The Effects of Salinity on the Anatomy and Gene Expression Patterns in Leaflets of Tomato cv. Micro-Tom
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Set-Up and Growth Conditions
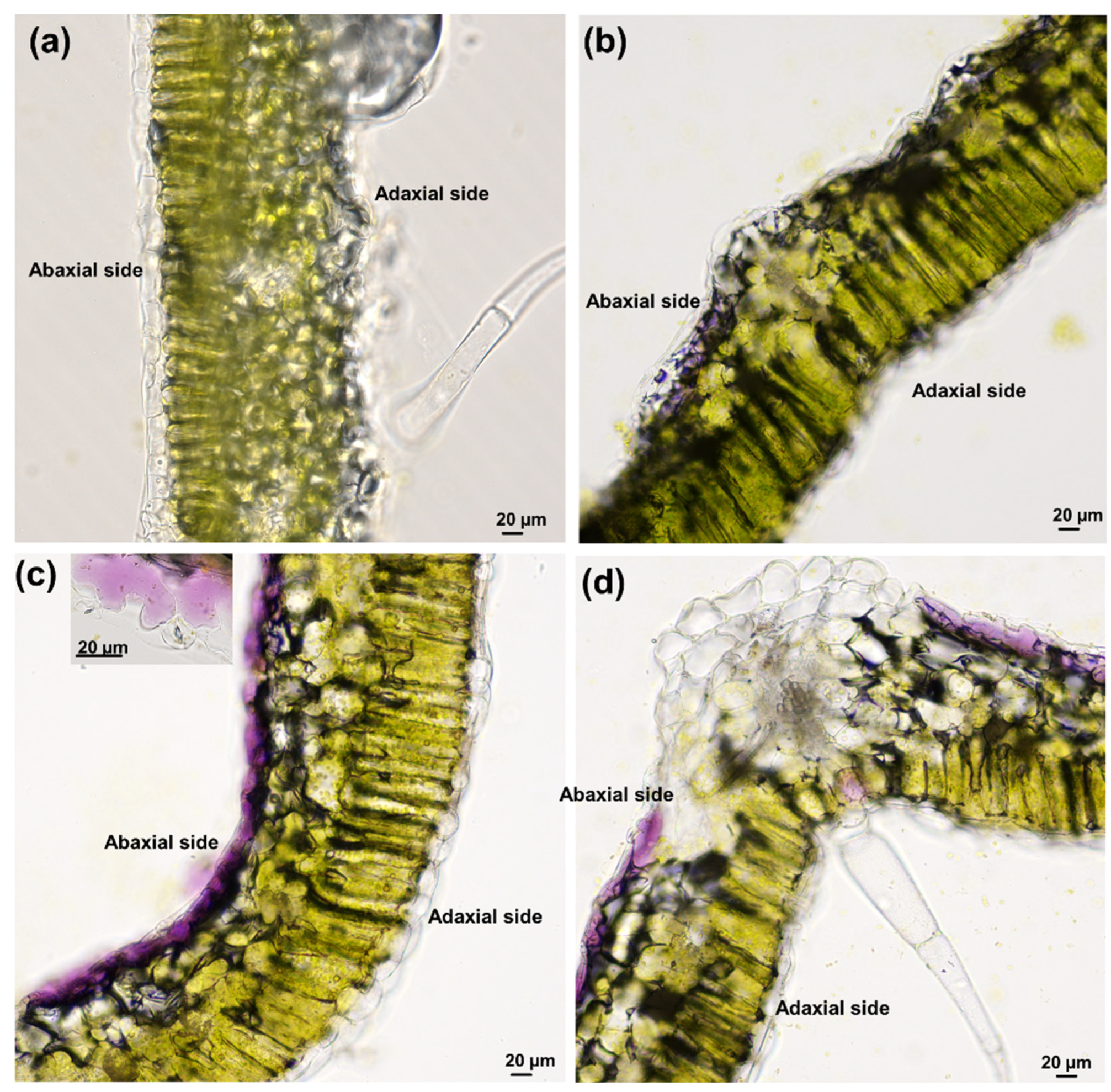
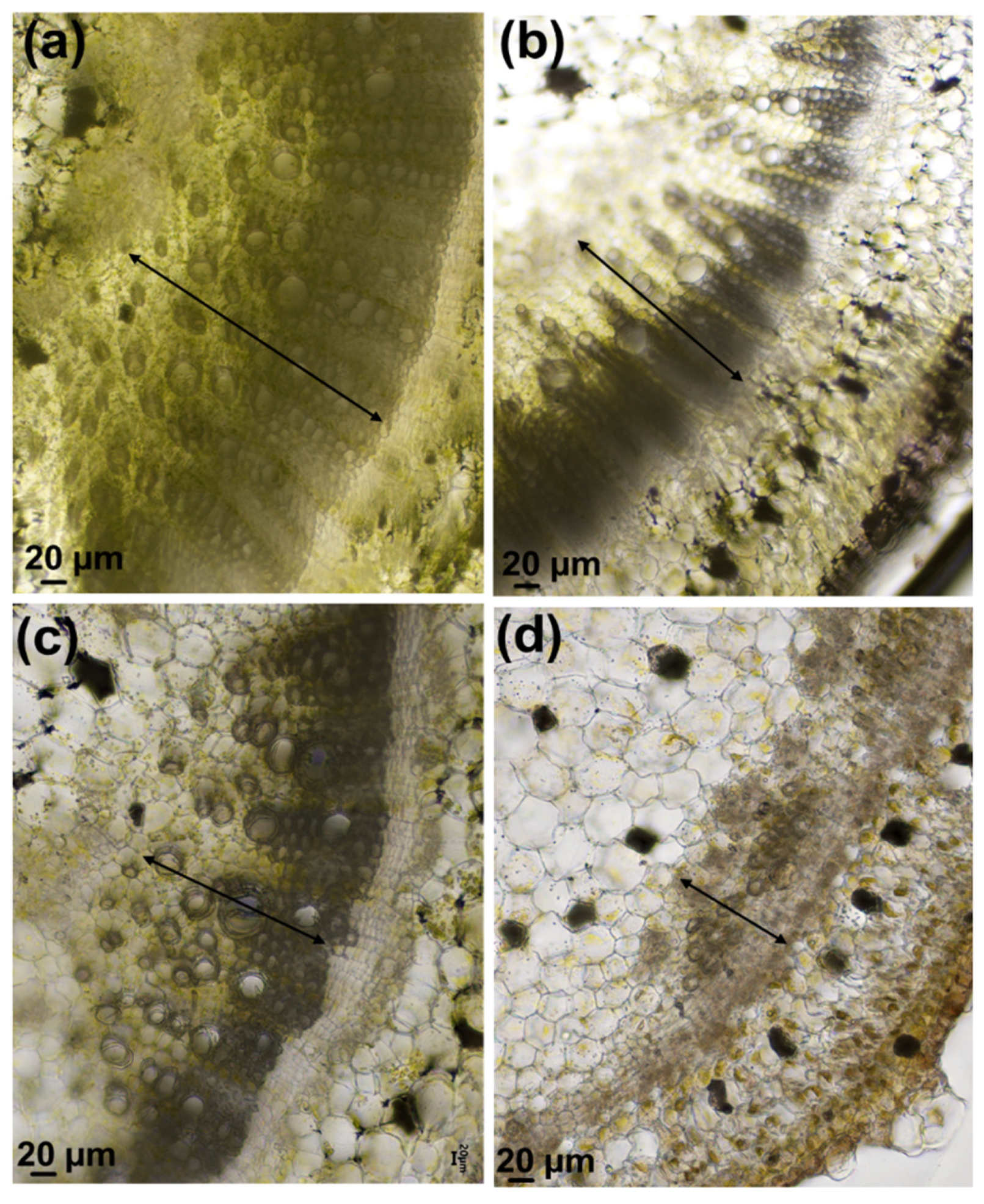
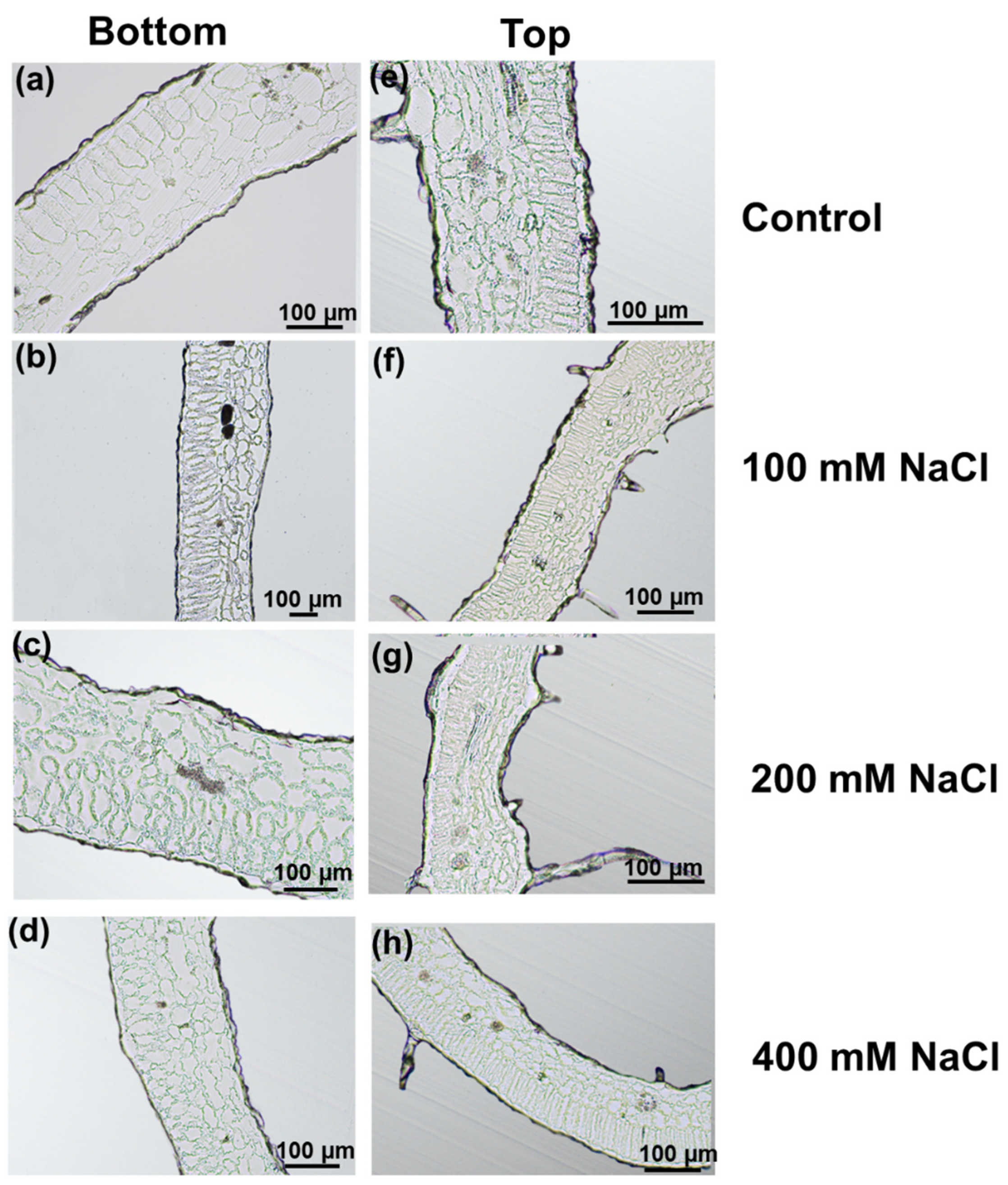
2.2. Sample Preparation and Microscopy
2.3. Design and Validation of Primers
2.4. RNA Extraction, cDNA Synthesis and qPCR
2.5. Gene Expression Analysis and Statistics
3. Results and Discussion
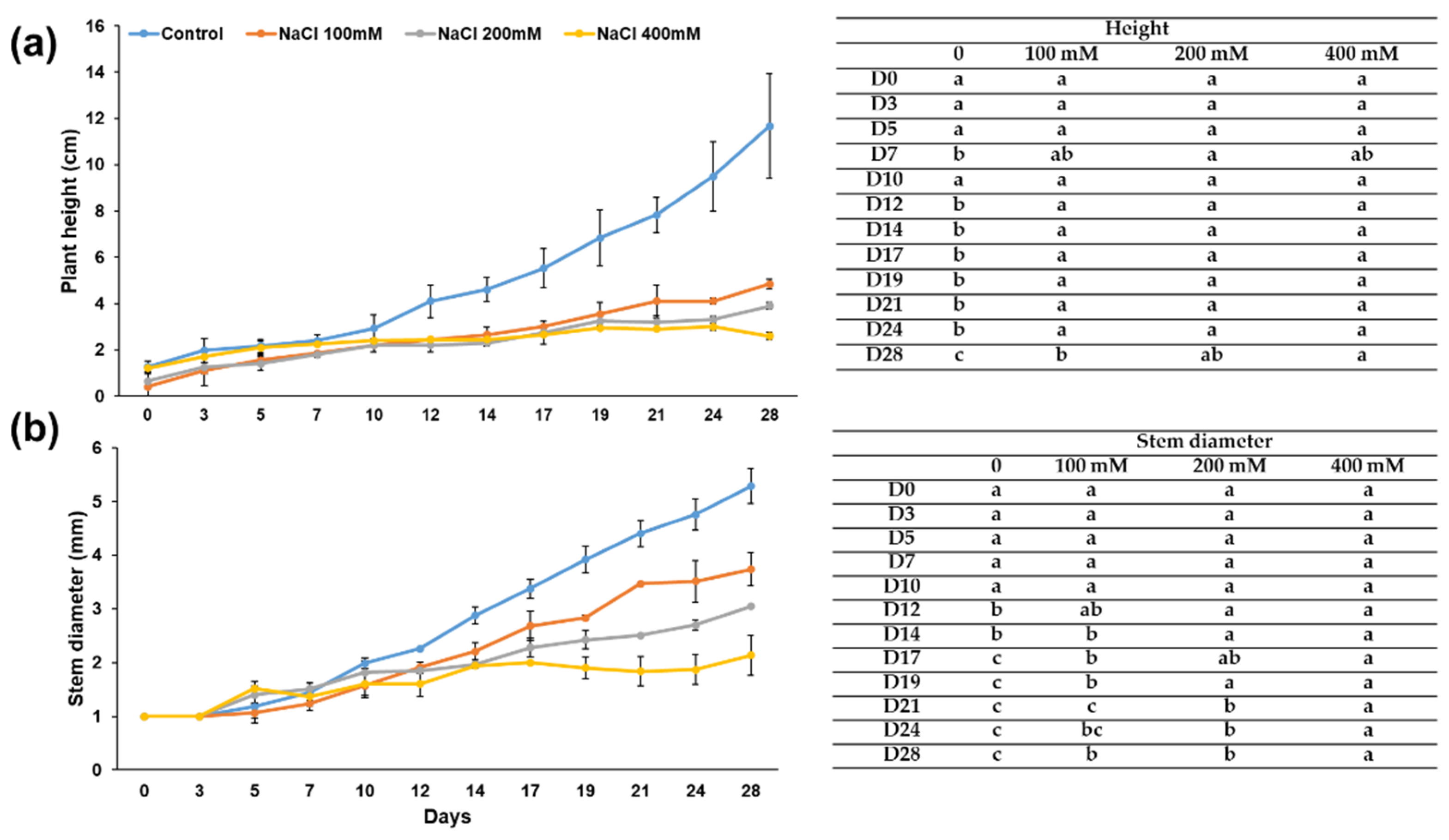
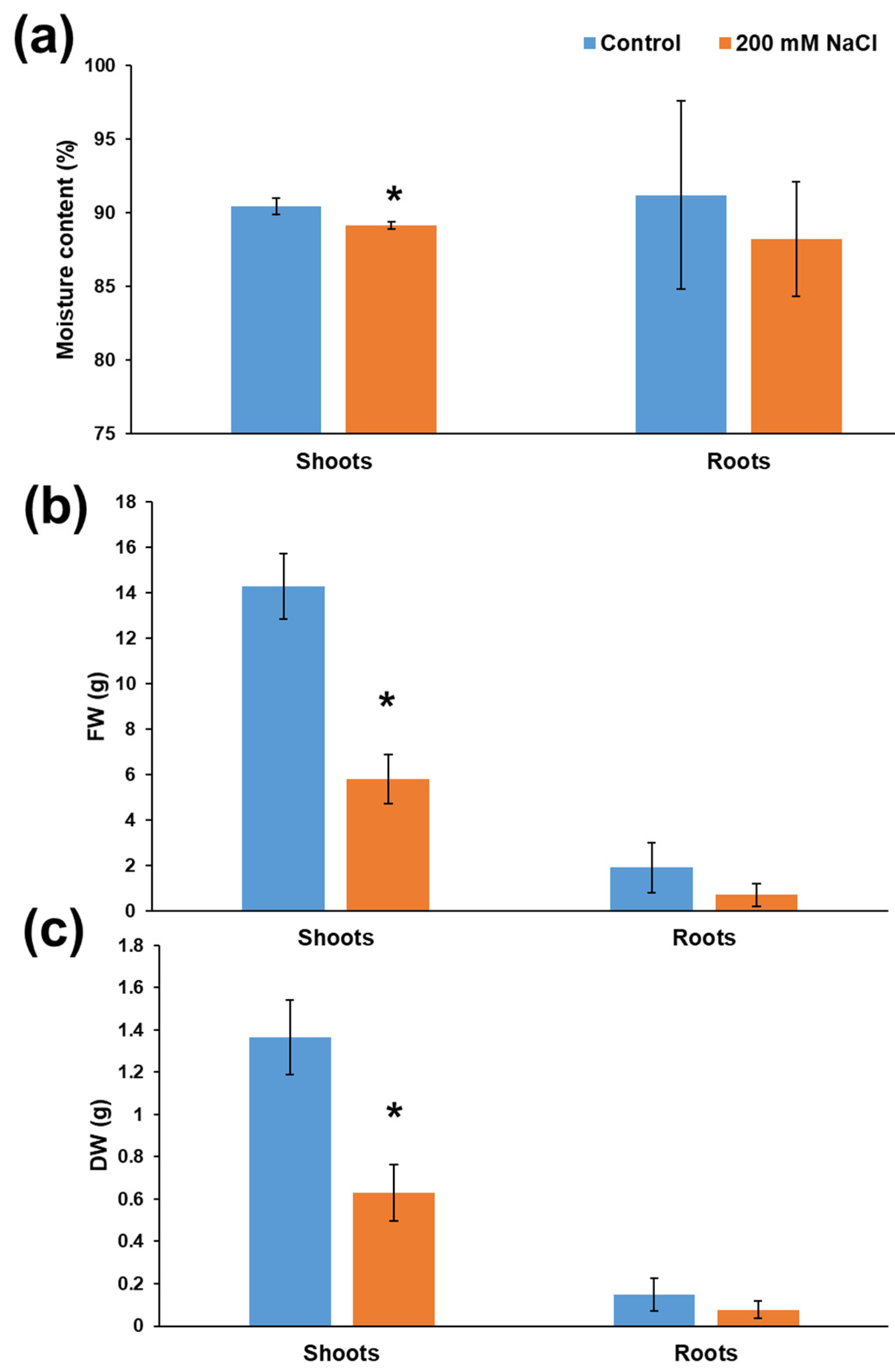
3.1. Effects of Increasing Salt Concentrations on Micro-Tom Leaves and Stems
3.2. Gene Expression Analysis in Leaflets of Micro-Tom Exposed to Salinity
3.2.1. Genes Involved in the Phenylpropanoid Pathway
3.2.2. Genes Involved in Cell Wall Biosynthesis and Remodelling
3.2.3. Genes Involved in Volatile Emission and Stress Response
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the Physiological and Biochemical Impacts of Salt Stress on Plant Growth and Development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Mishra, A.; Tanna, B. Halophytes: Potential Resources for Salt Stress Tolerance Genes and Promoters. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef]
- Dassanayake, M.; Larkin, J.C. Making Plants Break a Sweat: The Structure, Function, and Evolution of Plant Salt Glands. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moles, T.M.; de Brito Francisco, R.; Mariotti, L.; Pompeiano, A.; Lupini, A.; Incrocci, L.; Carmassi, G.; Scartazza, A.; Pistelli, L.; Guglielminetti, L.; et al. Salinity in Autumn-Winter Season and Fruit Quality of Tomato Landraces. Front. Plant. Sci 2019, 10. [Google Scholar] [CrossRef]
- Annex 1. Crop Salt Tolerance Data. Available online: http://www.fao.org/3/y4263e/y4263e0e.htm (accessed on 24 March 2021).
- Yu, J.; Li, Y.; Han, G.; Zhou, D.; Fu, Y.; Guan, B.; Wang, G.; Ning, K.; Wu, H.; Wang, J. The Spatial Distribution Characteristics of Soil Salinity in Coastal Zone of the Yellow River Delta. Environ. Earth Sci. 2014, 72, 589–599. [Google Scholar] [CrossRef] [Green Version]
- Gharbi, E.; Martínez, J.-P.; Benahmed, H.; Lepoint, G.; Vanpee, B.; Quinet, M.; Lutts, S. Inhibition of Ethylene Synthesis Reduces Salt-Tolerance in Tomato Wild Relative Species Solanum Chilense. J. Plant. Physiol. 2017, 210, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Martín-Davison, A.S.; Pérez-Díaz, R.; Soto, F.; Madrid-Espinoza, J.; González-Villanueva, E.; Pizarro, L.; Norambuena, L.; Tapia, J.; Tajima, H.; Blumwald, E.; et al. Involvement of SchRabGDI1 from Solanum Chilense in Endocytic Trafficking and Tolerance to Salt Stress. Plant. Sci. 2017, 263, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.P.; Prasanna, H.C.; Kumari, N.; Mishra, P.; Singh, B. Understanding Salt Tolerance Mechanism Using Transcriptome Profiling and de Novo Assembly of Wild Tomato Solanum Chilense. Sci. Rep. 2020, 10, 15835. [Google Scholar] [CrossRef] [PubMed]
- Žižková, E.; Dobrev, P.I.; Muhovski, Y.; Hošek, P.; Hoyerová, K.; Haisel, D.; Procházková, D.; Lutts, S.; Motyka, V.; Hichri, I. Tomato (Solanum Lycopersicum L.) SlIPT3 and SlIPT4 Isopentenyltransferases Mediate Salt Stress Response in Tomato. BMC Plant Biol. 2015, 15, 85. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Gupta, S.; Lindquist, I.E.; Cameron, C.T.; Mudge, J.; Rashotte, A.M. Transcriptome Analysis of Cytokinin Response in Tomato Leaves. PLoS ONE 2013, 8, e55090. [Google Scholar] [CrossRef] [Green Version]
- Bacha, H.; Tekaya, M.; Drine, S.; Guasmi, F.; Touil, L.; Enneb, H.; Triki, T.; Cheour, F.; Ferchichi, A. Impact of Salt Stress on Morpho-Physiological and Biochemical Parameters of Solanum Lycopersicum Cv. Microtom Leaves. S. Afr. J. Bot. 2017, 108, 364–369. [Google Scholar] [CrossRef]
- Palaniyandi, S.A.; Damodharan, K.; Yang, S.H.; Suh, J.W. Streptomyces Sp. Strain PGPA39 Alleviates Salt Stress and Promotes Growth of ‘Micro Tom’ Tomato Plants. J. Appl. Microbiol. 2014, 117, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Damodharan, K.; Palaniyandi, S.A.; Le, B.; Suh, J.W.; Yang, S.H. Streptomyces Sp. Strain SK68, Isolated from Peanut Rhizosphere, Promotes Growth and Alleviates Salt Stress in Tomato (Solanum Lycopersicum Cv. Micro-Tom). J. Microbiol. 2018, 56, 753–759. [Google Scholar] [CrossRef]
- Al Hassan, M.; Martínez Fuertes, M.; Ramos Sánchez, F.J.; Vicente, O.; Boscaiu, M. Effects of Salt and Water Stress on Plant Growth and on Accumulation of Osmolytes and Antioxidant Compounds in Cherry Tomato. Not. Bot. Horti Agrobot. Cluj-Napoca 2015, 43. [Google Scholar] [CrossRef]
- Shani, E.; Ben-Gera, H.; Shleizer-Burko, S.; Burko, Y.; Weiss, D.; Ori, N. Cytokinin Regulates Compound Leaf Development in Tomato. Plant Cell 2010, 22, 3206–3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.; Berni, R.; Hausman, J.-F.; Guerriero, G. A Review on the Beneficial Role of Silicon against Salinity in Non-Accumulator Crops: Tomato as a Model. Biomolecules 2020, 10, 1284. [Google Scholar] [CrossRef]
- Keshishian, E.A.; Hallmark, H.T.; Ramaraj, T.; Plačková, L.; Sundararajan, A.; Schilkey, F.; Novák, O.; Rashotte, A.M. Salt and Oxidative Stresses Uniquely Regulate Tomato Cytokinin Levels and Transcriptomic Response. Plant Direct 2018, 2, e00071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an Enhanced Web Interface to Primer3. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerriero, G.; Mangeot-Peter, L.; Hausman, J.-F.; Legay, S. Extraction of High Quality RNA from Cannabis Sativa Bast Fibres: A Vademecum for Molecular Biologists. Fibers 2016, 4, 23. [Google Scholar] [CrossRef] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. Open Access Method QBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- González-Aguilera, K.L.; Saad, C.F.; Chávez Montes, R.A.; Alves-Ferreira, M.; de Folter, S. Selection of Reference Genes for Quantitative Real-Time RT-PCR Studies in Tomato Fruit of the Genotype MT-Rg1. Front. Plant Sci. 2016, 7, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster Analysis and Display of Genome-Wide Expression Patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [Green Version]
- Saldanha, A.J. Java Treeview-Extensible Visualization of Microarray Data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [Green Version]
- Assaad, H.I.; Hou, Y.; Zhou, L.; Carroll, R.J.; Wu, G. Rapid Publication-Ready MS-Word Tables for Two-Way ANOVA. Springerplus 2015, 4, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalker-Scott, L. Do anthocyanins function as osmoregulators in leaf tissues? In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2002; Volume 37, pp. 103–127. [Google Scholar]
- Cirillo, V.; D’Amelia, V.; Esposito, M.; Amitrano, C.; Carillo, P.; Carputo, D.; Maggio, A. Anthocyanins Are Key Regulators of Drought Stress Tolerance in Tobacco. Biology 2021, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Sharmin, S.; Kogel, A.; Yu, D.; Kins, L.; Strijkstra, G.-J.; Polle, A. What Makes the Wood? Exploring the Molecular Mechanisms of Xylem Acclimation in Hardwoods to an Ever-Changing Environment. Forests 2019, 10, 358. [Google Scholar] [CrossRef] [Green Version]
- Berni, R.; Mandlik, R.; Hausman, J.F.; Guerriero, G. Silicon-Induced Mitigatory Effects in Salt-Stressed Hemp Leaves. Physiol. Plant. 2020, 171, 476–482. [Google Scholar] [CrossRef]
- Sánchez-Aguayo, I.; Rodríguez-Galán, J.M.; García, R.; Torreblanca, J.; Pardo, J.M. Salt Stress Enhances Xylem Development and Expression of S-Adenosyl-L-Methionine Synthase in Lignifying Tissues of Tomato Plants. Planta 2004, 220, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Rankenberg, T.; Geldhof, B.; van Veen, H.; Holsteens, K.; de Poel, B.V.; Sasidharan, R. Age-Dependent Abiotic Stress Resilience in Plants. Trends Plant Sci. 2021, 26, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-J.; Yang, H.-Y.; Bai, J.-P.; Liang, X.-Y.; Lou, Y.; Zhang, J.-L.; Wang, D.; Zhang, J.-L.; Niu, S.-Q.; Chen, Y. Ultrastructural and Physiological Responses of Potato (Solanum Tuberosum L.) Plantlets to Gradient Saline Stress. Front. Plant Sci. 2015, 5, 787. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Bañon, S.; Olmos, E.; Sánchez-Blanco, M.J. Effects of Sodium Chloride on Water Potential Components, Hydraulic Conductivity, Gas Exchange and Leaf Ultrastructure of Arbutus Unedo Plants. Plant Sci. 2007, 172, 473–480. [Google Scholar] [CrossRef]
- Guerriero, G.; Behr, M.; Hausman, J.F.; Legay, S. Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis. Genes 2017, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Shahid, S.A.; Zaman, M.; Heng, L. Introduction to Soil Salinity, Sodicity and Diagnostics Techniques. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Zaman, M., Shahid, S.A., Heng, L., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–42. ISBN 978-3-319-96190-3. [Google Scholar]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Cuong, D.M.; Kwon, S.-J.; Nguyen, B.V.; Chun, S.W.; Kim, J.K.; Park, S.U. Effect of Salinity Stress on Phenylpropanoid Genes Expression and Related Gene Expression in Wheat Sprout. Agronomy 2020, 10, 390. [Google Scholar] [CrossRef] [Green Version]
- Bouwmeester, H.; Schuurink, R.C.; Bleeker, P.M.; Schiestl, F. The Role of Volatiles in Plant Communication. Plant J. 2019, 100, 892–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Gayoso, C.; Pomar, F.; Novo-Uzal, E.; Merino, F.; Martínez de Ilárduya, Ó. The Ve-Mediated Resistance Response of the Tomato to Verticillium Dahliae Involves H2O2, Peroxidase and Lignins and Drives PAL Gene Expression. BMC Plant Biol. 2010, 10, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, K.C.; Mizuuchi, K.; Koshio, A.; Kaneko, K.; Mitsui, T.; Takeno, K. Stress Enhances the Gene Expression and Enzyme Activity of Phenylalanine Ammonia-Lyase and the Endogenous Content of Salicylic Acid to Induce Flowering in Pharbitis. J. Plant Physiol. 2014, 171, 895–902. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, L.; Chen, S.; Sun, H.; Ma, S. Transcription Profile Analysis of Lycopersicum Esculentum Leaves, Unravels Volatile Emissions and Gene Expression under Salinity Stress. Plant Physiol. Biochem. 2018, 126, 11–21. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, A.; Lim, M.-H.; Lee, S.-W.; Robb, E.J.; Nazar, R.N. Tomato Phenylalanine Ammonia-Lyase Gene Family, Highly Redundant but Strongly Underutilized. J. Biol. Chem. 2008, 283, 33591–33601. [Google Scholar] [CrossRef] [Green Version]
- Rigano, M.M.; Raiola, A.; Docimo, T.; Ruggieri, V.; Calafiore, R.; Vitaglione, P.; Ferracane, R.; Frusciante, L.; Barone, A. Metabolic and Molecular Changes of the Phenylpropanoid Pathway in Tomato (Solanum Lycopersicum) Lines Carrying Different Solanum Pennellii Wild Chromosomal Regions. Front. Plant Sci. 2016, 7, 1484. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four Isoforms of Arabidopsis 4-Coumarate:CoA Ligase Have Overlapping yet Distinct Roles in Phenylpropanoid Metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, D.J. Plant Expansins: Diversity and Interactions with Plant Cell Walls. Curr. Opin. Plant Biol 2015, 25, 162–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a Rose Expansin Gene, Modulates Leaf Growth and Confers Drought and Salt Tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef]
- Abuqamar, S.; Ajeb, S.; Sham, A.; Enan, M.R.; Iratni, R. A Mutation in the Expansin-like A2 Gene Enhances Resistance to Necrotrophic Fungi and Hypersensitivity to Abiotic Stress in Arabidopsis Thaliana. Mol. Plant Pathol. 2013, 14, 813–827. [Google Scholar] [CrossRef]
- Baumann, M.J.; Eklöf, J.M.; Michel, G.; Kallas, Å.M.; Teeri, T.T.; Czjzek, M.; Brumer, H. Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Re-Constructing Our Models of Cellulose and Primary Cell Wall Assembly. Curr. Opin. Plant Biol. 2014, 22, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Nishitani, K.; Tominaga, R. Endo-Xyloglucan Transferase, a Novel Class of Glycosyltransferase That Catalyzes Transfer of a Segment of Xyloglucan Molecule to Another Xyloglucan Molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar] [CrossRef]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan Endotransglycosylase, a New Wall-Loosening Enzyme Activity from Plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.E.; Fry, S.C. Restructuring of Wall-Bound Xyloglucan by Transglycosylation in Living Plant Cells. Plant J. 2001, 26, 23–34. [Google Scholar] [CrossRef]
- Zheng, S.J.; Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; et al. XTH31, Encoding an in Vitro XEH/XET-Active Enzyme, Regulates Aluminum Sensitivity by Modulating in Vivo XET Action, Cell Wall Xyloglucan Content, and Aluminum Binding Capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive Expression of CaXTH3, a Hot Pepper Xyloglucan Endotransglucosylase/Hydrolase, Enhanced Tolerance to Salt and Drought Stresses without Phenotypic Defects in Tomato Plants (Solanum Lycopersicum Cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef]
- Selmar, D.; Kleinwächter, M. Stress Enhances the Synthesis of Secondary Plant Products: The Impact of Stress-Related over-Reduction on the Accumulation of Natural Products. Plant Cell Physiol. 2013, 54, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Savvas, D.; Ntatsi, G. Biostimulant Activity of Silicon in Horticulture. Sci. Hortic. 2015, 196, 66–81. [Google Scholar] [CrossRef]
- Rubio, M.C.; Bustos-Sanmamed, P.; Clemente, M.R.; Becana, M. Effects of Salt Stress on the Expression of Antioxidant Genes and Proteins in the Model Legume Lotus Japonicus. New Phytol. 2009, 181, 851–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shavrukov, Y. Salt Stress or Salt Shock: Which Genes Are We Studying? J. Exp. Bot. 2013, 64, 119–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Mayor, A.; Pineda, B.; Garcia-Abellán, J.O.; Antón, T.; Garcia-Sogo, B.; Sanchez-Bel, P.; Flores, F.B.; Atarés, A.; Angosto, T.; Pintor-Toro, J.A.; et al. Overexpression of Dehydrin Tas14 Gene Improves the Osmotic Stress Imposed by Drought and Salinity in Tomato. J. Plant Physiol. 2012, 169, 459–468. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and Phylogenetic Analysis of Late Embryogenesis Abundant Proteins Family in Tomato (Solanum Lycopersicum). Planta 2014, 241, 757–772. [Google Scholar] [CrossRef]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A Mitochondrial Late Embryogenesis Abundant Protein Stabilizes Model Membranes in the Dry State. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1926–1933. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional Roles of Plant Dehydrins in Response to Environmental Stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, L.; Xing, X.; Sun, L.; Pan, J.; Kong, X.; Zhang, M.; Li, D. ZmLEA3, a Multifunctional Group 3 LEA Protein from Maize (Zea Mays L.), Is Involved in Biotic and Abiotic Stresses. Plant Cell Physiol. 2013, 54, 944–959. [Google Scholar] [CrossRef] [Green Version]
- Belver, A.; Olías, R.; Huertas, R.; Rodríguez-Rosales, M.P. Involvement of SlSOS2 in Tomato Salt Tolerance. Bioengineered 2012, 3. [Google Scholar] [CrossRef]
- Mosblech, A.; Feussner, I.; Heilmann, I. Oxylipins: Structurally Diverse Metabolites from Fatty Acid Oxidation. Plant Physiol. Biochem. 2009, 47, 511–517. [Google Scholar] [CrossRef]
- Andreou, A.; Feussner, I. Lipoxygenases–Structure and Reaction Mechanism. Phytochemistry 2009, 70, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, Z.M. Effect of Nano-Silicon Application on the Expression of Salt Tolerance Genes in Germinating Tomato (Solanum Lycopersicum L.) Seedlings under Salt Stress. POJ 2016, 9, 106–114. [Google Scholar]
- Yang, X.; Zhu, W.; Zhang, H.; Liu, N.; Tian, S. Heat Shock Factors in Tomatoes: Genome-Wide Identification, Phylogenetic Analysis and Expression Profiling under Development and Heat Stress. PeerJ 2016, 2016, e1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffmann, J.; Berni, R.; Sutera, F.M.; Gutsch, A.; Hausman, J.-F.; Saffie-Siebert, S.; Guerriero, G. The Effects of Salinity on the Anatomy and Gene Expression Patterns in Leaflets of Tomato cv. Micro-Tom. Genes 2021, 12, 1165. https://doi.org/10.3390/genes12081165
Hoffmann J, Berni R, Sutera FM, Gutsch A, Hausman J-F, Saffie-Siebert S, Guerriero G. The Effects of Salinity on the Anatomy and Gene Expression Patterns in Leaflets of Tomato cv. Micro-Tom. Genes. 2021; 12(8):1165. https://doi.org/10.3390/genes12081165
Chicago/Turabian StyleHoffmann, Jonas, Roberto Berni, Flavia Maria Sutera, Annelie Gutsch, Jean-Francois Hausman, Suzanne Saffie-Siebert, and Gea Guerriero. 2021. "The Effects of Salinity on the Anatomy and Gene Expression Patterns in Leaflets of Tomato cv. Micro-Tom" Genes 12, no. 8: 1165. https://doi.org/10.3390/genes12081165
APA StyleHoffmann, J., Berni, R., Sutera, F. M., Gutsch, A., Hausman, J.-F., Saffie-Siebert, S., & Guerriero, G. (2021). The Effects of Salinity on the Anatomy and Gene Expression Patterns in Leaflets of Tomato cv. Micro-Tom. Genes, 12(8), 1165. https://doi.org/10.3390/genes12081165







