Population Genetic Data of 30 Insertion-Deletion Markers in the Polish Population
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. PCR Amplification and InDel Genotyping
2.3. Statistical Analyses
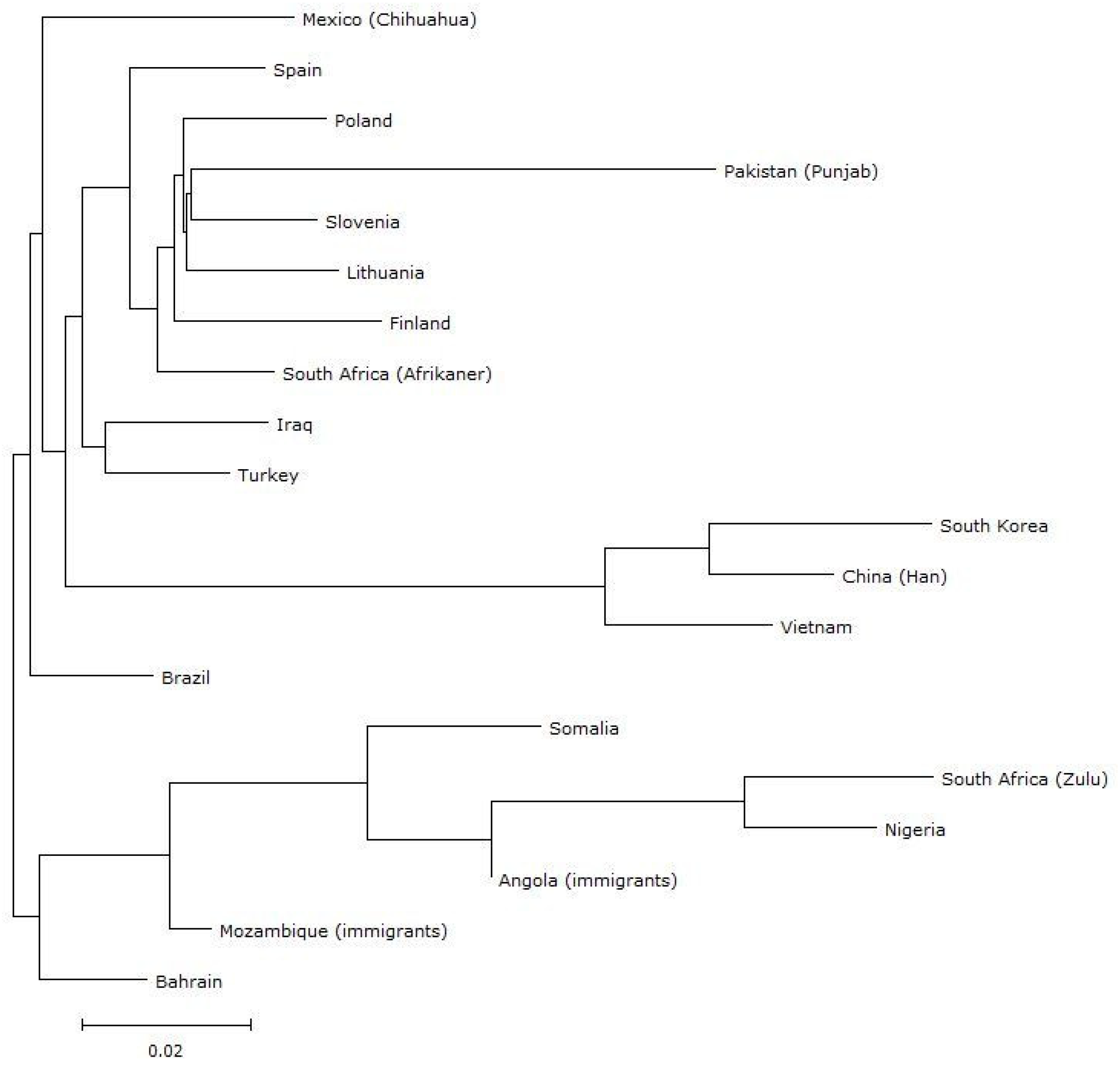
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mills, R.E.; Luttig, C.T.; Larkins, C.E.; Beauchamp, A.; Tsui, C.; Pittard, W.S.; Devine, S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006, 16, 1182–1190. [Google Scholar] [CrossRef]
- Pereira, R.; Phillips, C.; Alves, C.; Amorim, A.; Carracedo, Á.; Gusmão, L. A new multiplex for human identification insertion/deletion polymorphisms. Electrophoresis 2009, 30, 3682–3690. [Google Scholar] [CrossRef]
- Fondevila, M.; Phillips, C.; Santos, C.; Pereira, R.; Gusmão, L.; Carracedo, Á.; Butler, J.M.; Lareu, M.V.; Vallone, P.M. Forensic performance of two insertion-deletion marker assays. Int. J. Legal Med. 2012, 126, 725–737. [Google Scholar] [CrossRef]
- Pereira, R.; Phillips, C.; Alves, C.; Amorim, A.; Carracedo, Á.; Gusmão, L. Insertion/deletion polymorphisms: A multiplex assay and forensic applications. Forensic Sci. Int. 2009, 2, 513–515. [Google Scholar] [CrossRef]
- Pereira, R.; Phillips, C.; Pinto, N.; Santos, C.; Santos, S.; Amorim, A.; Carracedo, Á.; Gusmão, L. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS ONE 2012, 7, e29684. [Google Scholar] [CrossRef]
- Trzeciecki, M. The Past Societies; Institute of Archaeology and Ethnology: Warsaw, Poland, 2016; Volume 5, pp. 500AD–1000AD. [Google Scholar]
- Britannica. Available online: britannica.com/place/Poland (accessed on 10 August 2022).
- World Population Review. Available online: worldpopulationreview.com/countries/poland-population (accessed on 10 August 2022).
- Soltyszewski, I.; Pepinski, W.; Wolanska-Nowak, P.; Maciejewska, A.; Paszkowska, R.; Abreu-Glowacka, M.; Achrem, W.; Jonkisz, A.; Lebioda, A.; Konarzewska, M.; et al. Polish population data on 15 autosomal STRs of AmpFlSTR NGM PCR kit. Forensic Sci. Int. Genet. 2014, 9, 142–149. [Google Scholar] [CrossRef]
- Jarczak, J.; Grochowalski, L.; Marciniak, B.; Lach, J.; Słomka, M.; Sobalska-Kwapis, M.; Lorkiewicz, W.; Pułaski, L.; Strapagiel, D. Mitochondrial DNA variability of the Polish population. Eur. J. Hum. Genet. 2019, 27, 1304–1314. [Google Scholar] [CrossRef]
- Ploski, R.; Woźniak, M.; Pawlowski, R.; Monies, D.; Branicki, W.; Kupiec, T.; Kloosterman, A.; Dobosz, T.; Bosch, E.; Nowak, M.; et al. Homogeneity and distinctiveness of Polish paternal lineages revealed by Y chromosome microsatellite haplotype analysis. Hum. Genet. 2002, 110, 592–600. [Google Scholar] [CrossRef]
- Kayser, M.; Lao, O.; Anslinger, K.; Augustin, C.; Bargel, G.; Edelmann, J.; Elias, S.; Heinrich, M.; Henke, J.; Henke, L.; et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum. Genet. 2005, 117, 428–443. [Google Scholar] [CrossRef]
- Grochowalski, L.; Jarczak, J.; Urbanowicz, M.; Slomka, M.; Szargut, M.; Borowka, P.; Sobalska-Kwapis, M.; Marciniak, B.; Ossowski, A.; Lorkiewicz, W.; et al. Y-Chromosome genetic analysis of modern Polish population. Front. Genet. 2020, 11, 567309. [Google Scholar] [CrossRef]
- Schneider, P.M. Scientific standards for studies in forensic genetics. Forensic Sci. Int. 2007, 165, 238–243. [Google Scholar] [CrossRef]
- Gouy, A.; Zieger, M. STRAF—A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 2017, 30, 148–151. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol. Biol. Evol. 2010, 27, 747–752. [Google Scholar] [CrossRef]
- Latter, B.D.H. Selection in finite with multiple alleles. 3. Genetic divergence with centripetal selection and mutation. Genetics 1972, 70, 475–490. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Martín, P.; García, O.; Heinrichs, B.; Yurrebaso, I.; Aguirre, A.; Alonso, A. Population genetic data of 30 autosomal indels in Central Spain and the Basque Country populations. Forensic Sci. Int. Genet. 2013, 7, e27–e30. [Google Scholar] [CrossRef]
- Martínez-Cortés, G.; García-Aceves, M.; Favela-Mendoza, A.F.; Muñoz-Valle, J.F.; Velarde-Felix, J.S.; Rangel-Villalobos, H. Forensic parameters of the Investigator DIPplex kit (Qiagen) in six Mexican populations. Int. J. Legal Med. 2016, 130, 683–685. [Google Scholar] [CrossRef]
- Seong, K.M.; Park, J.H.; Hyun, Y.S.; Kang, P.W.; Choi, D.H.; Han, M.S.; Park, K.W.; Chung, K.W. Population genetics of insertion-deletion polymorphisms in South Koreans using Investigator DIPplex kit. Forensic Sci. Int. Genet. 2014, 8, 80–83. [Google Scholar] [CrossRef]
- Neuvonen, A.M.; Palo, J.U.; Hedman, M.; Sajantila, A. Discrimination power of Investigator DIPplex loci in Finnish and Somali populations. Forensic Sci. Int. Genet. 2012, 6, e99–e102. [Google Scholar] [CrossRef]
- Shahzad, M.; Hussain, M.; Shafique, M.; Perveen, R.; Sheikh, N. Population genetic data of 30 insertion-deletion markers in Punjabi population of Pakistan. Int. J. Legal Med. 2020, 134, 511–512. [Google Scholar] [CrossRef]
- Hefke, G.; Davison, S.; D’Amato, M.E. Forensic performance of Investigator DIPplex indels genotyping kit in native, immigrant, and admixed populations in South Africa. Electrophoresis 2015, 36, 3018–3025. [Google Scholar] [CrossRef]
- Tomas, C.; Poulsen, L.; Drobnič, K.; Ivanova, V.; Jankauskiene, J.; Bunokiene, D.; Børsting, C.; Morling, N. Thirty autosomal insertion-deletion polymorphisms analyzed using the Investigator® DIPplex Kit in populations from Iraq, Lithuania, Slovenia, and Turkey. Forensic Sci. Int. Genet. 2016, 25, 142–144. [Google Scholar] [CrossRef]
- Du, W.; Peng, Z.; Feng, C.; Zhu, B.; Wang, B.; Wang, Y.; Liu, C.; Chen, L. Forensic efficiency and genetic variation of 30 InDels in Vietnamese and Nigerian populations. Oncotarget 2017, 8, 88934–88940. [Google Scholar] [CrossRef]
- Al-Snan, N.R.; Shabbir, S.; Baksh, S.S.; AlQerainees, M.; Haidar, M.; Messaoudi, S.A.; Bakhiet, M. Population genetics of 30 insertion/deletion polymorphisms in the Bahraini population. Sci. Rep. 2021, 11, 6843. [Google Scholar] [CrossRef]
- Inácio, A.; Costa, H.A.; Da Silva, C.V.; Ribeiro, T.; Porto, M.J.; Santos, J.C.; Igrejas, G.; Amorim, A. Study of InDel genetic markers with forensic and ancestry informative interest in PALOP’s immigrant populations in Lisboa. Int. J. Legal Med. 2017, 131, 657–660. [Google Scholar] [CrossRef]
- Palha, T.D.J.B.F.; Rodrigues, E.M.R.; Cavalcante, G.C.; Marrero, A.; de Souza, I.R.; Uehara, C.J.S.; da Motta, C.H.A.S.; Koshikene, D.; da Silva, D.A.; de Carvalho, E.F.; et al. Population genetic analysis of insertion-deletion polymorphisms in a Brazilian population using the Investigator DIPplex kit. Forensic Sci. Int. Genet. 2015, 19, 10–14. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Zhao, S.; Hu, Z.; Sun, K.; Li, C. Population genetics of 30 insertion-deletion polymorphisms in two Chinese populations using Qiagen Investigator® DIPplex kit. Forensic Sci. Int. Genet. 2014, 11, e12–e14. [Google Scholar] [CrossRef]
- de Leeuw, J.; Mair, P. Multidimensional Scaling Using Majorization: SMACOF in R. J. Stat. Soft. 2009, 31, 1–30. Available online: statsoft.org/v31/i03/ (accessed on 15 January 2022). [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: R-project.org/ (accessed on 15 January 2022).
- John, S.E.; Antony, D.; Eaaswarkhanth, M.; Hebbar, P.; Channanath, A.M.; Thomas, D.; Devarajan, S.; Tuomilehto, J.; Al-Mulla, F.; Alsmadi, O.; et al. Assessment of coding region variants in Kuwaiti population: Implications for medical genetics and population genomics. Sci. Rep. 2018, 8, 16583. [Google Scholar] [CrossRef]
- Nachman, M.W.; Crowell, S.L. Estimate of the mutation rate per nucleotide in humans. Genetics 2000, 156, 297–304. [Google Scholar] [CrossRef]
- Pimenta, J.R.; Pena, S.D.J. Efficient human paternity testing with a panel of 40 short insertion-deletion polymorphisms. Genet. Mol. Res. 2010, 9, 601–607. [Google Scholar] [CrossRef]
- Gao, T.Z.; Yun, L.B.; He, W.; Gu, Y.; Hou, Y.P. The application of multi-InDel as supplementary in paternity cases with STR mutation. Forensic. Sci. Int. Genet. Suppl. Series 2015, 5, e218–e219. [Google Scholar] [CrossRef]
- Krawczak, M. Informativity assessment for biallelic single nucleotide polymorphisms. Electrophoresis 1999, 20, 1676–1681. [Google Scholar] [CrossRef]

| HLD | Chromosomal | GenBank | DIP(−) | DIP(+) | Ho | He | p HWE | PIC | PD | PE | TPI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | SNP ID | ||||||||||
| HLD77 | 7q31.1 | rs1611048 | 0.4422 | 0.5578 | 0.4937 | 0.4628 | 0.1224 | 0.3716 | 0.6348 | 0.1569 | 0.9307 |
| HLD45 | 2q31.1 | rs2307959 | 0.4739 | 0.5261 | 0.4990 | 0.5040 | 0.8096 | 0.3743 | 0.6216 | 0.1910 | 1.0080 |
| HLD131 | 7q36.2 | rs1611001 | 0.4564 | 0.5436 | 0.4966 | 0.4628 | 0.0934 | 0.3731 | 0.6377 | 0.1569 | 0.9307 |
| HLD70 | 6q16.1 | rs2307652 | 0.4731 | 0.5269 | 0.4989 | 0.4929 | 0.8100 | 0.3743 | 0.6270 | 0.1813 | 0.9859 |
| HLD6 | 16q13 | rs1610905 | 0.4707 | 0.5293 | 0.4987 | 0.4564 | 0.0384 | 0.3741 | 0.6422 | 0.1521 | 0.9198 |
| HLD111 | 17p11.2 | rs1305047 | 0.4976 | 0.5024 | 0.5004 | 0.4818 | 0.3837 | 0.3750 | 0.6336 | 0.1720 | 0.9648 |
| HLD58 | 5q14.1 | rs1610937 | 0.4461 | 0.5539 | 0.4946 | 0.4739 | 0.2955 | 0.3721 | 0.6312 | 0.1656 | 0.9503 |
| HLD56 | 4q25 | rs2308292 | 0.3352 | 0.6648 | 0.4460 | 0.4358 | 0.5868 | 0.3464 | 0.5966 | 0.1372 | 0.8862 |
| HLD118 | 20p11.1 | rs16438 | 0.5143 | 0.4857 | 0.5000 | 0.4707 | 0.1451 | 0.3748 | 0.6380 | 0.1631 | 0.9446 |
| HLD92 | 11q22.2 | rs17174476 | 0.5919 | 0.4081 | 0.4835 | 0.4707 | 0.5146 | 0.3664 | 0.6215 | 0.1631 | 0.9446 |
| HLD93 | 12q22 | rs2307570 | 0.4477 | 0.5523 | 0.4949 | 0.5436 | 0.0157 | 0.3722 | 0.5949 | 0.2286 | 1.0955 |
| HLD99 | 14q23.1 | rs2308163 | 0.4097 | 0.5903 | 0.4841 | 0.4929 | 0.6853 | 0.3667 | 0.6122 | 0.1813 | 0.9859 |
| HLD88 | 9q22.32 | rs8190570 | 0.5895 | 0.4105 | 0.4843 | 0.4881 | 0.8711 | 0.3669 | 0.6147 | 0.1773 | 0.9768 |
| HLD101 | 15q26.1 | rs2307433 | 0.4786 | 0.5214 | 0.4995 | 0.4913 | 0.7021 | 0.3745 | 0.6283 | 0.1800 | 0.9829 |
| HLD67 | 5q33.2 | rs1305056 | 0.3954 | 0.6046 | 0.4785 | 0.4834 | 0.8013 | 0.3638 | 0.6110 | 0.1734 | 0.9678 |
| HLD83 | 8p22 | rs2308072 | 0.5127 | 0.4873 | 0.5001 | 0.5119 | 0.5797 | 0.3748 | 0.6185 | 0.1981 | 1.0244 |
| HLD114 | 17p13.3 | rs2307581 | 0.6513 | 0.3487 | 0.4545 | 0.4469 | 0.7246 | 0.3510 | 0.6015 | 0.1451 | 0.9040 |
| HLD48 | 2q11.2 | rs28369942 | 0.4770 | 0.5230 | 0.4993 | 0.4628 | 0.0669 | 0.3745 | 0.6405 | 0.1569 | 0.9307 |
| HLD124 | 22q12.3 | rs6481 | 0.3574 | 0.6426 | 0.4597 | 0.4612 | 1.0000 | 0.3538 | 0.6015 | 0.1557 | 0.9279 |
| HLD122 | 21q22.11 | rs8178524 | 0.5388 | 0.4612 | 0.4974 | 0.5261 | 0.1444 | 0.3735 | 0.6079 | 0.2114 | 1.0552 |
| HLD125 | 22q11.23 | rs16388 | 0.4889 | 0.5111 | 0.5002 | 0.4897 | 0.6298 | 0.3749 | 0.6297 | 0.1786 | 0.9798 |
| HLD64 | 5q12.3 | rs1610935 | 0.4532 | 0.5468 | 0.4960 | 0.4501 | 0.0258 | 0.3728 | 0.6419 | 0.1474 | 0.9092 |
| HLD81 | 7q21.3 | rs17879936 | 0.5689 | 0.4311 | 0.4909 | 0.4437 | 0.0184 | 0.3702 | 0.6389 | 0.1428 | 0.8989 |
| HLD136 | 22q13.1 | rs16363 | 0.5475 | 0.4525 | 0.4959 | 0.4834 | 0.5776 | 0.3727 | 0.6284 | 0.1734 | 0.9678 |
| HLD133 | 3p22.1 | rs2067235 | 0.4461 | 0.5539 | 0.4946 | 0.4739 | 0.2933 | 0.3721 | 0.6312 | 0.1656 | 0.9503 |
| HLD97 | 13q12.3 | rs17238892 | 0.4493 | 0.5507 | 0.4952 | 0.4834 | 0.5686 | 0.3724 | 0.6278 | 0.1734 | 0.9678 |
| HLD40 | 1p32.3 | rs2307956 | 0.5634 | 0.4366 | 0.4924 | 0.4580 | 0.0871 | 0.3709 | 0.6353 | 0.1533 | 0.9225 |
| HLD128 | 1q31.3 | rs2307924 | 0.5563 | 0.4437 | 0.4941 | 0.4691 | 0.2199 | 0.3718 | 0.6327 | 0.1619 | 0.9418 |
| HLD39 | 1p22.1 | rs17878444 | 0.6268 | 0.3732 | 0.4682 | 0.4263 | 0.0265 | 0.3584 | 0.6216 | 0.1307 | 0.8715 |
| HLD84 | 8q24.12 | rs3081400 | 0.4532 | 0.5468 | 0.4960 | 0.4945 | 1.0000 | 0.3728 | 0.6234 | 0.1827 | 0.9890 |
| ESP | MEX | KOR | FIN | SOM | PAK | ZAZ | IRQ | LTU | SLO | TUR | VIE | NGA | BHR | ZAA | ANG | MOZ | BRA | CHN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POL | 0.037 | 0.064 | 0.124 | 0.044 | 0.107 | 0.079 | 0.163 | 0.049 | 0.036 | 0.033 | 0.045 | 0.106 | 0.155 | 0.053 | 0.035 | 0.098 | 0.063 | 0.050 | 0.116 |
| ESP | * | 0.056 | 0.130 | 0.047 | 0.090 | 0.090 | 0.144 | 0.043 | 0.039 | 0.038 | 0.039 | 0.108 | 0.135 | 0.043 | 0.034 | 0.082 | 0.051 | 0.043 | 0.122 |
| MEX | + | * | 0.129 | 0.062 | 0.098 | 0.127 | 0.142 | 0.056 | 0.064 | 0.063 | 0.050 | 0.116 | 0.128 | 0.052 | 0.057 | 0.085 | 0.059 | 0.045 | 0.120 |
| KOR | + | + | * | 0.136 | 0.166 | 0.193 | 0.224 | 0.135 | 0.140 | 0.127 | 0.122 | 0.059 | 0.227 | 0.125 | 0.132 | 0.172 | 0.135 | 0.125 | 0.041 |
| FIN | + | + | + | * | 0.103 | 0.090 | 0.147 | 0.056 | 0.044 | 0.041 | 0.054 | 0.121 | 0.137 | 0.058 | 0.042 | 0.088 | 0.056 | 0.046 | 0.125 |
| SOM | + | + | + | + | * | 0.149 | 0.071 | 0.084 | 0.108 | 0.106 | 0.088 | 0.138 | 0.067 | 0.070 | 0.098 | 0.050 | 0.059 | 0.073 | 0.146 |
| PAK | + | + | + | + | + | * | 0.198 | 0.105 | 0.081 | 0.078 | 0.103 | 0.176 | 0.192 | 0.111 | 0.077 | 0.137 | 0.107 | 0.099 | 0.178 |
| ZAZ | + | + | + | + | + | + | * | 0.132 | 0.166 | 0.161 | 0.147 | 0.213 | 0.038 | 0.107 | 0.148 | 0.052 | 0.076 | 0.109 | 0.214 |
| IRQ | + | + | + | + | + | + | + | * | 0.051 | 0.048 | 0.034 | 0.112 | 0.120 | 0.037 | 0.045 | 0.073 | 0.050 | 0.045 | 0.122 |
| LTU | + | + | + | + | + | + | + | * | 0.034 | 0.048 | 0.116 | 0.157 | 0.057 | 0.037 | 0.102 | 0.065 | 0.051 | 0.125 | |
| SLO | + | + | + | + | + | + | + | + | * | 0.044 | 0.108 | 0.151 | 0.052 | 0.033 | 0.096 | 0.061 | 0.048 | 0.117 | |
| TUR | + | + | + | + | + | + | + | + | + | * | 0.098 | 0.137 | 0.038 | 0.040 | 0.084 | 0.054 | 0.045 | 0.109 | |
| VIE | + | + | + | + | + | + | + | + | + | + | + | * | 0.214 | 0.107 | 0.109 | 0.158 | 0.119 | 0.109 | 0.047 |
| NGA | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.100 | 0.141 | 0.043 | 0.071 | 0.101 | 0.217 |
| BHR | + | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.046 | 0.060 | 0.039 | 0.039 | 0.116 |
| ZAA | + | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.087 | 0.054 | 0.042 | 0.119 | |
| ANG | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.042 | 0.060 | 0.162 |
| MOZ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.039 | 0.125 |
| BRA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | * | 0.115 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu-Glowacka, M.; Pepinski, W.; Michalak, E.; Konarzewska, M.; Zak, K.; Skawronska, M.; Niemcunowicz-Janica, A.; Soltyszewski, I.; Krajewski, P.; Zaba, C. Population Genetic Data of 30 Insertion-Deletion Markers in the Polish Population. Genes 2022, 13, 1683. https://doi.org/10.3390/genes13101683
Abreu-Glowacka M, Pepinski W, Michalak E, Konarzewska M, Zak K, Skawronska M, Niemcunowicz-Janica A, Soltyszewski I, Krajewski P, Zaba C. Population Genetic Data of 30 Insertion-Deletion Markers in the Polish Population. Genes. 2022; 13(10):1683. https://doi.org/10.3390/genes13101683
Chicago/Turabian StyleAbreu-Glowacka, Monica, Witold Pepinski, Eliza Michalak, Magdalena Konarzewska, Krzysztof Zak, Malgorzata Skawronska, Anna Niemcunowicz-Janica, Ireneusz Soltyszewski, Pawel Krajewski, and Czeslaw Zaba. 2022. "Population Genetic Data of 30 Insertion-Deletion Markers in the Polish Population" Genes 13, no. 10: 1683. https://doi.org/10.3390/genes13101683
APA StyleAbreu-Glowacka, M., Pepinski, W., Michalak, E., Konarzewska, M., Zak, K., Skawronska, M., Niemcunowicz-Janica, A., Soltyszewski, I., Krajewski, P., & Zaba, C. (2022). Population Genetic Data of 30 Insertion-Deletion Markers in the Polish Population. Genes, 13(10), 1683. https://doi.org/10.3390/genes13101683






