41, 543–559. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Lai, G.; Sun, L. Basement-Membrane-Related Gene Signature Predicts Prognosis in WHO Grade II/III Gliomas. Genes 2022, 13, 1810. https://doi.org/10.3390/genes13101810
Zhang Z, Lai G, Sun L. Basement-Membrane-Related Gene Signature Predicts Prognosis in WHO Grade II/III Gliomas. Genes. 2022; 13(10):1810. https://doi.org/10.3390/genes13101810
Chicago/Turabian StyleZhang, Zhaogang, Guichuan Lai, and Lingling Sun. 2022. "Basement-Membrane-Related Gene Signature Predicts Prognosis in WHO Grade II/III Gliomas" Genes 13, no. 10: 1810. https://doi.org/10.3390/genes13101810
APA StyleZhang, Z., Lai, G., & Sun, L. (2022). Basement-Membrane-Related Gene Signature Predicts Prognosis in WHO Grade II/III Gliomas. Genes, 13(10), 1810. https://doi.org/10.3390/genes13101810







