Molecular Cytogenetic and Y Copy Number Analysis of a Reciprocal ECAY-ECA13 Translocation in a Stallion with Complete Meiotic Arrest
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Animal and Samples
2.2. Chromosome Preparations for Molecular Cytogenetic Analysis
2.3. Selection of Probes for Fluorescence In Situ Hybridization (FISH)
2.4. Genomic and BAC DNA Isolation
2.5. Fluorescence In Situ Hybridization (FISH)
2.6. Digital Droplet PCR Analysis (ddPCR)
3. Results
3.1. Molecular Cytogenetic Analysis of ECAY and ECA13 Reciprocal Translocation
3.2. Copy Number Analysis of Horse MSY Multicopy Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nielsen, J.; Rasmussen, K. Y/autosomal translocations. Clin. Genet. 1976, 9, 609–617. [Google Scholar] [CrossRef]
- Pinton, A.; Letron, I.R.; Berland, H.; Bonnet, N.; Calgaro, A.; Garnier-Bonnet, A.; Yerle, M.; Ducos, A. Meiotic studies in an azoospermic boar carrying a Y;14 translocation. Cytogenet. Genome Res. 2008, 120, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Barasc, H.; Mary, N.; Letron, I.R.; Calgaro, A.; Dudez, A.; Bonnet, N.; Lahbib-Mansais, Y.; Yerle, M.; Ducos, A.; Pinton, A. Y-Autosome Translocation Interferes with Meiotic Sex Inactivation and Expression of Autosomal Genes: A Case Study in the Pig. Sex. Dev. 2012, 6, 143–150. [Google Scholar] [CrossRef]
- Villagómez, D.A.; Revay, T.; Donaldson, B.; Rezaei, S.; Pinton, A.; Palomino, M.; Junaidi, A.H.A.; Honaramooz, A.; King, W.A. Azoospermia and Testicular Hypoplasia in a Boar Carrier of a Novel Y-Autosome Translocation. Sex. Dev. 2017, 11, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Mary, N.; Villagomez, D.A.F.; Revay, T.; Rezaei, S.; Donaldson, B.; Pinton, A.; King, W.A. Meiotic Synapsis and Gene Expression Altered by a Balanced Y-Autosome Reciprocal Translocation issn an Azoospermic Pig. Sex. Dev. 2018, 12, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Switonski, M.; Szczerbal, I.; Krumrych, W.; Nowacka-Woszuk, J. A Case of Y-Autosome Reciprocal Translocation in a Holstein-Friesian Bull. Cytogenet. Genome Res. 2011, 132, 22–25. [Google Scholar] [CrossRef]
- Iannuzzi, L.; Molteni, L.; Di Meo, G.; De Giovanni, A.; Perucatti, A.; Succi, G.; Incarnato, D.; Eggen, A.; Cribiu, E. A case of azoospermia in a bull carrying a Y-autosome reciprocal translocation. Cytogenet. Cell Genet. 2001, 95, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.J.; Castaneda, C.; Raudsepp, T.; Tibary, A. Azoospermia and Y Chromosome–Autosome Translocation in a Friesian Stallion. J. Equine Veter. Sci. 2019, 82, 102781. [Google Scholar] [CrossRef] [PubMed]
- Janečka, J.E.; Davis, B.W.; Ghosh, S.; Paria, N.; Das, P.J.; Orlando, L.; Schubert, M.; Nielsen, M.K.; Stout, T.A.E.; Brashear, W.; et al. Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes. Nat. Commun. 2018, 9, 2945. [Google Scholar] [CrossRef]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.-J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef]
- Morel, F.; Duguépéroux, I.; McElreavey, K.; Le Bris, M.-J.; Herry, A.; Parent, P.; Le Martelot, M.-T.; Fellous, M.; De Braekeleer, M. Transmission of an unbalanced (Y;1) translocation in Brittany, France. J. Med. Genet. 2002, 39, 52e. [Google Scholar] [CrossRef]
- Benitez, J.; Rivera, L.; Ramos, C.; Tejedor, E.; Sanchez-Cascos, A. Translocation of a supernumerary Y to a 15: Study of six cases (three males and three females) in three generations. Qual. Life Res. 1979, 48, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Oliver-Bonet, M.; Turek, P.; Ko, E.; Martin, R. Meiotic studies in an azoospermic human translocation (Y;1) carrier. Mol. Hum. Reprod. 2005, 11, 361–364. [Google Scholar] [CrossRef]
- Wang, D.; Chen, R.; Kong, S.; Pan, Q.-Y.; Zheng, Y.-H.; Qiu, W.-J.; Fan, Y.; Sun, X.-F. Cytogenic and molecular studies of male infertility in cases of Y chromosome balanced reciprocal translocation. Mol. Med. Rep. 2017, 16, 2051–2054. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cribiu, E.P.; Di Berardino, D.; Di Meo, G.P.; Eggen, A.; Gallagher, D.S.; Gustavsson, I.; Hayes, H.; Iannuzzi, L.; Popescu, C.P.; Rubes, J.; et al. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet. Genome Res. 2001, 92, 283–299. [Google Scholar] [CrossRef]
- Raudsepp, T.; Chowdhary, B.P. The Eutherian Pseudoautosomal Region. Cytogenet. Genome Res. 2015, 147, 81–94. [Google Scholar] [CrossRef]
- Turner, J.M.; Mahadevaiah, S.K.; Ellis, P.J.; Mitchell, M.J.; Burgoyne, P.S. Pachytene Asynapsis Drives Meiotic Sex Chromosome Inactivation and Leads to Substantial Postmeiotic Repression in Spermatids. Dev. Cell 2006, 10, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Chowdhary, B.P. FISH for Mapping Single Copy Genes. Methods Mol. Biol. 2008, 422, 31–49. [Google Scholar]
- Raudsepp, T.; Gustafson-Seabury, A.; Durkin, K.; Wagner, M.L.; Goh, G.; Seabury, C.M.; Brinkmeyer-Langford, C.; Lee, E.-J.; Agarwala, R.; Rice, E.S.; et al. A 4103 marker integrated physical and comparative map of the horse genome. Cytogenet. Genome Res. 2008, 122, 28–36. [Google Scholar] [CrossRef]
- Raudsepp, T.; Chowdhary, B. The horse pseudoautosomal region (PAR): Characterization and comparison with the human, chimp and mouse PARs. Cytogenet. Genome Res. 2008, 121, 102–109. [Google Scholar] [CrossRef]
- Raudsepp, T.; Chowdhary, B.P. Construction of chromosome-specific paints for meta- and submetacentric autosomes and the sex chromosomes in the horse and their use to detect homologous chromosomal segments in the donkey. Chromosom. Res. 1999, 7, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S., Jr.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O’connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 197. [Google Scholar] [CrossRef]
- Castañeda, C.; Hillhouse, A.; Janecka, J.; Juras, R.; Ruiz, A.; Tibary, A.; Love, C.; Varner, D.; Raudsepp, T. Contribution of the Y Chromosome to Stallion Fertility. J. Equine Veter. Sci. 2018, 66, 30. [Google Scholar] [CrossRef]
- Bugno-Poniewierska, M.; Raudsepp, T. Horse Clinical Cytogenetics: Recurrent Themes and Novel Findings. Animals 2021, 11, 831. [Google Scholar] [CrossRef]
- Das, P.; Lyle, S.; Beehan, D.; Chowdhary, B.; Raudsepp, T. Cytogenetic and Molecular Characterization of Y Isochromosome in a 63XO/64Xi(Yq) Mosaic Karyotype of an Intersex Horse. Sex. Dev. 2011, 6, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Herzog, A.; Höhn, H.; Klug, E.; Hecht, W. A sex chromosome mosaic in male pseudohermaphroditism in a horse. Tierarztliche Prax. 1989, 17, 171–175. [Google Scholar]
- Raudsepp, T.; Durkin, K.; Lear, T.L.; Das, P.J.; Avila, F.; Kachroo, P.; Chowdhary, B.P. Molecular heterogeneity of XY sex reversal in horses. Anim. Genet. 2010, 41, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Barišić, A.; Tomljanović, A.B.; Čizmarević, N.S.; Ostojić, S.; Romac, P.; Vraneković, J. A rare Y-autosome translocation found in a patient with nonobstructive azoospermia: Case report. Syst. Biol. Reprod. Med. 2021, 67, 1–7. [Google Scholar] [CrossRef]
- Delobel, B.; Djlelati, R.; Gabriel-Robez, O.; Croquette, M.-F.; Rousseaux-Prévost, R.; Rousseaux, J.; Rigot, J.-M.; Rumpler, Y. Y-autosome translocation and infertility: Usefulness of molecular, cytogenetic and meiotic studies. Qual. Life Res. 1998, 102, 98–102. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Mahadevaiah, S.K.; Celeste, A.; Romanienko, P.J.; Camerini-Otero, R.; Bonner, W.M.; Manova, K.; Burgoyne, P.; Nussenzweig, A. H2AX Is Required for Chromatin Remodeling and Inactivation of Sex Chromosomes in Male Mouse Meiosis. Dev. Cell 2003, 4, 497–508. [Google Scholar] [CrossRef]
- Skinner, B.M.; Lachani, K.; Sargent, C.A.; Affara, N.A. Regions of XY homology in the pig X chromosome and the boundary of the pseudoautosomal region. BMC Genet. 2013, 14, 3. [Google Scholar] [CrossRef]
- Sciurano, R.; Rahn, M.; Rey-Valzacchi, G.; Solari, A.J. The asynaptic chromatin in spermatocytes of translocation carriers contains the histone variant γ-H2AX and associates with the XY body. Hum. Reprod. 2007, 22, 142–150. [Google Scholar] [CrossRef]
- Hsu, L.Y.F. Phenotype/Karyotype correlations of Y chromosome aneuploidy with emphasis on structural aberrations in postnatally diagnosed cases. Am. J. Med. Genet. 1994, 53, 108–140. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, P.; Cavigliano, P.M.; Boni, M.; Malcovati, L.; Calatroni, S.; Astori, C.; Caresana, M.; Bernasconi, C. A novel t(Y;11) translocation with MLL gene rearrangement in a case of acute myelomonocytic leukemia (AML-M4). Leukemia 1999, 13, 487–489. [Google Scholar] [CrossRef][Green Version]
- Royo, H.; Polikiewicz, G.; Mahadevaiah, S.K.; Prosser, H.; Mitchell, M.; Bradley, A.; de Rooij, D.; Burgoyne, P.S.; Turner, J.M. Evidence that Meiotic Sex Chromosome Inactivation Is Essential for Male Fertility. Curr. Biol. 2010, 20, 2117–2123. [Google Scholar] [CrossRef] [PubMed]
- Waters, P.D.; Ruiz-Herrera, A. Meiotic Executioner Genes Protect the Y from Extinction. Trends Genet. 2020, 36, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nat. Cell Biol. 2014, 508, 488–493. [Google Scholar] [CrossRef]
- Vernet, N.; Mahadevaiah, S.K.; Yamauchi, Y.; Decarpentrie, F.; Mitchell, M.J.; Ward, M.A.; Burgoyne, P.S. Mouse Y-Linked Zfy1 and Zfy2 Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2nd Meiotic Division. PLoS Genet. 2014, 10, e1004444. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Pyntikova, T.; Koutseva, N.; Raudsepp, T.; Brown, L.G.; Bellott, D.W.; Cho, T.-J.; Dugan-Rocha, S.; Khan, Z.; et al. Sequence analysis in Bos taurus reveals pervasiveness of X–Y arms races in mammalian lineages. Genome Res. 2020, 30, 1716–1726. [Google Scholar] [CrossRef]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.; Verduzco-Gómez, A.; Favetta, L.; Blondin, P.; King, W. Testis-Specific Protein Y-Encoded Copy Number Is Correlated to Its Expression and the Field Fertility of Canadian Holstein Bulls. Sex. Dev. 2012, 6, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Krausz, C.; Giachini, C.; Forti, G. TSPY and Male Fertility. Genes 2010, 1, 308–316. [Google Scholar] [CrossRef] [PubMed]
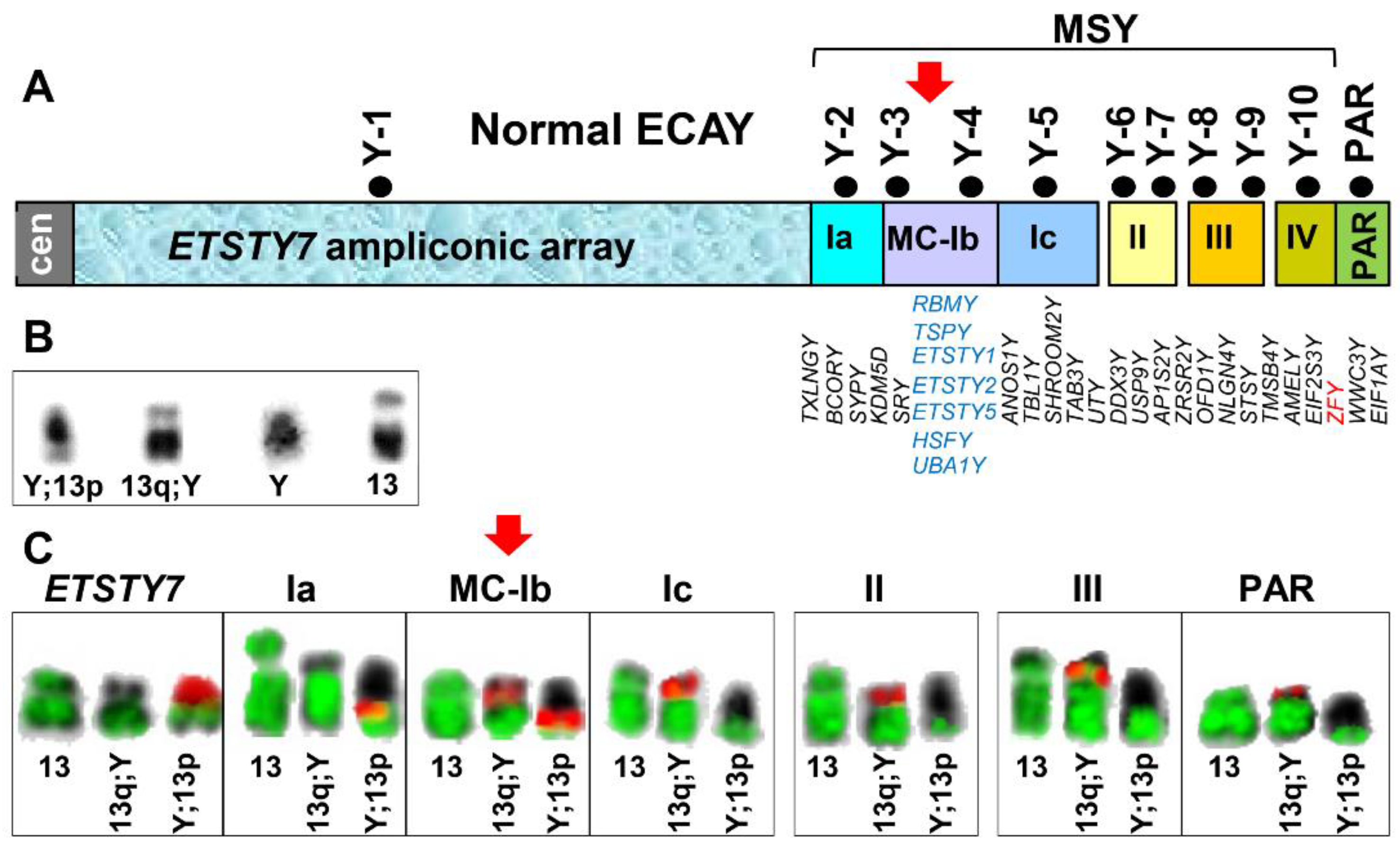


| Marker ID | CH241 BAC | Cytogenetic Location | Location in EquCab3 or ECAY BAC Contig Map | Reference Marker | Reference |
|---|---|---|---|---|---|
| Y-1 | 069E11 | Yq proximal 2/3 | Y and Xq heterochromatin | ETSTY7 ampliconic array | [9] |
| Y-2 | 022P7 | Yqdistal | Y Contig Ia, single copy | KDM5D | [9] |
| Y-3 | 140M23 | Yqdistal | Y Contig Ib, multicopy (MC) | SRY | [9] |
| Y-4 | 017D15 | Yqdistal | Y Contig Ib, multicopy (MC) | TSPY | [9] |
| Y-5 | 090G18 | Yqdistal | Y Contig Ic, single copy | n/a | [9] |
| Y-6 | 112E12 | Yqdistal | Y Contig II, single copy | NLGN4Y | [9] |
| Y-7 | 011B8 | Yqdistal | Y Contig II, single copy | n/a | [9] |
| Y-8 | 125H6 | Yqdistal | Y Contig III, single copy | TMSB4Y | [9] |
| Y-9 | 102J15 | Yqdistal | Y Contig III, single copy | TMSB4Y | [9] |
| Y-10 | 106F1 | Yqdistal | Y Contig IV single copy | ZFY | [9] |
| PAR | 194E12 | Xpter/Yqter | chrX:3945-246,703 | PLCXD1 | [20] |
| 13-1 | 078E13 | 13p15 | chr13:5,913,678-6,105,097 | GPER1 | This study |
| 13-2 | 060D24 | 13p13 | chr13:11,481,169-11,662,804 | ELN | This study |
| 13-3 | 158P20 | 13q12 | chr13:18,059,033-18,254,381 | LEX041 | [19] |
| Gene | Forward Primer 5′–3′ | Reverse Primer 5′–3′ | Probe Sequence 5′–3′ |
|---|---|---|---|
| TSPY | CATAGTGGAGGAAGAGGATGAAA | GGCAATGGTTTAACCCTGAAA | CTCTTTCTGGGAGACCTGCCCTTT |
| SRY | TTCTGTGATCTATGCTGGCG | TTACCCTCCGGACTTTCTCA | AACAGGGACTCTGCCGCCACCA |
| RBMY | GAAGCTCCACAACTTGAGGT | CTCTGACCTATGATGGAAGCA | TGTCTGCCACCATGCTCACGACCA |
| ETSTY1 | GACGGACGACCTTGTGTT | ACGCTCACAGATGACAGTAG | TGTCCCGGCCACCTCAGGGC |
| ETSTY2 | TTGTTGTTAGGCTACCTGGC | AAGGGCAAACCATAACCTCC | TGGGCAAGCTTCTCCATGGTTGCTGCA |
| ETSTY5 | GAGGCAGGTACTTCGTTACC | TCACTCACAAAGTCAACGCT | TGCCGTGAGCTTGAGGGCGAA |
| UBA1Y | TTTCTGTTGTCTGGACGGAG | CTCCACGGATGTAGTCAGAG | AGCAGAGGCCTCCTGTGTCTGAGCT |
| HSFY | AGGCTTTCTCCACTGGTTTC | GAGGCTGTCCCGAACTTTTA | CCCCTGCTCTAAAGTGCTTCCTGTCG |
| MYOZ1 | GACTTTCCAGATGCCCAAGT | ACCAGAACCTCTCCAACAGGCCTTCT | GCTCCTCTGTTTCTCCATCC |
| Sample ID | TSPY | ETSTY1 | ETSTY2 | ETSTY5 | SRY | RBMY | HSFY | UBA1Y |
|---|---|---|---|---|---|---|---|---|
| H787 | 5.6 | 1.9 | 3.8 | 2.6 | 0.8 | 1.9 | 0.9 | 3.1 |
| 23346 | 9.1 | 3.6 | 5.7 | 5.3 | 1 | 1.5 | 0.9 | 3.8 |
| 23348 | 8.2 | 7.4 | 5.7 | 4.8 | 1 | 2.3 | 0.9 | 3.5 |
| 70858 | 9.7 | 4.1 | 4.2 | 4.1 | 0.8 | 2 | 0.9 | 3.2 |
| 70980 | 10.8 | 3.9 | 5.1 | 4.4 | 0.9 | 1.7 | 0.9 | 3.8 |
| 70981 | 11.3 | 3.8 | 4.6 | 4.4 | 0.8 | 1.7 | 1.1 | 3 |
| 73901 | 8.6 | 3.9 | 4.1 | 3.9 | 1 | 2 | 1 | 3 |
| 74413 | 8.2 | 3.9 | 4.3 | 4.3 | 0.9 | 1.9 | 1 | 3.6 |
| 74836 | 8.2 | 3.7 | 4.6 | 4 | 0.8 | 1.7 | 1.2 | 3.3 |
| 74837 | 8.6 | 3.9 | 4.5 | 3.7 | 0.7 | 1.9 | 1 | 3.8 |
| 75052 | 9.2 | 3.6 | 4.6 | 4.2 | 0.9 | 2 | 1 | 3.7 |
| TR007 | 9 | 4.2 | 3.8 | 4.1 | 1 | 1.9 | 1 | 3.9 |
| TR008 | 9.5 | 4 | 3.7 | 4.1 | 0.9 | 2.1 | 1 | 4.3 |
| TR009 | 8.2 | 3.9 | 3.8 | 3.8 | 0.9 | 2 | 1 | 4 |
| H061 | 8 | 4.1 | 4.5 | 3.6 | 0.8 | 1.9 | 1.1 | 1.8 |
| H294 | 13.3 | 3.8 | 5.4 | 4.4 | 0.8 | 1.7 | 1.1 | 3.3 |
| Bravo | 8.5 | 4.9 | 5.6 | 3.6 | 0.9 | 2 | 1.1 | 1.5 |
| p-value | 0.0249 * | 0.0293 * | 0.2494 | 0.0038 ** | 0.4003 | 0.9756 | 0.2365 | 0.7583 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaneda, C.; Ruiz, A.J.; Tibary, A.; Raudsepp, T. Molecular Cytogenetic and Y Copy Number Analysis of a Reciprocal ECAY-ECA13 Translocation in a Stallion with Complete Meiotic Arrest. Genes 2021, 12, 1892. https://doi.org/10.3390/genes12121892
Castaneda C, Ruiz AJ, Tibary A, Raudsepp T. Molecular Cytogenetic and Y Copy Number Analysis of a Reciprocal ECAY-ECA13 Translocation in a Stallion with Complete Meiotic Arrest. Genes. 2021; 12(12):1892. https://doi.org/10.3390/genes12121892
Chicago/Turabian StyleCastaneda, Caitlin, Agustin J. Ruiz, Ahmed Tibary, and Terje Raudsepp. 2021. "Molecular Cytogenetic and Y Copy Number Analysis of a Reciprocal ECAY-ECA13 Translocation in a Stallion with Complete Meiotic Arrest" Genes 12, no. 12: 1892. https://doi.org/10.3390/genes12121892
APA StyleCastaneda, C., Ruiz, A. J., Tibary, A., & Raudsepp, T. (2021). Molecular Cytogenetic and Y Copy Number Analysis of a Reciprocal ECAY-ECA13 Translocation in a Stallion with Complete Meiotic Arrest. Genes, 12(12), 1892. https://doi.org/10.3390/genes12121892







