Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize
Abstract
:1. Introduction
2. Results
2.1. Identification, Phylogenetic Analysis, and Chromosomal Localization of Maize SWEET Genes
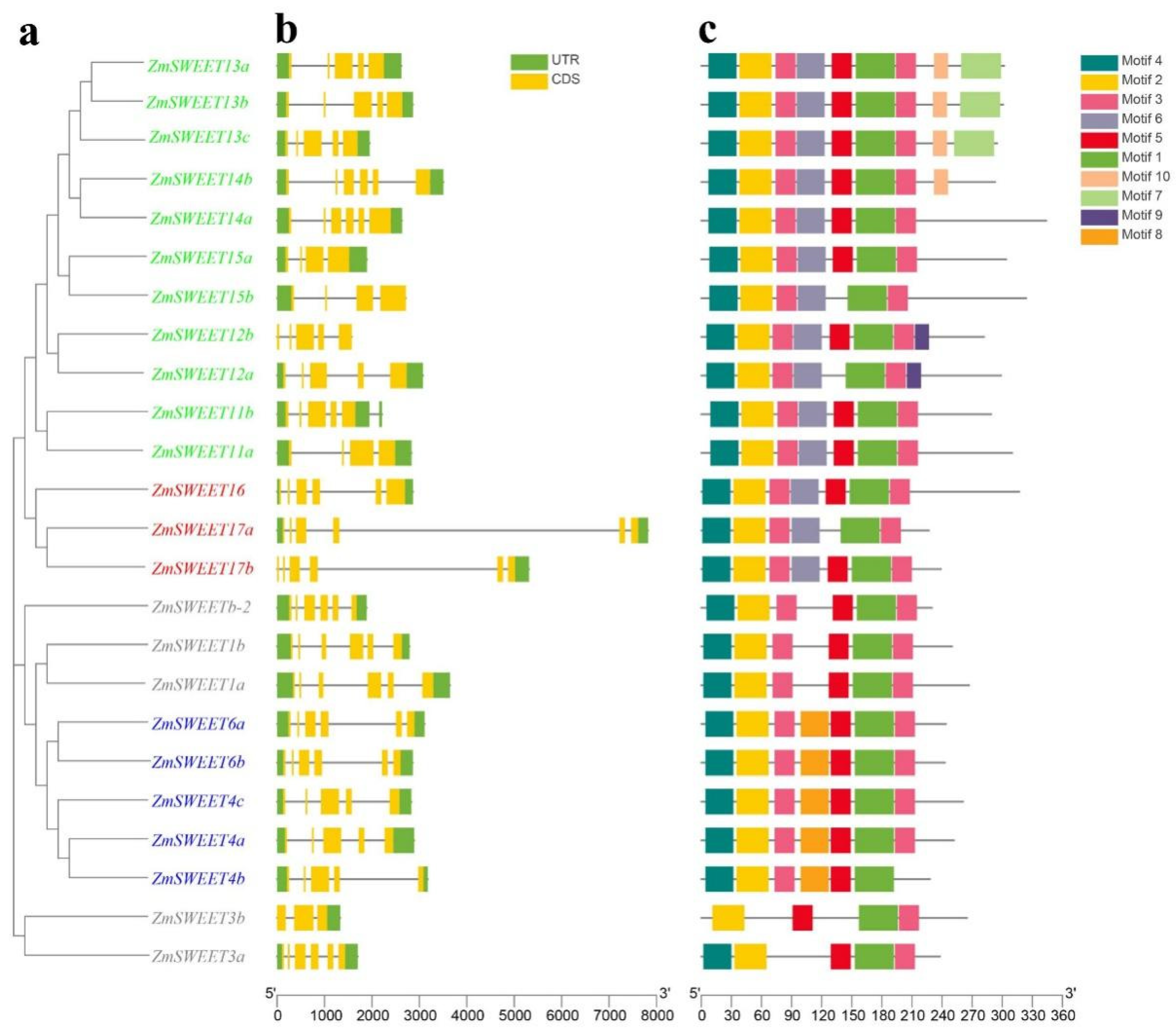
2.2. Gene Structure and Conserved Motif Analysis of ZmSWEET Gene Family
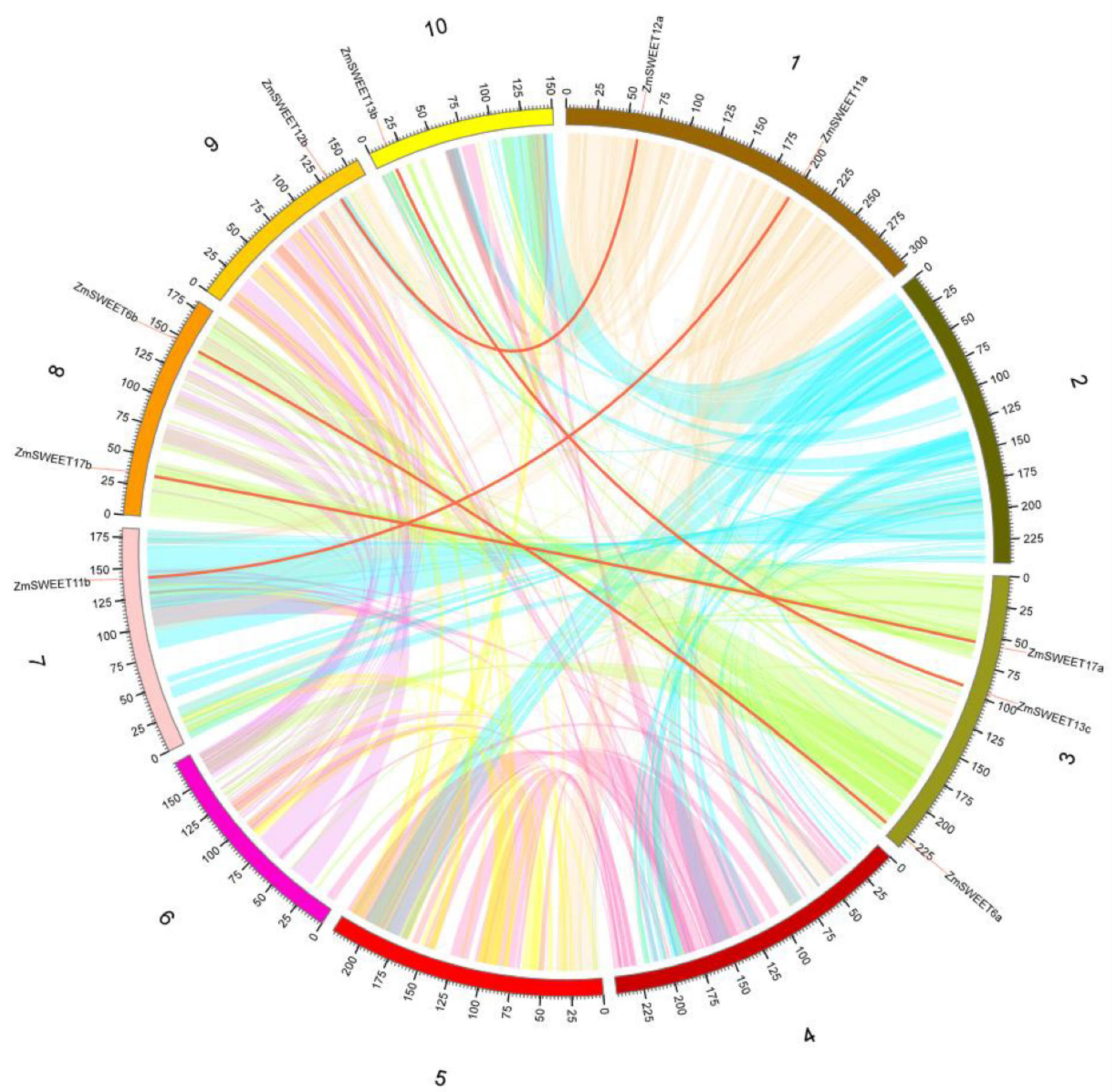
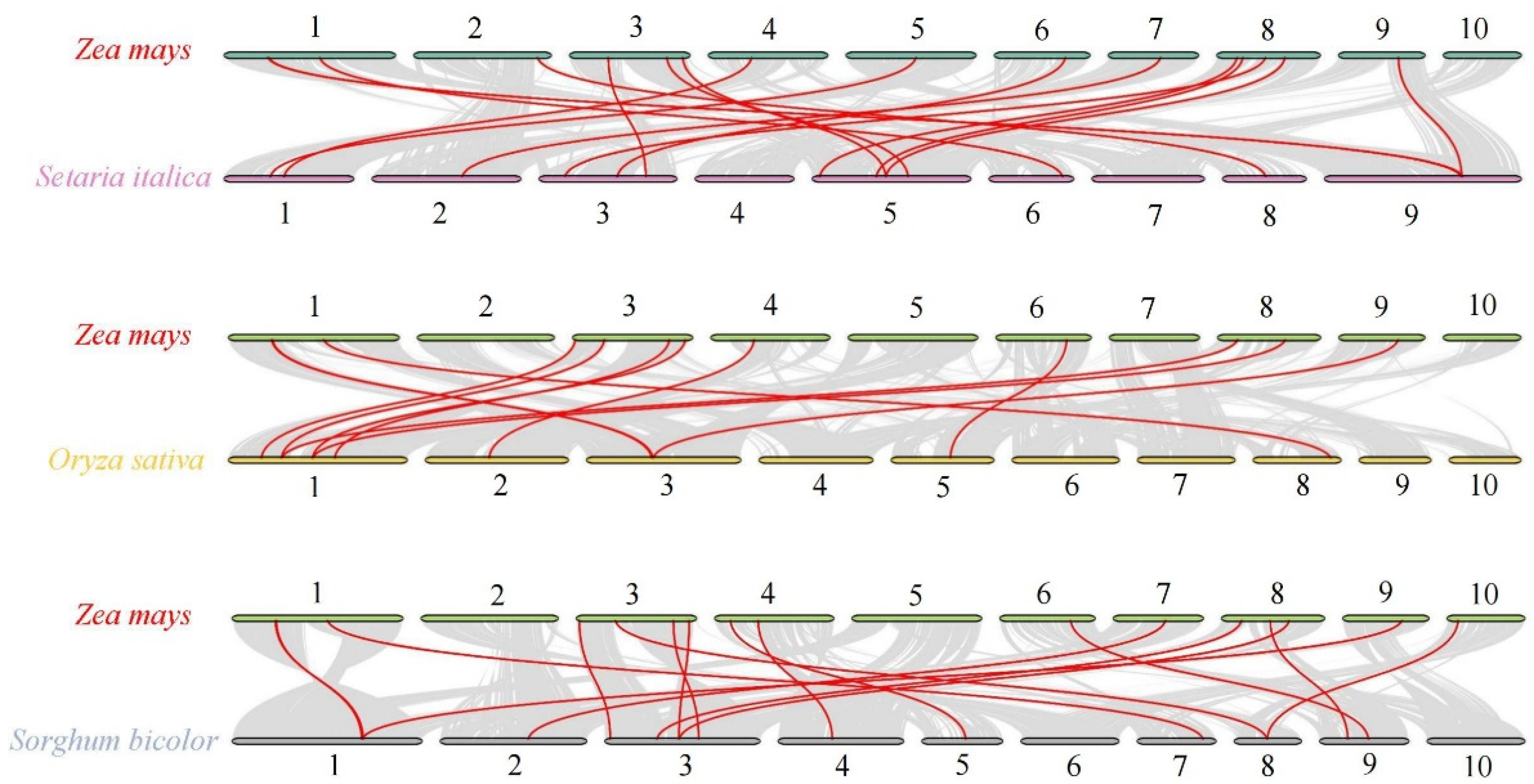
2.3. Evolutionary Differentiation and Collinearity Analysis of ZmSWEET Genes
2.4. Promoter Region Analysis of ZmSWEET Gene Family
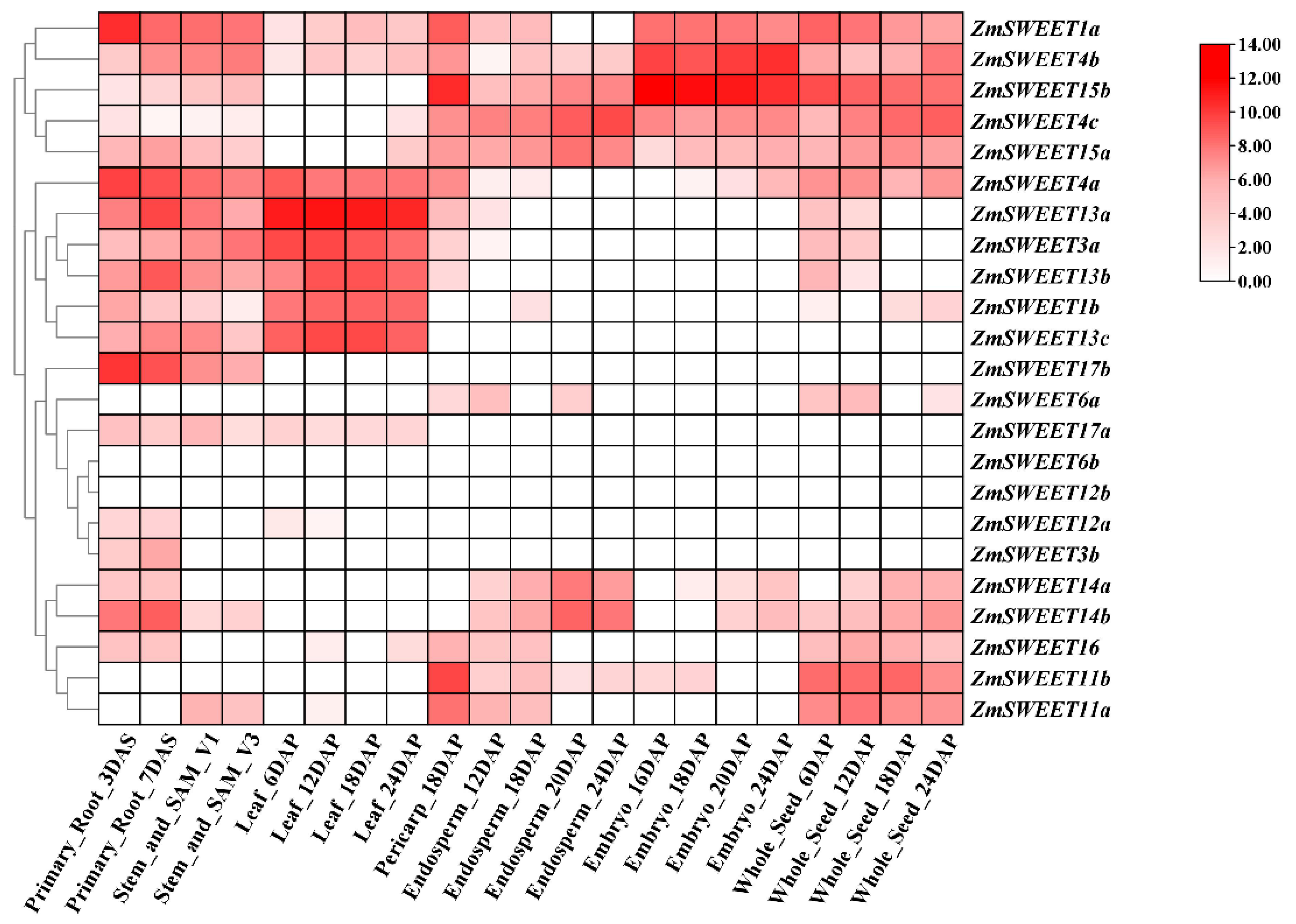
2.5. Expression Profiles of ZmSWEET Genes in Different Tissues and Expression Analysis of ZmSWEET Genes in Response to Abiotic Stresses and Exogenous ABA
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification and Evolutionary Analysis
4.3. Sequence Analysis
4.4. Replication Events and Ka/Ks Analysis of SWEET Genes
4.5. Quantitative Real-Time PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walmsley, A.R.; Barrett, M.P.; Bringaud, F.; Gould, G.W. Sugar transporters from bacteria, parasites and mammals: Structure–activity relationships. Trends Biochem. Sci. 1998, 23, 476–481. [Google Scholar] [CrossRef]
- Yamada, K.; Osakabe, Y. Sugar compartmentation as an environmental stress adaptation strategy in plants. Semin. Cell Dev. Biol. 2017, 83, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Pommerrenig, B.; Ludewig, F.; Cvetkovic, J.; Trentmann, O.; Klemens, P.A.W.; Neuhaus, H.E. In Concert: Orchestrated Changes in Carbohydrate Homeostasis Are Critical for Plant Abiotic Stress Tolerance. Plant Cell Physiol. 2018, 59, 1290–1299. [Google Scholar] [CrossRef]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Y.; Tian, J.; Zhu, Y.S.; Fan, J.J. Changes in sucrose metabolism in maize varieties with different cadmium sensitivities under cadmium stress. PLoS ONE 2020, 15, e0243835. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Gamas, P.; Fde, C.N.; Lescure, N.; Cullimore, J. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 1996, 9, 233–242. [Google Scholar] [CrossRef]
- Artero, R.D.; Terol-Alcayde, J.; Paricio, N.; Ring, J.; Bargues, M.; Torres, A.; Perez-Alonso, M. Saliva, a new Drosophila gene expressed in the embryonic salivary glands with homologues in plants and vertebrates. Mech. Dev. 1998, 75, 159–162. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londono, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Peng, W.M.H.; Xiong, F. Sucrose transport involves in disease response to Xanthomonas oryzae pathovar oryzae. Plant Signal. Behav. 2019, 14, 1656949. [Google Scholar] [CrossRef] [PubMed]
- Bezrutczyk, M.; Yang, J.; Eom, J.S.; Prior, M.; Sosso, D.; Hartwig, T.; Szurek, B.; Oliva, R.; Vera-Cruz, C.; White, F.F.; et al. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018, 93, 675–685. [Google Scholar] [CrossRef]
- Bezrutczyk, M.; Hartwig, T.; Horschman, M.; Char, S.N.; Yang, J.; Yang, B.; Frommer, W.B.; Sosso, D. Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 2018, 218, 594–603. [Google Scholar] [CrossRef]
- Lopez-Coria, M.; Sanchez-Sanchez, T.; Martinez-Marcelo, V.H.; Aguilera-Alvarado, G.P.; Flores-Barrera, M.; King-Diaz, B.; Sanchez-Nieto, S. SWEET Transporters for the Nourishment of Embryonic Tissues during Maize Germination. Genes 2019, 10, 780. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Chen, L.Q. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014, 201, 1150–1155. [Google Scholar] [CrossRef]
- Chu, Z.; Yuan, M.; Yao, J.; Ge, X.; Yuan, B.; Xu, C.; Li, X.; Fu, B.; Li, Z.; Bennetzen, J.L.; et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006, 20, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Romer, P.; Recht, S.; Strauss, T.; Elsaesser, J.; Schornack, S.; Boch, J.; Wang, S.; Lahaye, T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010, 187, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.W.; Patzke, K.; Krapp, A.; Chardon, F.; Neuhaus, H.E. SWEET16 and SWEET17, two novel vacuolar sugar carriers with impact on cellular sugar homeostasis and plant traits. Biochem. Cell Biol. 2014, 92, 589. [Google Scholar]
- Seo, P.J.; Park, J.M.; Kang, S.K.; Kim, S.G.; Park, C.M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [PubMed]
- Mickaël, D.; Benoît, P.; Nils, H.; Laurence, M.; Rémi, L.; Nathalie, P. Water Deficit Enhances C Export to the Roots in Arabidopsis thaliana Plants with Contribution of Sucrose Transporters in Both Shoot and Roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar]
- Du, Y.L.; Li, W.J.; Geng, J.; Li, S.Q.; Zhang, W.J.; Liu, X.X.; Hu, M.H.; Zhang, Z.N.; Fan, Y.R.; Yuan, X.K.; et al. Genome-wide identification of the SWEET gene family in Phaseolus vulgaris L. and their patterns of expression under abiotic stress. J. Plant Interact. 2022, 17, 390–403. [Google Scholar] [CrossRef]
- Xuan, C.Q.; Lan, G.P.; Si, F.F.; Zeng, Z.L.; Wang, C.X.; Yadav, V.; Wei, C.H.; Zhang, X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. Int. J. Mol. Sci. 2021, 22, 8407. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, S.; Ren, Y.; Gan, C.Y.; Li, B.B.; Fan, Y.Y.W.; Zhao, X.Q.; Yuan, Z.H. Identification, Analysis and Gene Cloning of the SWEET Gene Family Provide Insights into Sugar Transport in Pomegranate (Punica granatum). Int. J. Mol. Sci. 2022, 23, 2471. [Google Scholar] [CrossRef]
- Nobuhiro, S.; Elias, B.; Hamilton, J.S.; Inupakutika, M.A.; Izquierdo, Z.S.; Deesha, T.; Yuting, L.; Erin, D.; Ginga, F.; Ayana, K.; et al. ABA Is Required for Plant Acclimation to a Combination of Salt and Heat Stress. PLoS ONE 2016, 11, e0147625. [Google Scholar]
- Kovacs, Z.; Simon-Sarkadi, L.; Sovany, C.; Kirsch, K.; Galiba, G.; Kocsy, G. Differential effects of cold acclimation and abscisic acid on free amino acid composition in wheat. Plant Sci. 2011, 180, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.T.; Li, G.Y.; Islam, M.R.; Fu, G.F. Abscisic acid synergizes with sucrose to enhance grain yield and quality of rice by improving the source-sink relationship. BMC Plant Biol. 2019, 19, 525. [Google Scholar] [CrossRef] [PubMed]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant 2021, 171, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Emes, M.J. Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 2017, 7, 81. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, H.; Ma, Z.Y. Comparison of SWEET gene family between maize and foxtail millet through genomic, transcriptomic, and proteomic analyses. Plant Genome 2022, 15, e20226. [Google Scholar] [CrossRef]
- Ayaz, A.; Huang, H.D.; Zheng, M.L.; Zaman, W.; Li, D.H.; Saqib, S.; Zhao, H.Y.; Lü, S. Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef]
- Hu, Y.B.; Sosso, D.; Qu, X.Q.; Chen, L.Q.; Ma, L.; Chermak, D.; Zhang, D.C.; Frommer, W.B. Phylogenetic evidence for a fusion of archaeal and bacterial SemiSWEETs to form eukaryotic SWEETs and identification of SWEET hexose transporters in the amphibian chytrid pathogen Batrachochytrium dendrobatidis. FASEB J. 2016, 30, 3644–3654. [Google Scholar] [CrossRef]
- Song, J.; Ma, D.; Yin, J.; Yang, L.; He, Y.; Zhu, Z.; Tong, H.; Chen, L.; Zhu, G.; Liu, Y.; et al. Genome-Wide Characterization and Expression Profiling of Squamosa Promoter Binding Protein-Like (SBP) Transcription Factors in Wheat (Triticum aestivum L.). Agronomy 2019, 9, 527. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Jin, D.; Guo, H.; Lee, T.H.; Liu, T.; Paterson, A.H. Genome Alignment Spanning Major Poaceae Lineages Reveals Heterogeneous Evolutionary Rates and Alters Inferred Dates for Key Evolutionary Events. Mol. Plant 2015, 8, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, H.; Cao, P.B.; Fawal, N.; Mathé, C.; Azar, S.; Cassan-Wang, H.; Myburg, A.A.; Grima-Pettenati, J.; Marque, C.; et al. Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biol. Evol. 2015, 7, 1068–1081. [Google Scholar] [CrossRef] [Green Version]
- Hothem, S.D.; Marley, K.A.; Larson, R.A. Photochemistry in Hoagland’s Nutrient Solution. J. Plant Nutr. 2003, 26, 845–854. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. Cell Mol. Biol. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Baxter, L.; Hickman, R.; Beynon, J.; Moore, J.D.; Ott, S. MEME-LaB: Motif analysis in clusters. Bioinformatics 2013, 29, 1696–1697. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari Zabta, K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Yun, K.Y.; Park, M.; Mohanty, B.; Herath, V.; Xu, F.Y.; Mauleon, R.; Wijaya, E.; Bajic, V.; Bruskiewich, R.; de Los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010, 10, 16. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Shi, Y.; Miao, N.; Bian, Y.; Yin, Z. Diversification, phylogeny and evolution of auxin response factor (ARF) family: Insights gained from analyzing maize ARF genes. Mol. Biol. Rep. 2012, 39, 2401–2415. [Google Scholar] [CrossRef]
- Gaut, B.S.; Morton, B.R.; McCaig, B.C.; Clegg, M.T. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 1996, 93, 10274–10279. [Google Scholar] [CrossRef]
- Naeem, M.; Shahzad, K.; Saqib, S.; Asim, S.; Nasrullah, M.Y.; Muhammad, I.A. The Solanum melongena COP1LIKE manipulates fruit ripening and flowering time in tomato (Solanum lycopersicum). Plant Growth Regul. 2022, 96, 369–382. [Google Scholar] [CrossRef]
- Trick, A.Y.; Chen, F.E.; Schares, J.A.; Freml, B.E.; Lor, P.; Yun, Y.; Wang, T.H. High resolution estimates of relative gene abundance with quantitative ratiometric regression PCR (qRR-PCR). Analyst 2021, 146, 6463–6469. [Google Scholar] [CrossRef] [PubMed]



| Name | GeneID | Chrom | Transcript | No. of Amino Acids | TMHs | Theoretical pI | Molecular Weight (Average) |
|---|---|---|---|---|---|---|---|
| ZmSWEET1a | Zm00001d000222 | B73V4_ctg26 | 1 | 250 | 6 | 9.26 | 28,631.97 |
| ZmSWEET1b | Zm00001d038226 | Chrom06 | 1 | 267 | 7 | 8.9 | 26,773.47 |
| ZmSWEET3a | Zm00001d010440 | Chrom08 | 2 | 239 | 7 | 9.02 | 26,104 |
| ZmSWEET3b | Zm00001d039347 | Chrom03 | 1 | 265 | 7 | 9.43 | 28,672.58 |
| ZmSWEET4a | Zm00001d015905 | Chrom05 | 1 | 252 | 6 | 9.44 | 27,278.59 |
| ZmSWEET4b | Zm00001d015914 | Chrom05 | 6 | 255 | 6 | 9.48 | 24,756.56 |
| ZmSWEET4c | Zm00001d015912 | Chrom05 | 1 | 261 | 7 | 9.54 | 28,279.61 |
| ZmSWEET6a | Zm00001d044421 | Chrom03 | 1 | 244 | 6 | 9.03 | 27,133.44 |
| ZmSWEET6b | Zm00001d011299 | Chrom08 | 1 | 243 | 7 | 8.87 | 26,912.16 |
| ZmSWEET11a | Zm00001d031647 | Chrom01 | 1 | 289 | 7 | 9.47 | 33,490.68 |
| ZmSWEET11b | Zm00001d021064 | Chrom07 | 1 | 310 | 7 | 9.2 | 31,314.46 |
| ZmSWEET12a | Zm00001d029135 | Chrom01 | 1 | 306 | 7 | 6.82 | 32,686.56 |
| ZmSWEET12b | Zm00001d047487 | Chrom09 | 1 | 282 | 7 | 9.2 | 30,837.73 |
| ZmSWEET13a | Zm00001d023677 | Chrom10 | 1 | 302 | 7 | 9.57 | 32,950.4 |
| ZmSWEET13b | Zm00001d023673 | Chrom10 | 1 | 301 | 7 | 9.57 | 32,929.51 |
| ZmSWEET13c | Zm00001d041067 | Chrom03 | 1 | 295 | 7 | 9.64 | 32,280.66 |
| ZmSWEET14a | Zm00001d007365 | Chrom02 | 1 | 344 | 7 | 9.48 | 37,267.29 |
| ZmSWEET14b | Zm00001d049252 | Chrom04 | 1 | 293 | 7 | 9.28 | 31,679.58 |
| ZmSWEET15a | Zm00001d050577 | Chrom04 | 1 | 304 | 7 | 5.67 | 32,944.15 |
| ZmSWEET15b | Zm00001d016590 | Chrom05 | 1 | 333 | 6 | 5.1 | 34,340.3 |
| ZmSWEET16 | Zm00001d029098 | Chrom01 | 1 | 317 | 7 | 6.6 | 33,998.45 |
| ZmSWEET17a | Zm00001d040656 | Chrom03 | 3 | 238 | 6 | 6.71 | 17,560.88 |
| ZmSWEET17b | Zm00001d009071 | Chrom08 | 2 | 239 | 7 | 6.1 | 25,906.62 |
| ZmSWEETb-2 | Zm00001d043735 | Chrom03 | 1 | 230 | 7 | 8.75 | 25,165.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Zhou, L.; Li, T.; Ruan, Y.; Zhang, A.; Dong, X.; Zhu, Y.; Li, C.; Fan, J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes 2022, 13, 1682. https://doi.org/10.3390/genes13101682
Zhu J, Zhou L, Li T, Ruan Y, Zhang A, Dong X, Zhu Y, Li C, Fan J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes. 2022; 13(10):1682. https://doi.org/10.3390/genes13101682
Chicago/Turabian StyleZhu, Jialun, Lu Zhou, Tianfeng Li, Yanye Ruan, Ao Zhang, Xiaomei Dong, Yanshu Zhu, Cong Li, and Jinjuan Fan. 2022. "Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize" Genes 13, no. 10: 1682. https://doi.org/10.3390/genes13101682
APA StyleZhu, J., Zhou, L., Li, T., Ruan, Y., Zhang, A., Dong, X., Zhu, Y., Li, C., & Fan, J. (2022). Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes, 13(10), 1682. https://doi.org/10.3390/genes13101682





