Abstract
Sugar plays an important role in regulating the flowering of plants. However, studies of genes related to flowering regulation by the sugar pathway of Brassicaceae plants are scarce. In this study, we performed a comprehensive comparative genomics analysis of the flowering genes in the sugar pathway from seven members of the Brassicaceae, including: Arabidopsis thaliana, Arabidopsis lyrata, Astelia pumila, Camelina sativa, Brassica napus, Brassica oleracea, and Brassica rapa. We identified 105 flowering genes in the sugar pathway of these plants, and they were categorized into nine groups. Protein domain analysis demonstrated that the IDD8 showed striking structural variations in different Brassicaceae species. Selection pressure analysis revealed that sugar pathway genes related to flowering were subjected to strong purifying selection. Collinearity analysis showed that the identified flowering genes expanded to varying degrees, but SUS4 was absent from the genomes of Astelia pumila, Camelina sativa, Brassica napus, Brassica oleracea, and Brassica rapa. Tissue-specific expression of ApADG indicated functional differentiation. To sum up, genome-wide identification revealed the expansion, contraction, and diversity of flowering genes in the sugar pathway during Brassicaceae evolution. This study lays a foundation for further study on the evolutionary characteristics and potential biological functions of flowering genes in the sugar pathway of Brassicaceae.
1. Introduction
Polyploidy events have occurred frequently during plant speciation [1]. Dicotyledons experienced the (γ) event [2,3], and monocotyledons underwent the Tau (τ) event [4,5]. Polyploidy events can enhance the adaptability of plants to tolerate extreme environmental changes [6,7]. As the largest flowering plant family on Earth, the family Brassicaceae has undergone whole-genome duplications (At-α) [8,9], while Brassica L. crops have even undergone an additional genome-wide triploid duplication (Br-β) [10,11]. Comparative genomic analysis has shown that C. sativa is an allohexaploid plant, derived from the hybridization of three ancestral Arabidopsis species, and it has undergone an additional genome-wide triploid duplication [12,13].
Flowering is an important part of the life cycle of angiosperms, affecting the transmission of genetic information, the size of population and the genetic diversity of species [14]. The flowering process is mainly divided into three stages: flower induction, flowering transition, and flower organ differentiation. Flowering is comprehensively affected by both plant internal signals and environmental factors. In Arabidopsis, Bouché et al. [15] systematically identified eight pathways that promote flowering: aging, ambient temperature, circadian clock, hormones, autonomy, photoperiod, vernalization, and sugar pathways. The aging pathway is mainly regulated by the microRNAs (miRNAs) miR156-SPL and miR172-AP2 [16]. The ambient temperature pathway is mainly affected by the interaction between two spliceosome proteins of FLOWERING LOCUS M and SHORT VEGETATIVE PHASE [17]. The response of flowering in plants to light signals requires the interaction between the photoperiod and circadian clock pathways [18]. Studies on the hormone pathways have mainly focused on the gibberellin (GA) signaling pathway. GA not only promotes the development of plant organs, but also induces flowering. However, the effect of GA on different plants is not consistent [19,20]. An autonomous pathway mainly affects flowering through inhibiting the expression of FLOWERING LOCUS C [21]. The research on the vernalization pathway has primarily focused on FLOWERING LOCUS C and FRIGIDA [22].
Research on the sugar pathway and flowering has largely focused on the synthesis and transport of sugars, but the contribution of sugars to flowering regulation is still poorly understood. In the sugar pathway, the genes ADP GLUCOSE PYROPHOSPHORYLASE 1 (ADG1), PHOSPHOGLUCOSE ISOMERASE 1 (PGI1), and PHOSPHOGLUCOMUTASE (PGM1) promote starch synthesis under active photosynthesis [23]. SUCROSE SYNTHASE 4 (SUS4) can also promote starch synthesis [24] and INDETERMINATE DOMAIN 8 (IDD8) regulates the expression of SUS4 [25]. SNF1 KINASE HOMOLOG 10 (SNRK1.1) inactivates IDD8 through protein phosphorylation, thereby delaying flowering [26]. Carbohydrates respond to HEXOKINASE 1 (HXK1) through the SYMPORTER 9 (SUC9) protein as a signaling molecule in the form of sucrose [27], thereby repressing miR156 expression and promoting the flowering transition [28], or after conversion of sucrose to trehalose-6-phosphate (T6P), with the involvement of TREHALOSE-6-PHOSPHATE SYNTHASE 1 (TPS1) [29].
Although the effect of sugar on flowering time has been studied many times, how sugar regulates flowering time is unclear. Either sugar promotes flowering or represses flowering depends on sucrose concentration [30], localization of invertases [31], and T6P levels [32,33]. In addition, sugar stimulates the expression levels of genes involved in regulating the circadian clock, such as GIGANTEA, TIMING OF CAB EXPRESSION 1, and CIRCADIAN CLOCK ASSOCIATED 1 [34,35]. So, sugar affects the flowering transition in multiple ways. However, the molecular mechanism by which sugars regulate flowering and fruiting in members of the Brassicaceae is still unclear.
To date, the genomes of more than 1000 species, including the model plant A. thaliana, have been sequenced and assembled [36,37,38], including those of other members of the Brassicaceae, such as A. lyrata [39], C. sativa [12], B. napus [40], B. oleracea [41], and B. rapa [42]. A. pumila, an ephemeral plant of the Brassicaceae and a distant relative of A. thaliana [43,44], is mainly distributed in the deserts of the Junggar Basin in northern Xinjiang in China [45,46]. To analyze its molecular mechanism for adaptation to the arid desert environment, we have performed a transcriptomic study of leaf tissues of A. pumila in response to continuous salt stress [46]. In recent years, although there have been reports of gene members in the sugar pathway that regulate flowering, there has been little research conducted on the identification and comparative genomic analysis of gene members of this pathway regulating the transition to flowering in the Brassicaceae, especially in the recently sequenced species. In this study, we performed genome-wide identification of flowering genes in the sugar pathway from seven Brassicaceae species: A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa. Bioinformatics analyses, such as protein domain, gene structure, and comparative evolution, were carried out to reveal their characteristics during evolution. This study provides a theoretical basis for further study of interspecific phylogeny and variation and the mining of flowering genes in the sugar pathway of Brassicaceae.
2. Materials and Methods
2.1. Plant Materials
Seeds of A. pumila were collected in May 2012 from the southern margin (44°20′ N and 87°46′ E) of the Gurbantunggut Desert in Xinjiang, China, as described by Yang et al. [47]. A. pumila seeds were surface sterilized and planted as described by Jin et al. [48]. Seven-day-old seedlings were transplanted into pots containing peat soil and vermiculite (1:1) and placed in an illuminated incubator at 22 °C under long-day (LD) conditions (16 h light/8 h dark) with 300 μmol m−2 s−1 light intensity. A. pumila tissue samples were collected at different stages, from embryos (at 2 d after sowing), young roots (8 d), mature roots (30 d), rosette leaves (30 d), cauline leaves (60 d), buds (60 d), flowers (65 d), siliques (70 d), and seeds (70 d), for gene expression analysis. Immediately after collection, the samples were snap-frozen with liquid nitrogen and then stored in a −80 °C freezer.
2.2. Identification and Naming of Gene Members
We referred to FLOR-ID to identify flowering gene members in the sugar pathway of A. thaliana (http://www.phytosystems.ulg.ac.be/florid/, accessed on 10 November 2021) [15], and genomic data were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 10 November 2021) (Table S1). A. lyrata and C. sativa genomic data were downloaded from Ensemble Plant (http://plants.ensembl.org/index.html, accessed on 10 November 2021). B. napus, B. oleracea, and B. rapa genomic data were downloaded from BRAD (http://brassicadb.cn/#/, accessed on 10 November 2021). The coding sequence (CDS) and protein sequencer files of A. pumila were provided by our groups. Algorithm-based BLASTP was performed using the amino acid sequences of A. thaliana SUS and SUC proteins as queries in the protein databases of A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa, with an E < 1 × 10−5 and other parameters as default values. The candidate protein sequences were compared with Pfam (http://pfam.xfam.org/, accessed on 27 April 2022) database using HMMER (http://www.hmmer.org/, accessed on 27 April 2022). Identification of the orthologous genes between A. thaliana and the other six species used the Ortho Venn2 web tool (http://www.bioinfogenome.net/OrthoVenn/, accessed on 27 April 2022) for analysis. The flowering gene members in the sugar pathway, the SUS gene family members, and the SUC gene family members of six plant species were identified based on the clustering of homologous genes with those of A. thaliana. Flowering gene members in the sugar pathway were also named according to the homologous genes with A. thaliana. Names of the SUS and SUC gene family members are listed in Tables S2 and S3. The physicochemical properties of the protein encoded were analyzed by the ProtParam tool of ExPASy (https://web.expasy.org/protparam/, accessed on 28 April 2022). The subcellular localization of flowering proteins was predicted by Cell-PLOC 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/, accessed on 28 April 2022).
2.3. Phylogenetic Tree Construction, Gene Structure, and Protein Motif Analysis
The ClusterW program [49] was used to perform multiple sequence alignments between the flowering protein sequences in the sugar pathway of A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa with default parameters. MEGAX [50] was used to construct Neighbor-Joining phylogenetic trees and to explore the evolutionary relationship between sugar pathway flowering genes among different species. The bootstraps test was carried out with 1000 iterations. The phylogenetic trees were visualized using the Interactive Tree of Life (iTOL) program (https://itol.embl.de/, accessed on 29 April 2022). Based on the genome gff3 files of A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/, accessed on 29 April 2022) visualization server, the distribution of gene exons-introns was analyzed. The conserved motifs of protein were identified using the MEME website (http://meme-suite.org/tools/meme, accessed on 29 April 2022). The motif length range was set to 10–60 amino acid residues, and the maximum number of motif discoveries was set to 10, while other parameters were set to default values.
2.4. Select Pressure and Duplication Type Analysis
MCscanX [51] was used to calculate the collinearity of homologous genes between species, with a threshold of E < 10−5, while its downstream program duplicate_gene_classifier was used to analyze the duplication type of genes. Easy_KaKs program was used to calculate the synonymous substitution ratio (Ks), non-synonymous substitution ratio (Ka), and Ka/Ks ratio; the evolution mode was judged according to the size of the Ka/Ks ratio [52]. Tbtools [53] was used to visualize the collinearity relationship among gene members of A. thaliana and the other six species.
2.5. Gene Expression Analysis and Quantitative Real-Time-PCR (qPCR) Validation
A. pumila transcriptome sequencing data were downloaded from the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject, accessed on 8 June 2021) (Table S4), and the transcriptome accession number of the nine tissues was PRJNA721579. Sequencing samples were collected from the following tissues of A. pumila: embryos (2 d after sowing), young roots (8 d), mature roots (30 d), rosette leaves (30 d), cauline leaves (60 d), buds (60 d), flowers (65 d), siliques (70 d), and seeds (70 d).
The fastp tools were used to filter and compare sequencing data [54], and the comparison was achieved using the Bowtie2 tool [55]; parameters are set to default values. The results were standardized using the fragments per kilobase of transcript per million mapped reads (FPKM) of a gene. After the FPKM value was converted by Log2(FPKM + 1), a heatmap was created using the R package ‘Pheatmap’ (https://CRAN.R-project.org/package=pheatttmap, accessed on 1 May 2022), and the expression of flowering genes in the sugar pathway among different tissue was analyzed.
qPCR primers were designed according to the gene CDS sequence, and the ApGADPH gene was used as the internal reference gene (Table S5) [48]. RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) was used to extract RNA. M-MLV RTase cDNA Synthesis Kit (Takara, Dalian, China) was used to reverse transcription. RNA extraction and reverse transcription followed the methods of Huang et al. [45]. qPCR was carried out with the SYBR Green PCR Master Mix system (Takara) on an ABI ViiA7 real-time fluorescence quantitative PCR machine (Life Technologies, New York, NY, USA). The procedure of qPCR and the analysis method of experimental data were carried out by referring to the method of Jin et al. [48]. Three biological replicates were performed, with RNA isolated independently, and each qPCR reaction had three replicates.
2.6. Protein Interaction Network Prediction
The Ortho Venn2 tool (http://www.bioinfogenome.net/OrthoVenn/, accessed on 27 April 2022) was used to identify the orthologous genes between A. pumila and A. thaliana. Then, interaction networks of A. pumila flowering genes in the sugar pathway were identified based on the orthologous genes between A. pumila and A. thaliana, using STRING (https://string-db.org/, accessed on 3 May 2022) databases, and the predicted protein-protein interaction network was displayed Cytoscape software [56].
3. Result
3.1. Homology-Based Identification of Flowering Genes in the Sugar Pathway in Seven Species of the Brassicaceae and their Evolution
Genome-wide analyses identified 105 genes from seven species of the Brassicaceae associated with flowering in the sugar pathway, based on their homology with the nine known genes of this pathway in A. thaliana. These included 8 in A. lyrata, 16 in A. pumila, 22 in C. sativa, 24 in B. napus, 13 in B. oleracea, and 13 in B. rapa (Table S6), the number of homologs of each A. thaliana gene varying between the species (Figure 1). The highest numbers of homologs were for genes SNRK1.1 and ADG1, whereas SUS4 and SUC9 exhibited the fewest homologs (Figure S1). Phylogenetic analysis of the SUS and SUC gene families in the Brassicaceae showed that SUS4 was absent from all species except A. thaliana and A. lyrata, while only A. thaliana contained the SUC9 gene (Figure S2). Phylogenetic analysis also revealed that, with the exception of ADG1, SNRK1.1, and PGI1, the sugar pathway gene members formed two well-separated clusters, the first composed of sequences from A. thaliana, A. lyrata, A. pumila, and C. sativa and the second of sequences from B. napus, B. oleracea, and B. rapa (Figure 1 and Figure 2A). The physicochemical properties and subcellular localization details of their corresponding proteins are summarized in Table S6.
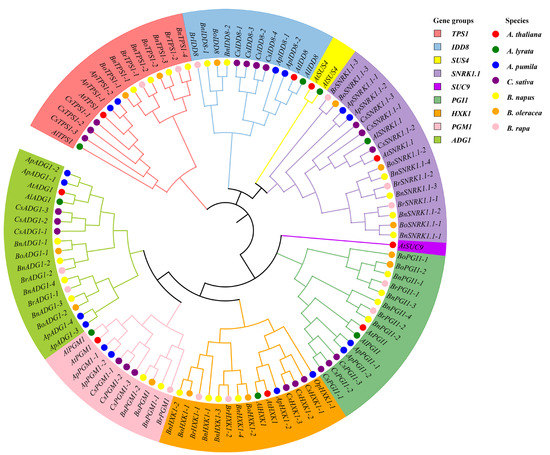
Figure 1.
Neighbor-joining phylogenetic tree of the flowering genes in the sugar pathway from seven members of Brassicaceae.
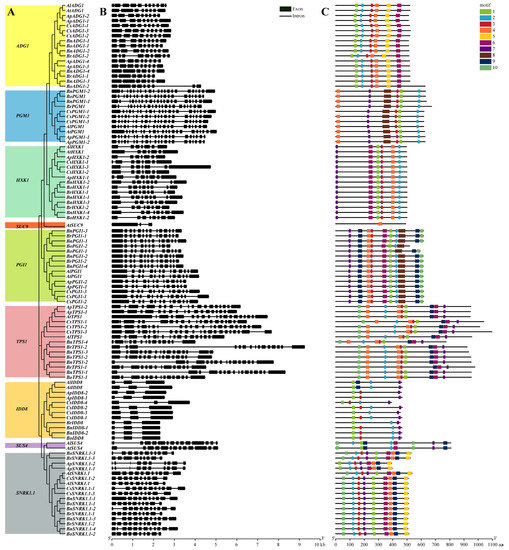
Figure 2.
Phylogenetic tree, exon-intron structures, and motif distributions of the flowering genes in the sugar pathway in A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa. (A) Phylogenetic tree; (B) exon-intron distribution; (C) distribution characteristics of the conserved motifs; aa, amino acid.
3.2. Gene Structure and Protein Conserved Protein Motifs
Gene structure comparisons showed that the flowering genes in the sugar pathway were relatively highly conserved but that there were substantial differences in the number of exons in different genes (Figure 2B), ranging from three in IDD8 to 22 in PGM1. The number of exons in PGM1, SUC9, IDD8, and SUS4 genes in the seven species studied was conserved among the species, whereas the number of exons in other genes showed species-related differences. For example, AtHXK1, AlHXK1, ApHXK1, and CsHXK1 all contain seven exons, while BnHXK1, BoHXK1, and BrHXK1 contain eight or nine exons. Based on the differences in exon number, the seven species of the Brassicaceae could be broadly clustered into two groups, one consisting of A. thaliana, A. lyrata, A. pumila, and C. sativa, and the other consisting of B. napus, B. oleracea, and B. rapa.
Comparative analysis of motif structure showed that most proteins of each species are highly conserved (Figure 2C). The distributions and numbers of motifs in ADG1 and HXK1 of the seven species are completely consistent, but differences were observed in other proteins. Relative to the majority consensus protein structures in the seven species, CsPGM1-1 has no motif 4; BoPGI1-1 has no motif 2; BoPGI1-2 lacks both motif 9 and motif 10; BrTPS1-1 lacks motif 1; ApSNRK1.1 shows a loss of motif 10 and an altered position for motif 1. The IDD8 homologs displayed the most striking differences between the species, involving loss or gain of motifs with substantial differences in inter-motif distances.
3.3. Collinear Relationship of Flowering Gene Members
To investigate the contraction and expansion of flowering gene members in the sugar pathway during evolution, the relationship of orthologous genes between A. thaliana and six species of the Brassicaceae was explored using MCScanX software [51]. The gene members with collinear relationships in each plant species were only distributed on some chromosomes (Figure 3). A. thaliana has 8, 14, 20, 20, 11, and 12 collinear genes with A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa, respectively (Table S7). Except for SUC9, the eight flowering genes of A. thaliana show collinearity with A. lyrata (Figure 3A). In A. pumila, except for the two missing genes, namely AtSUS4 and AtSUC9, each of the other seven genes has two collinear gene pairs with A. thaliana. In C. sativa, the IDD8 gene has two collinear gene pairs with A. thaliana, whereas each of the other six flowering genes of the sugar pathway has three collinear gene pairs with A. thaliana. The comparison shows that B. oleracea and B. rapa showed the same number of homologs of their A. thaliana counterparts (minus SUS4 and SUC9), three for SNRK1.1, two for TPS1 and HXK1, and one for each of the others (Figure 3B). However, in PGI1, only one collinear gene pair occurred between A. thaliana and B. oleracea, with two collinear gene pairs between A. thaliana and B. rapa (Figure 3B). Compared with A. thaliana, there were four collinear gene pairs of TPS1, SNRK1.1, PGI1, and HXK1, two of IDD8, and one each of ADG1 and PGM1 in B. napus. Overall, the number of flowering genes in B. napus was approximately three times that of A. thaliana and twice that of B. oleracea or B. rapa (Figure 3B).
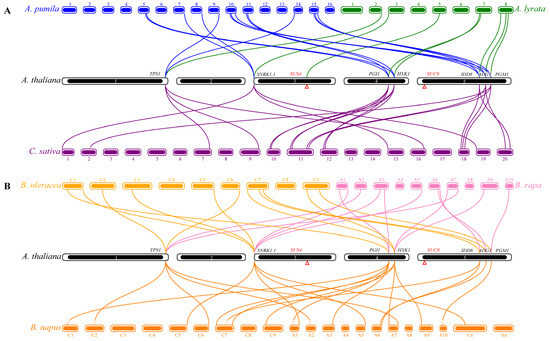
Figure 3.
Syntenic relationships of sugar pathway genes associated with flowering in A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa. Different lines represent collinear gene pairs between different species. Blocks of different colors represent the chromosomes of different plant species. Red triangles in chromosomes 4 and 5 of A. thaliana represent the locations of SUS4 and SUC9 genes. (A) Syntenic relationships of sugar pathway genes associated with flowering in A. thaliana, A. lyrata, A. pumila, and C. sativa; (B) Syntenic relationships of sugar pathway genes associated with flowering in A. thaliana, B. napus, B. oleracea, and B. rapa.
3.4. Selection Pressure
The Ka/Ks ratios of the homologous genes between A. thaliana and each of the other six plant species are less than 1 (Figure 4), indicating that these homologous genes are subject to strong purifying selection. The average Ka/Ks value of the TPS1 collinear gene pairs was the smallest, indicating that they suffer from relatively strong purifying selection, whereas the average Ka/Ks value of the IDD8 gene pair was the highest, indicating a weaker purifying selection (Table S7). However, only A. thaliana and A. lyrata had Ka/Ks values for SUS4 genes. The Ka/Ks values for TPS1, HXK1, and IDD8 genes pairs were similar between the species, while the Ka/Ks values for SNRK1.1, PGI1, ADG1, and PGM1 gene pairs exhibited clearly larger variations among the species, suggesting that these genes may have undergone functional divergence during species evolution.
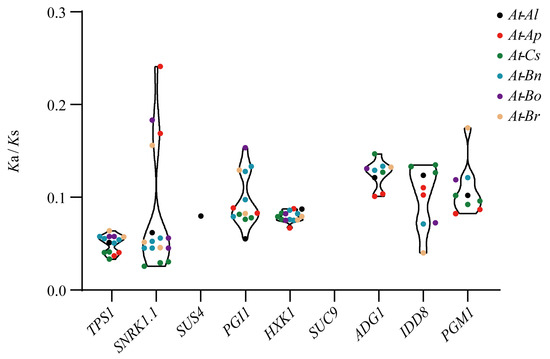
Figure 4.
Ka/Ks values exhibited by flowering genes in the sugar pathway among species. At, A. thaliana; Al, A. lyrata; Ap, A. pumila; Cs, C. sativa; Bn, B. napus; Bo, B. oleracea; Br, B. rapa.
3.5. Gene Duplication Type
The duplication types of the 105 genes in the seven species of Brassicaceae were determined. The results showed that the duplication types of these genes could be divided into three types: whole-genome duplication/segmental duplication (WGD/S), dispersed duplication, and tandem duplication (Table 1). WGD/S events in the seven species, namely A. thaliana, A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa, were 6 (66.7%), 4 (50%), 18 (100%), 24 (96%), 9 (85.7%), 10 (69.2%), and 24 (79.6%), respectively. To summarize, with regard to flowering genes of the sugar pathway, the main gene duplication events in A. lyrata were dispersed duplication and WGD/S, whereas the main process underlying gene duplications in the other six species was WGD/S.

Table 1.
Types of duplication of the flowering genes in the sugar pathway in the seven species of Brassicaceae.
3.6. Tissue Expression Profiles of the Flowering Genes in the Sugar Pathway in A. pumila
Using the RNA-Seq data from nine tissues of A. pumila, the tissue expression heatmap of 16 A. pumila genes was constructed (Figure 5A). The results showed that most genes showed a trend of high expression in all tissues at the early stages of growth, whereas the number of genes with high expression at the late stage of growth was relatively small. Moreover, the expression patterns of most gene duplicates were the same. For example, ApADG1-3/4 and ApPGM1-1/2 were both highly expressed in the rosette and cauline leaves, ApPGI1-1/2 and ApIDD8-1/2 were highly expressed in young and mature roots, and ApSNRK1.1-1/2 was highly expressed in both flowers and siliques (Figure 5). However, some duplicate gene pairs were differentially expressed in the same tissue. For example, ApADG1-1 was highly expressed in siliques, whereas ApADG1-2 expression was highest in embryos, and ApAHX1.2 was more highly expressed in the rosette and cauline leaves than ApHXK1-1.
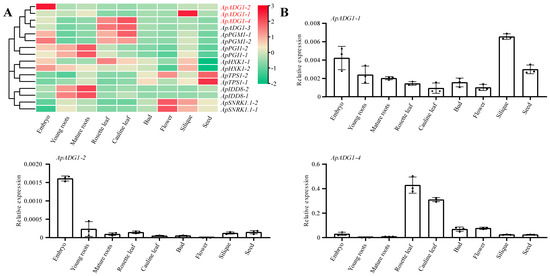
Figure 5.
Tissue expression profiles of the flowering genes in the sugar pathway in A. pumila. (A) Tissue expression heatmap of flowering genes; (B) qPCR analyses of the relative gene expression levels of ApADG1 homologs in different tissues. The data points represented mean ± SD.
qPCR was further used to validate the expression determined from transcriptomics of ApADG1-1/2/4 in different tissues (Figure 5B). The results from the two methods consistently confirmed the accuracy of the transcriptomic data.
3.7. Protein Interaction Network
In order to better understand the biological function of the sugar pathway proteins, we next predicted the interaction network of the proteins associated with flowering regulation in the sugar pathway. Arabidopsis PGM2 and PGM3 proteins can interconvert glucose-6-phosphate (G6P) and glucose-1-phosphate (G1P) (Figure 6). The Arabidopsis pgm2 pgm3 double mutant has been reported to markedly damage the function of male and female gametophytes [57]. ENOLASE 2 (ENO2) maintains normal growth, development, and successful reproduction in plants and is also able to help plants respond to abiotic stresses such as drought and cold [58,59,60]. ENOLASE 1 (ENO1) plays a crucial role in the development of gametophytes and sporophytes [61]. TRIOSE PHOSPHATE ISOMERASE (TPI) promotes the transition of plants from heterotrophic growth to autotrophic growth [62]. The Arabidopsis snf4 mutation disrupts the interaction of SNF4 with SNRK1 and thus affects the role of SNRK1 in the T6P-mediated signaling pathway [63].
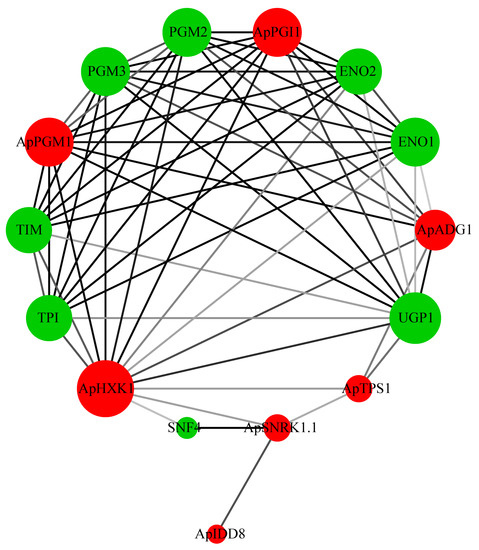
Figure 6.
Interaction network diagram of flowering regulatory proteins in the sugar pathway. The darkness of the lines is proportional to the combined_score (between 0 and 1, where the higher the value, the greater the reliability of the predicted interaction between the two proteins). The node size increases with the number of interacting proteins. Red circles are proteins that are members of the A. pumila sugar pathway; green circles are interacting with A. thaliana proteins.
4. Discussion
In this study, an evolutionary analysis of those genes of the sugar pathway that were also involved in the regulation of flowering in seven species in the Brassicaceae revealed that a total of nine genes were identified: TPS1, SNRK1.1, SUS4, PGI1, HXK1, SUC9, IDD8, ADG1, and PGM1 (Figure 1). Analysis showed a large number of genes in SNRK1.1 and ADG1 (Figure S1), indicating that these contributed largely to the gene expansion in the sugar pathway observed in the Brassicaceae species studied, other than A. thaliana and A. lyrata. In contrast, SUS4 and SUC9 were found only in A. thaliana and A. lyrata (Figure 1). However, SUS1 and SUS4 in A. thaliana both belong to the SUS I subfamily and have very similar gene structures and amino acid sequences [64]. Therefore, we speculate that the loss of function caused by the deletion of SUS4 in these five plant species may be complemented by the increased expression of SUS1, a hypothesis that needs to be further tested. In the SUC gene family, the six Brassicaceae species share common ancestors in AtSUC6, AtSUC7, AtSUC8, and AtSUC9 of the SUT1 subfamily [65]. In A. thaliana, both AtSUC8 and AtSUC9 were expressed in flowers, and the corresponding loss-of-function mutants Atsuc8 and Atsuc9 did not differ from the wild type in terms of flowering time and rosette size [66]. It is suggested that there may be genes that functionally substitute for or exhibit functional redundancy with AtSUC9 in A. lyrata, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa, but with different evolutionary branches from that of A. thaliana (Figure S2B).
The number of exons in each sugar pathway gene was largely similar in each of the plant species, indicating that these genes were relatively conserved during their evolution/expansion in the Brassicaceae. However, the organization and loss/gain of IDD8 motifs was seen to vary greatly between the different species, suggesting that IDD8 may have undergone neofunctionalization during evolution (Figure 2). The sugar pathway genes of the seven species were shown to have been subjected to strong purifying selection, which further supports the finding that these genes were highly conserved during evolution (Figure 4, Table S7).
WGD events have been observed to occur frequently during plant evolution [67,68]. In the current study, we found that the number of members of the sugar pathway genes had expanded in some Brassicaceae species. We identified eight, 16, 13, 13, 24, and 22 flowering genes in the sugar pathway from the genomes of A. lyrata, A. pumila, B. oleracea, B. rapa, B.napus, and C. sativa, respectively. After removing the missing genes SUS4 and SUC9 from consideration, the number of the flowering genes in the sugar pathway in these six spp. is 1.0, 2.3, 1.9, 1.9, 3.4, and 3.1 times that of A. thaliana, respectively. The results showed that, with the occurrence of WGD events, the number of flowering genes in the sugar pathway had also increased. In A. lyrata, the main duplication type of the flowering genes was dispersed duplication, which is different from that in A. thaliana, A. pumila, C. sativa, B. napus, B. oleracea, and B. rapa, with WGD/S as the main duplications (Table 1), indicating that WGD/S played a leading role in the expansion of flowering genes in the sugar pathway of the Brassicaceae.
The genome size of A. lyrata is about twice that of A. thaliana [36,39], but the number of genes identified in these two species is similar. The number of genes identified in C. sativa is about three times that of A. thaliana, indicating a triploidy event relative to A. thaliana. This finding is consistent with the research results of Kagale et al. [12]. However, the number of genes in the sugar pathway of B. napus is inconsistent with the fact that this species is an allotetraploid [69], and the numbers of genes in B. oleracea and B. rapa are also inconsistent with the current evidence that they have experienced ancient triploidy events [10,11]. The results of collinearity analysis showed that the number of BoSNRK1.1 and BrSNRK1.1 genes was three times that of AtSNRK1.1, whereas other genes showed no three-fold greater number than that in A. thaliana, indicating that flowering genes in the sugar pathway of B. oleracea and B. rapa may have been lost after the triploidy event (Figure 3). This result indicated that SNRK1.1 plays an important role in the sugar metabolism of B. oleracea and B. rapa, which is in agreement with the previous findings of A. thaliana [70]. There is no six-fold relationship in the number of any of the flowering regulatory genes between B. napus and A. thaliana, which also indicates that some of the duplicated flowering genes in the sugar pathway of B. napus were lost after the duplication event (Figure 3). B. napus is formed by the natural crossing of B. rapa (AA) and B. oleracea (CC) and subsequent tetraploidy [69]. Furthermore, 11 and 9 sugar pathway genes were located in the AA and the CC subgenome, respectively, suggesting that they played a similarly vital role in both ancestral species.
The diversity of gene expression patterns in different tissues suggests that flowering genes in the sugar pathway are involved in many aspects of the growth and development of A. pumila in addition to regulating flowering transition (Figure 5). For example, ApPGI1-1/2 and ApIDD8-1/2 were specifically expressed in roots during vegetative growth, a finding that is consistent with the trend of high expression of AtPGI and ApIDD8 in roots in Arabidopsis DNA microarray data (http://www.genevestigator.com, accessed on 30 May 2022). The result indicated that the functions of ApPGI1-1/2 and ApIDD8-1/2 are similar to those of AtPGI1 and AtIDDD8 [23,26]. In addition, some orthologous genes showed different expression profiles in the same tissues (Figure 5), indicating that, although there are certain functional similarities between the orthologous genes, functional differentiation has occurred during evolution. For example, ApADG1-1 is highly expressed in siliques, while ApADG1-2 is highly expressed in embryos. Similarly, ApADG1-4 is highly expressed in the rosette and cauline leaves, compared with ApADG-1/2 (Figure 5). In A. thaliana, ADG1 promotes the transition from a vegetative-to-reproductive phase of development [71], indicating that ApADG1-1/2 underwent functional differentiation during evolution. ApADG1-2 may promote seed germination, whereas ApADG1-1 appears to promote the maturation of siliques, a finding that needs further experimental verification.
Sugar distribution is the basis of plant growth and development and also plays an important role in flowering transition [72]. The analysis of protein interactions revealed that, in A. pumila, the proteins interacting with flowering regulatory proteins are mainly functional in the metabolism of carbohydrates and signaling, suggesting that the distribution of sugars plays key roles in plant growth and development, flowering transition, and tolerance of abiotic stresses (Figure 6). Four of these proteins, namely ApPGI1, ApPGM1, ApHXK1, and ApADG1, were located at the core of the protein interaction network, suggesting that they may be involved in the transcriptional regulation of more genes and hence in more metabolic regulation in plants (Figure 6).
In summary, we comprehensively identified the flowering gene members of the sugar pathway from seven Brassicaceae species and analyzed their structural characteristics and evolutionary characteristics. The results provide clues to further exploring the molecular mechanism(s) underlying the regulation of flowering by sugar metabolism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13101749/s1. Supplementary Materials contained Table S1: Genes related to the flowering transition in the sugar pathway in A. thaliana; Table S2: Names of SUS gene family members in seven Brassicaceae species; Table S3: Names of members of the SUC gene family members in seven Brassicaceae species; Table S4: Accession numbers of RNA-Seq data of A. pumila nine different tissues; Table S5: Primer sequences used in the qPCR assay; Table S6: Information on the identified sugar pathway gene members identified to be associated with flowering from seven Brassicaceae species; Table S7: The non-synonymous (Ka) and synonymous (Ks) of orthologous gene pairs of the sugar pathway members in seven Brassicaceae spp.; Figure S1: The number of the sugar pathway gene members associated with flowering from seven Brassicaceae species; Figure S2A: Phylogenetic trees of SUS gene family; Figure S2B: Phylogenetic trees of SUC gene family.
Author Contributions
X.H. designed the work; Y.Z. carried out the data analysis and wrote the original manuscript. Q.Z. carried out the qPCR analysis; H.A. contributed to analysis of experimental data; X.H. and T.F. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (NSFC) (grant numbers U1303302 and 31060149) and Anhui Science and Technology University Discipline Leader Introduction Talent Start-up Funds (grant number NXYJ202001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Flowering gene members in the sugar pathway of A. thaliana were downloaded from FLOR-ID (http://www.phytosystems.ulg.ac.be/florid/, accessed on 10 November 2021). A. thaliana genomic data were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 10 November 2021). A. lyrata and C. sativa genomic data were downloaded from Ensemble Plant (http://plants.ensembl.org/index.html, accessed on 10 November 2021). B. napus, B. oleracea, and B. rapa genomic data were downloaded from BRAD (http://brassicadb.cn/#/, accessed on 10 November 2021). A. pumila transcriptome sequencing data were downloaded from the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject, accessed on 8 June 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, K.; Wang, X.W.; Cheng, F. Plant polyploidy: Origin, evolution, and its influence on crop domestication. Acta Hortic. 2019, 5, 231–239. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Jiao, Y.N.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.B.; Bowers, J.E.; Wang, X.Y.; Paterson, A.H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 2010, 107, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.N.; Li, J.P.; Tang, H.B.; Paterson, A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.F.; van den Bergh, E.; Zeng, P.; Zhong, X.; Xu, J.J.; Liu, X.; Hofberger, J.; de Bruijn, S.; Bhide, A.S.; Kuelahoglu, C.; et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 2013, 25, 2813–2830. [Google Scholar] [CrossRef]
- Nikolov, L.A. Brassicaceae flowers: Diversity amid uniformity. J. Exp. Bot. 2019, 70, 2623–2635. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.C.; Pedersen, B.; Freeling, M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006, 16, 934–946. [Google Scholar] [CrossRef]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, N.; Wolf, E.M.; Lysak, M.A.; Koch, M.A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 2015, 27, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, 1167–1171. [Google Scholar] [CrossRef]
- Hyun, Y.; Richter, R.; Coupland, G. Competence to flower: Age-controlled sensitivity to environmental cues. Plant Physiol. 2017, 173, 36–46. [Google Scholar] [CrossRef]
- Posé, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef]
- Yanovsky, M.J.; Kay, S.A. Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 2001, 4, 429–435. [Google Scholar] [CrossRef]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Gottschalk, C.; van Nocker, S. Genetic mechanisms in the repression of flowering by gibberellins in apple (Malus × domestica Borkh.). BMC Genom. 2019, 20, 747. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 2004, 7, 570–574. [Google Scholar] [CrossRef]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis. Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.S.; Lue, W.L.; Wang, S.M.; Chen, J. Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol. 2000, 123, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Baroja-Fernández, E.; Muñoz, F.J.; Li, J.; Bahaji, A.; Almagro, G.; Montero, M.; Etxeberria, E.; Hidalgo, M.; Sesma, M.T.; Pozueta-Romero, J. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. USA 2012, 109, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Ryu, J.; Kang, S.K.; Park, C.M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011, 65, 418–429. [Google Scholar] [CrossRef]
- Jeong, E.Y.; Seo, P.J.; Woo, J.C.; Park, C.M. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 2015, 15, 110. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Reinders, A.; Johnson, M.E.; Krentz, A.D.; Grof, C.P.; Perroux, J.M.; Ward, J.M. Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 2007, 143, 188–198. [Google Scholar] [CrossRef]
- Yang, L.; Xu, M.L.; Koo, Y.; He, J.; Poethig, R.S. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2013, 2, e00260. [Google Scholar] [CrossRef]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef]
- Ohto, M.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef]
- Heyer, A.G.; Raap, M.; Schroeer, B.; Marty, B.; Willmitzer, L. Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. Plant J. 2004, 39, 161–169. [Google Scholar] [CrossRef]
- Gómez, L.D.; Gilday, A.; Feil, R.; Lunn, J.E.; Graham, I.A. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010, 64, 1–13. [Google Scholar] [CrossRef]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef]
- Knight, H.; Thomson, A.J.; McWatters, H.G. Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 2008, 148, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Dalchau, N.; Baek, S.J.; Briggs, H.M.; Robertson, F.C.; Dodd, A.N.; Gardner, M.J.; Stancombe, M.A.; Haydon, M.J.; Stan, G.B.; Gonçalves, J.M.; et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 2011, 108, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Sun, Y.Q.; Shang, L.G.; Zhu, Q.H.; Fan, L.J.; Guo, L.B. Twenty years of plant genome sequencing: Achievements and challenges. Trends Plant Sci. 2022, 27, 391–401. [Google Scholar] [CrossRef]
- Hu, T.T.; Pattyn, P.; Bakker, E.G.; Cao, J.; Cheng, J.F.; Clark, R.M.; Fahlgren, N.; Fawcett, J.A.; Grimwood, J.; Gundlach, H.; et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011, 43, 476–481. [Google Scholar] [CrossRef]
- Chen, X.Q.; Tong, C.B.; Zhang, X.T.; Song, A.X.; Hu, M.; Dong, W.; Chen, F.; Wang, Y.P.; Tu, J.X.; Liu, S.Y.; et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 2021, 19, 615–630. [Google Scholar] [CrossRef]
- Cai, X.; Wu, J.; Liang, J.L.; Lin, R.M.; Zhang, K.; Cheng, F.; Wang, X.W. Improved Brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theor. Appl. Genet. 2020, 133, 3187–3199. [Google Scholar] [CrossRef]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y.; Bai, Y.Q.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. Brassica rapa Genome Sequencing Project Consortium The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.M.; Aan, Z.X.; Zhou, G.L.; Yang, C.Y.; Han, Y.L.; Li, X.Y.; Zhang, Y.F. Flora of Xinjiang; Xinjiang Science and Technology Health Press: Urumqi, China, 1995; Volume 2, Part 2; pp. 145–146. [Google Scholar]
- Koch, M.A. The plant model system Arabidopsis set in an evolutionary, systematic, and spatio-temporal context. J. Exp. Bot. 2019, 70, 55–67. [Google Scholar] [CrossRef]
- Huang, X.Z.; Yang, L.F.; Jin, Y.H.; Lin, J.; Liu, F. Generation, annotation, and analysis of a large-scale expressed sequence tag library from Arabidopsis pumila to explore salt-responsive genes. Front. Plant Sci. 2017, 8, 955. [Google Scholar] [CrossRef]
- Jin, Y.H.; Guo, L.; Liu, D.Q.; Ai, H.; Huang, X.Z. Techniques for the regeneration and genetic transformation of Arabidopsis pumila: An ephemeral plant suitable for investigating the mechanisms for adaptation to desert environments. PCTOC 2022, 150, 237–246. [Google Scholar] [CrossRef]
- Yang, L.F.; Jin, Y.H.; Huang, W.; Sun, Q.; Liu, F.; Huang, X.Z. Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genom. 2018, 19, 717. [Google Scholar] [CrossRef]
- Jin, Y.H.; Liu, F.; Huang, W.; Sun, Q.; Huang, X.Z. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2002, 1, 2–3. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.Q.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 49. [Google Scholar] [CrossRef]
- Nekrutenko, A.; Makova, K.D.; Li, W.H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Egli, B.; Kölling, K.; Köhler, C.; Zeeman, S.C.; Streb, S. Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol. 2010, 154, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Guo, Y.; Ohta, M.; Xiong, L.; Stevenson, B.; Zhu, J.K. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002, 21, 2692–2702. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Zhang, Y.H.; Ma, X.F.; Ye, P.; Gao, F.; Li, X.F.; Zhou, Y.J.; Shi, Z.H.; Cheng, H.M.; Zheng, C.X.; et al. Biological functions of Arabidopsis thaliana MBP-1-like protein encoded by ENO2 in the response to drought and salt stresses. Physiol. Plant 2020, 168, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Rozhon, W.; Yang, S.Q.; Poppenberger, B. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1. Plant J. 2015, 81, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, V.; Löttgert, T.; Geimer, S.; Dörmann, P.; Krüger, S.; Vijayakumar, V.; Schreiber, L.; Göbel, C.; Feussner, K.; Feussner, I.; et al. Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 2010, 22, 2594–2617. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Thelen, J.J. The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 2010, 22, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, V.; Ponnu, J.; Crepin, N.; Langenecker, T.; Hagmann, J.; Skorzinski, N.; Musialak-Lange, M.; Wahl, V.; Rolland, F.; Schmid, M. Impaired KIN10 function restores developmental defects in the Arabidopsis trehalose 6-phosphate synthase1 (tps1) mutant. New Phytol. 2022, 235, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Vaultier, M.N.; Rochat, C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 2004, 55, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Kühn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 13, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Sauer, N.; Ludwig, A.; Knoblauch, A.; Rothe, P.; Gahrtz, M.; Klebl, F. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 2004, 40, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.N.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.Y.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Bouly, J.P.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Grahame, H.D.; Thomas, M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, I.G.; Massiah, A.J.; Thomas, B. Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ. 2013, 36, 1802–1811. [Google Scholar] [CrossRef]
- Choudhary, A.; Kumar, A.; Kaur, N.; Kaur, H. Molecular cues of sugar signaling in plants. Physiol. Plant 2022, 174, 13630. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).