Abstract
In this study, we investigated the clinical and genetic characteristics of 19 Korean patients with congenital stationary night blindness (CSNB) at two tertiary hospitals. Clinical evaluations, including fundus photography, spectral-domain optical coherence tomography, and electroretinography, were performed. Genetic analyses were conducted using targeted panel sequencing or whole exome sequencing. The median age was 5 (3–21) years at the initial examination, 2 (1–8) years at symptom onset, and 11 (5–28) years during the final visit. Genetic mutations were identified as CNGB1 and GNAT1 for the Riggs type (n = 2), TRPM1 and NYX for the complete type (n = 3), and CACNA1F (n = 14) for the incomplete type. Ten novel variants were identified, and best-corrected visual acuity (BCVA) and spherical equivalents (SE) were related to each type of CSNB. The Riggs and TRPM1 complete types presented mild myopia and good BCVA without strabismus and nystagmus, whereas the NYX complete and incomplete types showed mixed SE and poor BCVA with strabismus and nystagmus. This is the first case series of Korean patients with CSNB, and further studies with a larger number of subjects should be conducted to correlate the clinical and genetic aspects of CSNB.
1. Introduction
Congenital stationary night blindness (CSNB) is a group of non-progressive inherited retinal diseases (IRDs) with dysfunction of rod photoreceptors or signal transduction between photoreceptor cells and bipolar cells, accompanied by various clinical features and diverse genetic mutations [1]. It has been discovered that defects in the visual signal pathway related to the rod photoreceptors, rod ON bipolar cell synapses, or retinoid recycling in the retinal pigment epithelium cause CSNB [2]. Visual symptoms primarily include night blindness with decreased visual acuity, refractive errors, nystagmus, or strabismus [3]. A recent study suggested that children with CSNB may present without complaints of night blindness [4]. In addition, these symptoms can overlap with other progressive IRDs, such as cone–rod dystrophies; thus, accurate diagnosis is essential to predict future visual outcomes.
CSNB is categorized into four types according to electroretinography (ERG) and fundus abnormalities: Riggs type, Schubert–Bornschein type, fundus albipunctatus, and Oguchi [3]. Unlike fundus albipunctatus and Oguchi disease, the Riggs and Schubert–Bornschein types present normal fundus appearances, so they are frequently misdiagnosed as pathologic myopia or infantile nystagmus. The Riggs type shows a reduced wave in the dark-adapted (scotopic) response using ERG [5], whereas the Schubert–Bornschein type features a characteristic electronegative ERG pattern, detected as a normal A wave in the dark-adapted response with a drastically reduced B wave [6]. The Schubert–Bornschein type is divided into two subtypes: a complete type and an incomplete type [7].
The Riggs type genetic mutations have been previously reported, and the known pathogenic genes are autosomal dominant GNAT1 (rod-transducing α subunit) [8,9,10], PDE6B (phosphodiesterase β subunit) [11,12], RHO (rhodopsin) [13,14,15,16], and autosomal recessive SLC24A1 (sodium–calcium exchanger) [17]. Studies have found that Riggs type patients present relatively mild visual symptoms, such as restricted night blindness but normal photopic visual acuity, slight myopia, and no nystagmus [3]. The complete form of the Schubert–Bornschein type involves some notable pathogenic genes, such as X-linked recessive NYX (leucine-rich proteoglycan nyctalopin) [18,19,20], autosomal recessive GRM6 [21,22], TRPM1 [4,23,24,25,26,27,28,29,30], GPR179 [31,32], and LRIT3 [33], affecting signal transduction in the selective rod ON bipolar cell postsynaptic signal loss pathway. Typically, these complete type patients show moderate visual symptoms, including decreased visual acuity and high myopia. On the other hand, the incomplete form of the Schubert–Bornschein type is characterized by both ON and OFF response presynaptic signal dysfunction, and the light-adapted (photopic) responses are more severely reduced compared to the complete type. In addition, the light-adapted 30-Hz flicker response shows reduced amplitude with a double peak sign. The known pathogenic gene is X-linked recessive CACNA1F (calcium-channel alpha1 subunit) [3,7,34,35,36,37,38], and incomplete type patients present with varying degrees of visual symptoms, but commonly worse daylight symptoms than those with the complete type.
To our knowledge, this study, for the first time, documents a case series of 19 Korean CSNB patients using clinical observations and genetic analyses. In Korea, CSNB-related studies with a large number of subjects and investigations of both clinical and genetic aspects have not been performed. Hence, we described the specific details of each case in this paper and were therefore able to comprehend the overall characteristics and causative gene mutations in Korean CSNB patients.
2. Materials and Methods
2.1. Patients and Clinical Data Collection
We enrolled 19 Korean patients with CSNB who visited two tertiary hospitals, Seoul National University Bundang Hospital and Gangnam Severance Hospital, between January 2009 and December 2018 (a 10-year period), and the final follow-up was performed until December 2020. This study was approved by the Institutional Review Board (IRB) of Seoul National University Bundang Hospital (IRB No. B-2101/663-102) and adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from all patients prior to the genetic analyses.
All subjects underwent full ophthalmic examinations, including best-corrected visual acuity (BCVA), fundus photography, spectral-domain optical coherence tomography (SD-OCT; Spectralis OCT; Heidelberg Engineering, Heidelberg, Germany), Goldmann perimetry, full-field standard ERG, and multifocal ERG. The type and frequency of the nystagmus was determined by visual inspection. Full-field ERG was performed using procedures based on the International Society for Clinical Electrophysiology of Vision (ISCEV) [39].
2.2. Genetic Analyses
A comprehensive custom gene panel of 295 known and candidate genes or a 429-gene targeted panel linked to IRDs was performed for genetic analyses as previously described in our reports [40,41]. Targeted next-generation sequencing (Illumina NextSeq 550 system; San Diego, CA, USA) or whole exome sequencing (Illumina NovaSeq 6000 system) were performed. Target enrichment was performed using custom-designed RNA oligonucleotide probes and a target enrichment kit (Celemics, Seoul, South Korea). Whole exome sequencing was performed using xGen Exome Research Panel v1.0 (Integrated DNA Technologies, Inc., Coraville, IA, USA) and SureSelect Human All Exon v6 enrichment kit (Agilent Technologies, Santa Clara, CA, USA).
Burrows–Wheeler Aligner software was used to align the sequence reads in the human hg19 reference genome. Single nucleotide variants and small insertions or deletions were called and crosschecked using Genome Analysis Toolkit version 3.8.0 with HaplotypeCaller and VarScan version 2.4.0. Split-read-based detection of large structural variations was conducted using Pindel and Manta. Read-depth-based detection of copy number variation was conducted using ExomeDepth version 1.1.10 [42], followed by visualization using a base-level read depth normalization algorithm designed by the authors. The variants were annotated by ANNOVAR. The 1000 Genomes Project database; Single Nucleotide Polymorphism database build 137 (dbSNP147); Genome Aggregation Database (gnomAD); the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project; and the Korean Reference Genome Database were used to identify common variants. The Human Gene Mutation Database was searched to identify known pathogenic mutations. The variants were selected if they were not reported or had low frequencies (<1%) in the 1000 Genomes Project, dbSNP147, gnomAD, NHLBI Exome Sequencing Project, or Korean Reference Genome Database, and <30% heterozygous reads or <80% homozygous reads were excluded.
The clinical importance of each variant was categorized based on the latest recommendations of the American College of Medical Genetics and Genomics standards for the interpretation and reporting of sequence variations: pathogenic, likely pathogenic, uncertain significance, benign, and likely benign variant [43,44]. We utilized the automated classification system by Intervar and Varsome, and reviewed the significance of these variants.
3. Results
In this study, the demographics and clinical features of the 19 Korean patients with CSNB are documented in Table 1 and Table 2. The age at the first examination, age at symptom onset, age during the last visit, sex, SEs, initial and final BCVAs, genetic profiles, and confirmed diagnosis were presented in all 19 cases. Whether the identified genetic mutations were novel variants or previously reported is described in Table 3. The median age at initial examination was 5 years with a range of 3–21 years, the median age at symptom onset was 2 years with a range of 1–8 years, and the median age during the final visit was 11 years with a range of 5–28 years. The mean follow-up period was 5.6 ± 3.1 years. Among the 19 enrolled patients, two were male Riggs type CSNB patients, identified as harboring autosomal dominant CNGB1 and GNAT1 gene mutations, one was male complete type with autosomal recessive TRPM1 gene mutation, two were male complete types with X-linked recessive NYX gene mutations, and 14 were male incomplete types with X-linked recessive CACNA1F gene mutations. The representative multimodal images of the Riggs, complete, and incomplete type patients are presented in Figure 1, Figure 2, Figure 3 and Figure 4, which include initial fundus photography, SD-OCT, and full-field standard ERG.

Table 1.
Summary of Clinical Characteristics of Congenital Stationary Night Blindness in 19 Patients.

Table 2.
Clinical Characteristics of Congenital Stationary Night Blindness patients based on classification types.

Table 3.
Pathogenic or likely pathogenic mutations identified in 19 patients with congenital stationary night blindness.
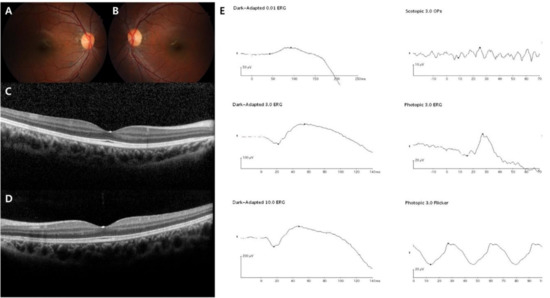
Figure 1.
Representative images of the Riggs type CSNB patient (case 1). The patient was diagnosed with CSNB at age 9, and the last visit was at age 12. GNAT1 was sequenced as the pathogenic gene. (A–D) Normal fundus structures observed using initial fundus photography and SD-OCT. (E) Full-field standard ERG showing significantly reduced dark-adapted rod responses (both the A and B waves) and normal light-adapted cone responses in the right eye.
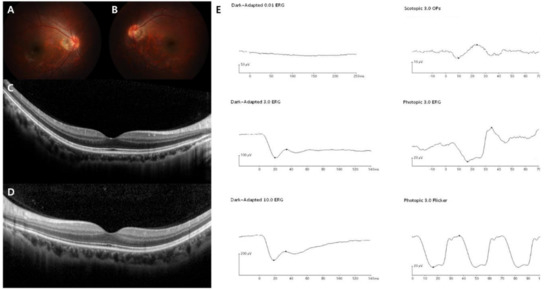
Figure 2.
Representative images of the complete form of CSNB (case 3). The patient was diagnosed with CSNB at age 19, and the last visit was at age 28. TRPM1 was sequenced as the pathogenic gene. (A–D) Normal fundus structures observed using initial fundus photography and SD-OCT. (E) Full-field standard ERG showing relatively preserved dark-adapted 3.0 A wave, while markedly reduced B wave, suggesting “electronegative ERG” pattern. Light-adapted 3.0 cone responses are slightly decreased, but normal 30-Hz flicker responses are detected in the right eye.
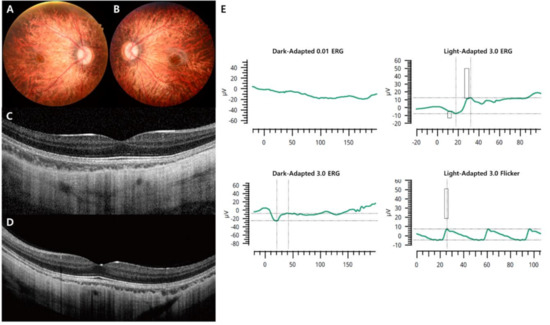
Figure 3.
Representative images of the complete form of CSNB caused by an NYX mutation (case 5). (A,B) Fundus photographs showing myopic tigroid fundus. (C,D) OCT revealing normal fovea and outer retina structures but thinning of the choroidal thickness. (E) There was no detectable ERG in the 0.01 dark-adapted response, and electronegative ERG was observed for the 3.0 dark-adapted response. Decreased light-adapted cone responses in the right eye were detected.
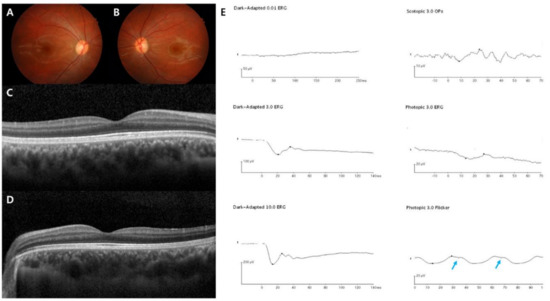
Figure 4.
Representative images of the incomplete form of the Schubert–Bornschein type (case 16). The patient was diagnosed with CSNB at age 7, and the last visit was at age 18. CACNA1F was sequenced as the pathogenic gene. (A–D) Normal fundus structures observed using initial fundus photography and SD-OCT. (E) Full-field standard ERG showing typical electronegative ERG pattern. Unlike the complete type, significantly decreased light-adapted 3.0 cone responses and 30-Hz flicker responses were detected with a double peak sign in the right eye (blue arrow), suggesting both severe cone and rod dysfunction.
Two male Riggs type patients with an autosomal dominant CNGB1 and GNAT1 gene mutations were enrolled. The patients all presented with emmetropic features and relatively good BCVAs. The pathogenic variants were CNGB1 [NM_001297, c.2544delG: p.(G848fs)], (NM_001297, c.1035-1G>A) and GNAT1 [NM_000172, c.753C>A:p.(Asp251Lys)]. The complete-type patient with autosomal recessive TRPM1 gene mutation showed high myopia with relatively good BCVAs in both eyes. The pathogenic variants were previously reported TRPM1 [NM_002420.6, c.3280C>T:p.(Arg1094*)], [NM_002420.6, c.3794delA:p.(Asn1265Ilefs*42)] [30]. The two male complete-type patients with X-linked recessive NYX gene mutations showed high myopic features (mean, −7.5D) with poor BCVAs (mean, logMAR 0.7) in both eyes. One patient was too young to measure visual acuity; thus, we documented the BCVA as “poor.” The pathogenic variants were NYX [NM_022567.2, c.182_183insT:p.(Cys62Valfs*53)], and NYX (NM_022567.2, c.38-1_38delGCinsTT) mutations [45]. The patient with the novel mutation had 3–4 Hz left beating jerk-pendular nystagmus.
The 14 male incomplete type patients with X-linked recessive CACNA1F gene mutations show mixed SE features (three highly myopic, seven slightly myopic, and four hyperopic cases) with poor BCVAs in both eyes maintained from the initial examination to the final visit: initial BCVA 0.76 ± 0.21 logMAR, final BCVA 0.71 ± 0.19 logMAR, median SE −1.12, and mean ± SD SE −2.4 ± 4.3 (Table 1 and Table 2). We discovered six novel pathogenic variants with one overlapping mutation: (1) NM_005183.2, exon (13–23) deletion; (2) NM_005183.2, c.2175_2179delins:p.(Gly726IIefs*61); (3) NM_005183.2, c.1910+1G>A; (4) NM_005183.2, c.4042-1G>T; (5) NM_005183.2, c.2761C>A:p.(Leu921IIe); (6) NM_005183.2, c.2767-1G>C. Seven other pathogenic variants have been described in previous literature [46,47,48,49,50,51]. Among the 14 patients with CACNA1F mutations, 10 (71%) had nystagmus.
In addition, we analyzed the ERG patterns of each CSNB type and summarized them in Table 2. The Riggs and TRPM1 complete types showed low dark-adapted 3.0 ERG A-wave and B-wave amplitudes, and preserved light-adapted 3.0, and 30-Hz flicker ERG amplitude with typical electronegative ERG (B/A ratio ≤ 1) compared to age-matched normal control values: Riggs dark-adapted 3.0 A-wave amplitude 66.9 ± 8.3, B-wave amplitude 54.1 ± 4.7, B/A ratio 0.81 ± 0.05, light-adapted 3.0 A-wave amplitude 19.5 ± 2.1, B-wave amplitude 55.9 ± 3.7, and 30-Hz flicker amplitude 51.8 ± 4.9; TRPM1 dark-adapted 3.0 A-wave amplitude 184.1 ± 38.3, B-wave amplitude 86.3 ± 1.2, B/A ratio 0.49 ± 0.09, light-adapted 3.0 A-wave amplitude 46.8 ± 4.1, B-wave amplitude 85.6 ± 0.3, and 30-Hz flicker amplitude 74.5 ± 5.1. On the other hand, the NYX complete and CACNA1F incomplete types showed substantially reduced A-wave and B-wave amplitude in dark-adapted 3.0-and light-adapted 3.0 as well as markedly decreased 30-Hz flicker ERG amplitude with electronegative ERG: NYX dark-adapted 3.0 A-wave amplitude 31.8 ± 6.2, B-wave amplitude 18.9 ± 1.1, B/A ratio 0.61 ± 0.08, light-adapted 3.0 A-wave amplitude 6.6 ± 0.7, B-wave amplitude 12.7 ± 3.8, and 30-Hz flicker amplitude 11.7 ± 2.6; CACNA1F dark-adapted 3.0 A-wave amplitude 43.7 ± 6.0, B-wave amplitude 25.3 ± 3.3, B/A ratio 0.58 ± 0.02, light-adapted 3.0 A-wave amplitude 3.8 ± 0.5, B-wave amplitude 6.5 ± 0.3, and 30-Hz flicker amplitude 5.7 ± 0.9. The characteristic “double peak” 30-Hz flicker ERG pattern (Figure 4) was observed in 9 incomplete type patients (64%).
4. Discussion
Our study investigated the clinical and genetic characteristics of 19 Korean patients with CSNB. We identified three types of CSNB, including Riggs and the complete and incomplete forms of Schubert–Bornschein, and their pathogenic genetic profiles. CACNA1F mutations appeared to be the major genetic cause in our cohort. The clinical characteristics according to CSNB type were similar to those reported in the literature; the Riggs type presented better BCVA with mild myopia compared to the Schubert–Bornschein type [3,7,52]. Among the Schubert–Bornschein type subjects, complete type patients with TRPM1 gene mutations presented better BCVA than the others. This genotype–phenotype correlation suggests the importance of genetic analyses for predicting visual outcomes in patients with CSNB.
The pathogenic genetic mutations of the Riggs type affect the phototransduction cascade and retinoid recycling pathway [1,3]. The pathogenic genetic mutations of the complete type, NYX and TRPM1, affect glutamate-induced signaling. In the dark-adapted environment, rod photoreceptors release glutamate, which binds to GRM6 (mGluR6 receptor) and influences TRPM1 ion channel closure [53]. If the TRPM1 channel is dysregulated, constant depolarization of the ON bipolar cells leads to decreased photoreceptor sensitivity [54]. The NYX gene consists of two exons encoding nyctalopin and is considered to localize the TRPM1 receptor in the proper position at the dendritic tip of ON bipolar cells [18,19]. The TRPM1 gene consists of 27 exons encoding 1642 amino acids, and the mutants displayed dysfunction of the TRPM1 channel [7,23]. The pathogenic genetic mutations of the incomplete type, CACNA1F, affect glutamate release, which plays a role in signal transmission from the photoreceptors to bipolar cells [55]. The CACNA1F gene consists of 48 exons encoding 1966 amino acids and is widely known for encoding the α1-subunit of an L-type voltage-dependent Ca2+ channel [56]. Dysregulation of glutamate release affects both ON and OFF bipolar cells; thus, the incomplete type is likely to present more severe visual symptoms than the complete type by influencing both rod and cone cell responses.
Clinical studies evaluating the genotype–phenotype correlation in CSNB patients have been conducted since the identification of pathogenic genes. Previous studies involving novel mutations revealed that the autosomal dominant Riggs type CSNB patients did not show typical myopia, nystagmus, strabismus, or severe visual impairment [8,9,10,11,12,13,14,15,57]. According to our case with a CNGB1 and GNAT1 mutation, mild visual symptoms, including good BCVAs, emmetropic to mild myopia, and night blindness, were documented, which were similar to prior clinical findings [58,59]. Electronegative ERGs with relatively preserved cone responses were also consistent with previous investigations [3]. In our cohort, compound heterozygous variants of CNGB1 and a heterozygous variant of GNAT1 were identified in two male Riggs type patients. Bi-allelic pathogenic variants of CNGB1 are known to cause retinitis pigmentosa. Recently, isolated rod dysfunction associated with CNGB1 was reported [60]. Although retinal pigmentary change may appear at a late age, we included this patient as CSNB based on the clinical features at the time of study enrollment, since all the ophthalmic examinations were completely normal with mild visual symptom of night blindness.
There have also been numerous investigations focusing on the Schubert–Bornschein type. Miyake et al. subdivided Schubert–Bornschein type CSNB into complete and incomplete depending on the dark-adapted ERG pattern [61]. The authors found that the refractive errors were different between the complete (mostly myopic) and incomplete (variation from highly myopic to hyperopic) types. Allen et al. reported that 11 X-linked CSNB families displayed either NYX or CACNA1F gene mutations and suggested that the complete or incomplete phenotypes do not correlate with genotype, whereas some ERG features, such as oscillatory potentials and ON/OFF bipolar responses, may be helpful for indicating genotypes [62]. Bijveld et al. studied 101 Dutch patients with NYX, TRPM1, GRM6, and GPR179 mutations in the complete type and CACNA1F and CABP4 mutations in the incomplete type [7]. The authors pointed out that the photopic ERG pattern was the most important clinical test to distinguish between the complete and incomplete types, and complete type patients showed better visual acuity than those with the incomplete type. In addition, the incomplete type showed both rod- and cone-related visual symptoms and only half of the patients suffered night blindness compared to the complete type, which was mainly rod-related and showed 100% night blindness. The authors also found that even though there were various genetic mutations in the complete type, there was only one unique phenotype, whereas in the incomplete type, CACNA1F and CABP4 mutations showed distinct visual symptoms. According to our three cases of the complete type, two cases of NYX gene mutations showed high myopia and poor visual acuity, whereas one case of TRPM1 gene mutations showed high myopia but retained visual acuity. This suggests that in the complete form of the Schubert–Bornschein type, genotype confirmation may be relevant to clinical phenotypes. In our 14 cases of the incomplete type, all CACNA1F gene mutations presented various features of SE from high myopic to hyperopic; however, all enrolled subjects showed poor visual acuity, which is consistent with prior findings [7,37,38,60]. Moreover, according to our ERG analysis, complete TRPM1 patients showed relatively preserved cone responses, whereas complete NYX and incomplete CACNA1F patients showed profoundly decreased cone responses, which is consistent with previous studies [3,7]. In addition, as Miyake et al. [59] reported, a 30-Hz flicker ERG double peak occurrence was detected in nine patients with incomplete ERG (64%).
This study had some limitations. First, despite enrolling CSNB patients from two tertiary hospitals, only 19 subjects (two Riggs type, three complete types, and 14 incomplete types) were included in the study. Our study may not reflect all patients with CSNB in Korea. Due to the small study population, we could not apply the appropriate statistical genotype–phenotype correlation analysis. Additional multicenter clinical trials with larger enrollment numbers should be conducted. Moreover, potential patients with CSNB may have been missed because of diagnostic complexity. Finally, the age of symptom onset may be inaccurate in adults, and the follow-up period may not be sufficient to conclude long-term visual acuity impairment.
Nonetheless, our study has some strengths that should be mentioned. To the best of our knowledge, this is the first multicenter study to analyze and document Korean patients with CSNB as a case series. A few reports regarding novel mutations in Korea have been published; however, evaluation of the genotype–phenotype correlation in 19 Korean CSNB cases may be valuable. Moreover, we identified 13 novel mutations in this article, which are summarized in Table 3.
5. Conclusions
This is the first Korean CSNB case series on the genotype–phenotype correlation. The Riggs and complete types of TPRM1 gene mutations presented good visual acuity, whereas the incomplete and complete types of NYX gene mutations were frequently associated with poor visual acuity and nystagmus. Clinicians should be aware of the possibility of diagnosing CSNB when examining children with symptoms of poor visual acuity, strabismus, nystagmus, and no definite retinal structural abnormalities [4]. Moreover, genetic analysis with next-generation sequencing is recommended as a frontline diagnostic tool for CSNB patients. Future studies with larger populations should be conducted to assess the statistical genotype–phenotype correlation in Korean patients with CSNB.
Author Contributions
Conceptualization, S.-J.W.; Writing—original draft preparation, H.-M.K. and K.J.; Writing—review and editing, H.-M.K., K.J., and J.H.; Supervision, S.-J.W.; Project administration, S.-J.W.; Funding acquisition, S.-J.W., J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Research Foundation grants (Nos. 2020R1F1A1072795 and 2020R1C1C1007965) from the Korean government (MSIT), the Korea Disease Control and Prevention Agency (Grant Nos. 2018-ER6902-02 and 2019-NG-051-01), and a research grant from Seoul National University Bundang Hospital (No. 02-2020-013). The funding organizations had no role in the design or conduct of this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zeitz, C.; Forster, U.; Neidhardt, J.; Feil, S.; Kalin, S.; Leifert, D.; Flor, P.J.; Berger, W. Night blindness-associated mutations in the ligand-binding, cysteine-rich, and intracellular domains of the metabotropic glutamate receptor 6 abolish protein trafficking. Hum. Mutat. 2007, 28, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Labs, S.; Lorenz, B.; Forster, U.; Uksti, J.; Kroes, H.Y.; De Baere, E.; Leroy, B.P.; Cremers, F.P.; Wittmer, M.; et al. Genotyping microarray for CSNB-associated genes. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5919–5926. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Robson, A.G.; Audo, I. Congenital stationary night blindness: An analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog. Retin. Eye Res. 2015, 45, 58–110. [Google Scholar] [CrossRef] [PubMed]
- Miraldi Utz, V.; Pfeifer, W.; Longmuir, S.Q.; Olson, R.J.; Wang, K.; Drack, A.V. Presentation of TRPM1-Associated Congenital Stationary Night Blindness in Children. JAMA Ophthalmol 2018, 136, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Riggs, L.A. Electroretinography in cases of night blindness. Am. J. Ophthalmol. 1954, 38, 70–78. [Google Scholar] [CrossRef]
- Schubert, G.; Bornschein, H. Analysis of the human electroretinogram. Ophthalmologica 1952, 123, 396–413. [Google Scholar] [CrossRef]
- Bijveld, M.M.; Florijn, R.J.; Bergen, A.A.; van den Born, L.I.; Kamermans, M.; Prick, L.; Riemslag, F.C.; van Schooneveld, M.J.; Kappers, A.M.; van Genderen, M.M. Genotype and phenotype of 101 dutch patients with congenital stationary night blindness. Ophthalmology 2013, 120, 2072–2081. [Google Scholar] [CrossRef]
- Dryja, T.P.; Berson, E.L.; Rao, V.R.; Oprian, D.D. Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat. Genet. 1993, 4, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Szabo, V.; Kreienkamp, H.J.; Rosenberg, T.; Gal, A.P. Gln200Glu, a putative constitutively active mutant of rod α-transducin (GNAT1) in autosomal dominant congenital stationary night blindness. Hum. Mutat. 2007, 28, 741–742. [Google Scholar] [CrossRef]
- Naeem, M.A.; Chavali, V.R.; Ali, S.; Iqbal, M.; Riazuddin, S.; Khan, S.N.; Husnain, T.; Sieving, P.A.; Ayyagari, R.; Riazuddin, S.; et al. GNAT1 associated with autosomal recessive congenital stationary night blindness. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1353–1361. [Google Scholar] [CrossRef]
- Gal, A.; Orth, U.; Baehr, W.; Schwinger, E.; Rosenberg, T. Heterozygous missense mutation in the rod cGMP phosphodiesterase β-subunit gene in autosomal dominant stationary night blindness. Nat. Genet. 1994, 7, 551. [Google Scholar] [CrossRef] [PubMed]
- Manes, G.; Cheguru, P.; Majumder, A.; Bocquet, B.; Senechal, A.; Artemyev, N.O.; Hamel, C.P.; Brabet, P. A truncated form of rod photoreceptor PDE6 β-subunit causes autosomal dominant congenital stationary night blindness by interfering with the inhibitory activity of the γ-subunit. PLoS ONE 2014, 9, e95768. [Google Scholar] [CrossRef]
- Rao, V.R.; Cohen, G.B.; Oprian, D.D. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature 1994, 367, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Sieving, P.A.; Richards, J.E.; Naarendorp, F.; Bingham, E.L.; Scott, K.; Alpern, M. Dark-light: Model for nightblindness from the human rhodopsin Gly-90-->Asp mutation. Proc. Natl. Acad. Sci. USA 1995, 92, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Al-Jandal, N.; Farrar, G.J.; Kiang, A.S.; Humphries, M.M.; Bannon, N.; Findlay, J.B.; Humphries, P.; Kenna, P.F. A novel mutation within the rhodopsin gene (Thr-94-Ile) causing autosomal dominant congenital stationary night blindness. Hum. Mutat. 1999, 13, 75–81. [Google Scholar] [CrossRef]
- Zeitz, C.; Gross, A.K.; Leifert, D.; Kloeckener-Gruissem, B.; McAlear, S.D.; Lemke, J.; Neidhardt, J.; Berger, W. Identification and functional characterization of a novel rhodopsin mutation associated with autosomal dominant CSNB. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4105–4114. [Google Scholar] [CrossRef]
- Riazuddin, S.A.; Shahzadi, A.; Zeitz, C.; Ahmed, Z.M.; Ayyagari, R.; Chavali, V.R.; Ponferrada, V.G.; Audo, I.; Michiels, C.; Lancelot, M.E.; et al. A mutation in SLC24A1 implicated in autosomal-recessive congenital stationary night blindness. Am. J. Hum. Genet. 2010, 87, 523–531. [Google Scholar] [CrossRef]
- Bech-Hansen, N.T.; Naylor, M.J.; Maybaum, T.A.; Sparkes, R.L.; Koop, B.; Birch, D.G.; Bergen, A.A.; Prinsen, C.F.; Polomeno, R.C.; Gal, A.; et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat. Genet. 2000, 26, 319–323. [Google Scholar] [CrossRef]
- Pusch, C.M.; Zeitz, C.; Brandau, O.; Pesch, K.; Achatz, H.; Feil, S.; Scharfe, C.; Maurer, J.; Jacobi, F.K.; Pinckers, A.; et al. The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat. Genet. 2000, 26, 324–327. [Google Scholar] [CrossRef]
- Ivanova, M.E.; Zolnikova, I.V.; Gorgisheli, K.V.; Atarshchikov, D.S.; Ghosh, P.; Barh, D. Novel frameshift mutation in NYX gene in a Russian family with complete congenital stationary night blindness. Ophthalmic. Genet. 2019, 40, 558–563. [Google Scholar] [CrossRef]
- Dryja, T.P.; McGee, T.L.; Berson, E.L.; Fishman, G.A.; Sandberg, M.A.; Alexander, K.R.; Derlacki, D.J.; Rajagopalan, A.S. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc. Natl. Acad. Sci. USA 2005, 102, 4884–4889. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; van Genderen, M.; Neidhardt, J.; Luhmann, U.F.; Hoeben, F.; Forster, U.; Wycisk, K.; Matyas, G.; Hoyng, C.B.; Riemslag, F.; et al. Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15-Hz flicker electroretinogram. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4328–4335. [Google Scholar] [CrossRef] [PubMed]
- Audo, I.; Kohl, S.; Leroy, B.P.; Munier, F.L.; Guillonneau, X.; Mohand-Said, S.; Bujakowska, K.; Nandrot, E.F.; Lorenz, B.; Preising, M.; et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2009, 85, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sergouniotis, P.I.; Michaelides, M.; Mackay, D.S.; Wright, G.A.; Devery, S.; Moore, A.T.; Holder, G.E.; Robson, A.G.; Webster, A.R. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am. J. Hum. Genet. 2009, 85, 711–719. [Google Scholar] [CrossRef]
- Van Genderen, M.M.; Bijveld, M.M.; Claassen, Y.B.; Florijn, R.J.; Pearring, J.N.; Meire, F.M.; McCall, M.A.; Riemslag, F.C.; Gregg, R.G.; Bergen, A.A.; et al. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am. J. Hum. Genet. 2009, 85, 730–736. [Google Scholar] [CrossRef]
- Al-Hujaili, H.; Taskintuna, I.; Neuhaus, C.; Bergmann, C.; Schatz, P. Long-term follow-up of retinal function and structure in TRPM1-associated complete congenital stationary night blindness. Mol. Vis. 2019, 25, 851–858. [Google Scholar] [PubMed]
- AlTalbishi, A.; Zelinger, L.; Zeitz, C.; Hendler, K.; Namburi, P.; Audo, I.; Sheffer, R.; Yahalom, C.; Khateb, S.; Banin, E.; et al. TRPM1 Mutations are the Most Common Cause of Autosomal Recessive Congenital Stationary Night Blindness (CSNB) in the Palestinian and Israeli Populations. Sci. Rep. 2019, 9, 12047. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, Y.; Zeevi, D.A.; Lam, B.L.; Scher, S.Y.; Bringer, R.; Cherki, B.; Cohen, C.C.; Muallem, H.; Chiang, J.P.; Pantrangi, M.; et al. A founder deletion in the TRPM1 gene associated with congenital stationary night blindness and myopia is highly prevalent in Ashkenazi Jews. Hum. Genome Var. 2019, 6, 45. [Google Scholar] [CrossRef]
- Hayashi, T.; Mizobuchi, K.; Kikuchi, S.; Nakano, T. Novel biallelic TRPM1 variants in an elderly patient with complete congenital stationary night blindness. Doc. Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Lee, Y.J.; Joo, K.; Seong, M.W.; Park, K.H.; Park, S.S.; Woo, S.J. Congenital Stationary Night Blindness due to Novel TRPM1 Gene Mutations in a Korean Patient. Korean J. Ophthalmol. 2020, 34, 170–172. [Google Scholar] [CrossRef]
- Audo, I.; Bujakowska, K.; Orhan, E.; Poloschek, C.M.; Defoort-Dhellemmes, S.; Drumare, I.; Kohl, S.; Luu, T.D.; Lecompte, O.; Zrenner, E.; et al. Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012, 90, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Peachey, N.S.; Ray, T.A.; Florijn, R.; Rowe, L.B.; Sjoerdsma, T.; Contreras-Alcantara, S.; Baba, K.; Tosini, G.; Pozdeyev, N.; Iuvone, P.M.; et al. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012, 90, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Jacobson, S.G.; Hamel, C.P.; Bujakowska, K.; Neuille, M.; Orhan, E.; Zanlonghi, X.; Lancelot, M.E.; Michiels, C.; Schwartz, S.B.; et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2013, 92, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Bech-Hansen, N.T.; Naylor, M.J.; Maybaum, T.A.; Pearce, W.G.; Koop, B.; Fishman, G.A.; Mets, M.; Musarella, M.A.; Boycott, K.M. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat. Genet. 1998, 19, 264–267. [Google Scholar] [CrossRef]
- Strom, T.M.; Nyakatura, G.; Apfelstedt-Sylla, E.; Hellebrand, H.; Lorenz, B.; Weber, B.H.; Wutz, K.; Gutwillinger, N.; Ruther, K.; Drescher, B.; et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat. Genet. 1998, 19, 260–263. [Google Scholar] [CrossRef]
- Boycott, K.M.; Pearce, W.G.; Bech-Hansen, N.T. Clinical variability among patients with incomplete X-linked congenital stationary night blindness and a founder mutation in CACNA1F. Can. J. Ophthalmol. 2000, 35, 204–213. [Google Scholar] [CrossRef]
- Nakamura, M.; Ito, S.; Terasaki, H.; Miyake, Y. Novel CACNA1F mutations in Japanese patients with incomplete congenital stationary night blindness. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1610–1616. [Google Scholar]
- Mahmood, U.; Mejecase, C.; Ali, S.M.A.; Moosajee, M.; Kozak, I. A Novel Splice-Site Variant in CACNA1F Causes a Phenotype Synonymous with Aland Island Eye Disease and Incomplete Congenital Stationary Night Blindness. Genes 2021, 12, 17. [Google Scholar] [CrossRef]
- Marmor, M.F.; Fulton, A.B.; Holder, G.E.; Miyake, Y.; Brigell, M.; Bach, M. International Society for Clinical Electrophysiology of, V. ISCEV Standard for full-field clinical electroretinography (2008 update). Doc. Ophthalmol. 2009, 118, 69–77. [Google Scholar] [CrossRef]
- Seong, M.W.; Seo, S.H.; Yu, Y.S.; Hwang, J.M.; Cho, S.I.; Ra, E.K.; Park, H.; Lee, S.J.; Kim, J.Y.; Park, S.S. Diagnostic application of an extensive gene panel for Leber congenital amaurosis with severe genetic heterogeneity. J. Mol. Diagn. 2015, 17, 100–105. [Google Scholar] [CrossRef]
- Kim, M.S.; Joo, K.; Seong, M.W.; Kim, M.J.; Park, K.H.; Park, S.S.; Woo, S.J. Genetic Mutation Profiles in Korean Patients with Inherited Retinal Diseases. J. Korean. Med. Sci. 2019, 34, e161. [Google Scholar] [CrossRef]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef]
- ClinVar. NM_022567.2(NYX):c.38-1_38delinsTT. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/496942/ (accessed on 13 March 2021).
- Hove, M.N.; Kilic-Biyik, K.Z.; Trotter, A.; Gronskov, K.; Sander, B.; Larsen, M.; Carroll, J.; Bech-Hansen, T.; Rosenberg, T. Clinical Characteristics, Mutation Spectrum, and Prevalence of Aland Eye Disease/Incomplete Congenital Stationary Night Blindness in Denmark. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6861–6869. [Google Scholar] [CrossRef] [PubMed]
- Rim, J.H.; Lee, S.T.; Gee, H.Y.; Lee, B.J.; Choi, J.R.; Park, H.W.; Han, S.H.; Han, J. Accuracy of Next-Generation Sequencing for Molecular Diagnosis in Patients With Infantile Nystagmus Syndrome. JAMA Ophthalmol. 2017, 135, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Zito, I.; Allen, L.E.; Patel, R.J.; Meindl, A.; Bradshaw, K.; Yates, J.R.; Bird, A.C.; Erskine, L.; Cheetham, M.E.; Webster, A.R.; et al. Mutations in the cacna1f and nyx genes in british csnbx families. Hum. Mutat. 2003, 21, 169. [Google Scholar] [CrossRef]
- Wutz, K.; Sauer, C.; Zrenner, E.; Lorenz, B.; Alitalo, T.; Broghammer, M.; Hergersberg, M.; de la Chapelle, A.; Weber, B.H.; Wissinger, B.; et al. Thirty distinct CACNA1F mutations in 33 families with incomplete type of XLCSNB and Cacna1f expression profiling in mouse retina. Eur. J. Hum. Genet. 2002, 10, 449–456. [Google Scholar] [CrossRef]
- Wang, X.; Zein, W.M.; D’Souza, L.; Roberson, C.; Wetherby, K.; He, H.; Villarta, A.; Turriff, A.; Johnson, K.R.; Fann, Y.C. Applying next generation sequencing with microdroplet pcr to determine the disease-causing mutations in retinal dystrophies. BMC Ophthalmol. 2017, 17, 157. [Google Scholar] [CrossRef]
- Sun, W.; Huang, L.; Xu, Y.; Xiao, X.; Li, S.; Jia, X.; Gao, B.; Wang, P.; Guo, X.; Zhang, Q. Exome Sequencing on 298 Probands With Early-Onset High Myopia: Approximately One-Fourth Show Potential Pathogenic Mutations in RetNet Genes. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8365–8372. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, F.; Almeshari, N.; Ahmad, K.; Magliyah, M.S.; Schatz, P. Congenital stationary night blindness: An update and review of the disease spectrum in Saudi Arabia. Acta Ophthalmol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Koike, C.; Obara, T.; Uriu, Y.; Numata, T.; Sanuki, R.; Miyata, K.; Koyasu, T.; Ueno, S.; Funabiki, K.; Tani, A.; et al. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc. Natl. Acad. Sci. USA 2010, 107, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Orhan, E.; Prezeau, L.; El Shamieh, S.; Bujakowska, K.M.; Michiels, C.; Zagar, Y.; Vol, C.; Bhattacharya, S.S.; Sahel, J.A.; Sennlaub, F.; et al. Further insights into GPR179: Expression, localization, and associated pathogenic mechanisms leading to complete congenital stationary night blindness. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8041–8050. [Google Scholar] [CrossRef]
- Specht, D.; Wu, S.B.; Turner, P.; Dearden, P.; Koentgen, F.; Wolfrum, U.; Maw, M.; Brandstatter, J.H.; tom Dieck, S. Effects of presynaptic mutations on a postsynaptic Cacna1s calcium channel colocalized with mGluR6 at mouse photoreceptor ribbon synapses. Investig. Ophthalmol. Vis. Sci. 2009, 50, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kerov, V.; Haeseleer, F.; Majumder, A.; Artemyev, N.; Baker, S.A.; Lee, A. Dysregulation of Ca(v)1.4 channels disrupts the maturation of photoreceptor synaptic ribbons in congenital stationary night blindness type 2. Channels (Austin) 2013, 7, 514–523. [Google Scholar] [CrossRef]
- Muradov, K.G.; Granovsky, A.E.; Artemyev, N.O. Mutation in rod PDE6 linked to congenital stationary night blindness impairs the enzyme inhibition by its γ-subunit. Biochemistry 2003, 42, 3305–3310. [Google Scholar] [CrossRef]
- Marmor, M.F.; Zeitz, C. Riggs-type dominant congenital stationary night blindness: ERG findings, a new GNAT1 mutation and a systemic association. Doc. Ophthalmol. 2018, 137, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Méjécase, C.; Stévenard, M.; Michiels, C.; Audo, I.; Marmor, M.F. A Novel Heterozygous Missense Mutation in GNAT1 Leads to Autosomal Dominant Riggs Type of Congenital Stationary Night Blindness. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Ba-Abbad, R.; Holder, G.E.; Robson, A.G.; Neveu, M.M.; Waseem, N.; Arno, G.; Webster, A.R. Isolated rod dysfunction associated with a novel genotype of CNGB1. Am. J. Ophthalmol. Case Rep. 2019, 14, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Yagasaki, K.; Horiguchi, M.; Kawase, Y.; Kanda, T. Congenital stationary night blindness with negative electroretinogram. A new classification. Arch. Ophthalmol. 1986, 104, 1013–1020. [Google Scholar] [CrossRef]
- Allen, L.E.; Zito, I.; Bradshaw, K.; Patel, R.J.; Bird, A.C.; Fitzke, F.; Yates, J.R.; Trump, D.; Hardcastle, A.J.; Moore, A.T. Genotype-phenotype correlation in British families with X linked congenital stationary night blindness. Br. J. Ophthalmol. 2003, 87, 1413–1420. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).