New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Strains and Genome Assemblies
2.2. Illumina Reads and Testis Transcriptome Assembly
2.3. Identification of Y-Linked Scaffolds and Annotation of Y-Linked Genes
2.4. Timeline of Gene Acquisitions by the Y Chromosome
2.5. Measurement of Gene Expression
2.6. Detection of Segmental Duplications in the Y Chromosome
3. Results
3.1. Identification of Y-Linked Sequences of D. willistoni
3.2. Identification of Y-Linked Protein Coding Genes
3.3. Y-Linked Segmental Duplications from Autosomes
3.4. Mechanism of Duplications to the Y: DNA or RNA-Based?
3.5. When Did the D. willistoni Y Chromosome Acquire Its Genes?
4. Discussion
4.1. D. willistoni Y-Linked Genes: Comparison with Other Drosophila Species
4.2. Gene Movements to the Y Chromosome and Reproductive Isolation
4.3. How Do Male Genes Move to the Y Chromosome?
4.4. Why Do Male Genes Move to the Y Chromosome?
4.5. Concluding Remarks and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Bridges, C.B. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1916, 1, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J. Evolution of Sex Determining Mechanisms; Benjamin/Cummings Pub. Co. Advanced Book Program: Menlo Park, CA, USA, 1983; p. xx, 316. [Google Scholar]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Meccariello, A.; Salvemini, M.; Primo, P.; Hall, B.; Koskinioti, P.; Dalikova, M.; Gravina, A.; Gucciardino, M.A.; Forlenza, F.; Gregoriou, M.E.; et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 2019, 365, 1457–1460. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.; Celniker, S.; Holt, R.; Evans, C.; Gocayne, J.; Amanatides, P.; Scherer, S.; Li, P.; Hoskins, R.; Galle, R. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Muller, H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool 1914, 17, 325–336. [Google Scholar] [CrossRef]
- Rice, W.R. Evolution of the Y sex chromosome in animals. Bioscience 1996, 46, 331–343. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1563–1572. [Google Scholar] [CrossRef]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Sturgill, D.; Zhang, Y.; Parisi, M.; Oliver, B. Demasculinization of X chromosomes in the Drosophila genus. Nature 2007, 450, 238–241. [Google Scholar] [CrossRef]
- Parisi, M.; Nuttall, R.; Naiman, D.; Bouffard, G.; Malley, J.; Andrews, J.; Eastman, S.; Oliver, B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 2003, 299, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Rozen, S. Genomics and genetics of human and primate Y chromosomes. Annu. Rev. Genom. Hum. Genet. 2012, 13, 83–108. [Google Scholar] [CrossRef]
- Carvalho, A.B.; Clark, A.G. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013, 23, 1894–1907. [Google Scholar] [CrossRef]
- Koerich, L.B.; Wang, X.; Clark, A.G.; Carvalho, A.B. Low conservation of gene content in the Drosophila Y chromosome. Nature 2008, 456, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Vicoso, B.; Russo, C.A.; Swenor, B.; Clark, A.G. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, 12450–12455. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Koerich, L.B.; Clark, A.G. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009, 25, 270–277. [Google Scholar] [CrossRef]
- Mahajan, S.; Bachtrog, D. Convergent evolution of Y chromosome gene content in flies. Nat. Commun. 2017, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- Gepner, J.; Hays, T.S. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc. Natl. Acad. Sci. USA 1993, 90, 11132–11136. [Google Scholar] [CrossRef]
- Carvalho, A.B. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 2002, 12, 664–668. [Google Scholar] [CrossRef]
- Hall, A.B.; Qi, Y.; Timoshevskiy, V.; Sharakhova, M.V.; Sharakhov, I.V.; Tu, Z. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genom. 2013, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Dobo, B.A.; Vibranovski, M.D.; Clark, A.G. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2001, 98, 13225–13230. [Google Scholar] [CrossRef]
- Long, M.; VanKuren, N.W.; Chen, S.; Vibranovski, M.D. New gene evolution: Little did we know. Annu. Rev. Genet. 2013, 47, 307–333. [Google Scholar] [CrossRef]
- Bhutkar, A.; Russo, S.M.; Smith, T.F.; Gelbart, W.M. Genome-scale analysis of positionally relocated genes. Genome Res. 2007, 17, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Reugels, A.M.; Kurek, R.; Lammermann, U.; Bunemann, H. Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant Threads loops in primary spermatocytes of Drosophila hydei. Genetics 2000, 154, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.D.; Koerich, L.B.; Carvalho, A.B.; Clark, A.G. Positive and purifying selection on the Drosophila Y chromosome. Mol. Biol. Evol. 2014, 31, 2612–2623. [Google Scholar] [CrossRef]
- Carvalho, A.B.; Clark, A.G. Intron size and natural selection. Nature 1999, 401, 344. [Google Scholar] [CrossRef]
- Diaz-Castillo, C.; Golic, K.G. Evolution of gene sequence in response to chromosomal location. Genetics 2007, 177, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Vibranovski, M.D.; Zhang, Y.; Long, M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009, 19, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Lazzaro, B.P.; Clark, A.G. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl. Acad. Sci. USA 2000, 97, 13239–13244. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The evolution of dominance. Biol. Rev. 1931, 6, 345–368. [Google Scholar] [CrossRef]
- Lynch, M.; Katju, V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004, 20, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Maclennan, J.; Kim, K.W.; Rambaut, A.; O’Grady, P.M.; Jiggins, F.M. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 2012, 29, 3459–3473. [Google Scholar] [CrossRef]
- Tamura, K.; Subramanian, S.; Kumar, S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 2004, 21, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.G.; Eisen, M.B.; Smith, D.R.; Bergman, C.M.; Oliver, B.; Markow, T.A.; Kaufman, T.C.; Kellis, M.; Gelbart, W.; Iyer, V.N.; et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef]
- Mardiros, X.B.; Park, R.; Clifton, B.; Grewal, G.; Khizar, A.K.; Markow, T.A.; Ranz, J.M.; Civetta, A. Postmating reproductive isolation between strains of Drosophila willistoni. Fly 2016, 10, 162–171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, B.Y.; Wang, J.R.; Miller, D.E.; Barmina, O.; Delaney, E.; Thompson, A.; Comeault, A.A.; Peede, D.; D’Agostino, E.R.; Pelaez, J.; et al. Highly contiguous assemblies of 101 drosophilid genomes. Elife 2021, 10, e66405. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Shu, S.Q.; Mungall, C.J.; Karpen, G.H. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 2007, 316, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Smith, C.D.; Carlson, J.W.; Carvalho, A.B.; Halpern, A.; Kaminker, J.S.; Kennedy, C.; Mungall, C.J.; Sullivan, B.A.; Sutton, G.G.; et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002, 3, 1–16. [Google Scholar] [CrossRef]
- Dupim, E.G.; Goldstein, G.; Vanderlinde, T.; Vaz, S.C.; Krsticevic, F.; Bastos, A.; Pinhao, T.; Torres, M.; David, J.R.; Vilela, C.R.; et al. An investigation of Y chromosome incorporations in 400 species of Drosophila and related genera. PLoS Genet. 2018, 14, e1007770. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jaime, M.; Polihronakis, M.; Kanegawa, K.; Markow, T.; Kaneshiro, K.; Oliver, B. Re-annotation of eight Drosophila genomes. Life Sci. Alliance 2018, 1, e201800156. [Google Scholar] [CrossRef] [PubMed]
- Betran, E. The “life histories” of genes. J. Mol. Evol. 2015, 80, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Tobler, R.; Nolte, V.; Schlotterer, C. High rate of translocation-based gene birth on the Drosophila Y chromosome. Proc. Natl. Acad. Sci. USA 2017, 114, 11721–11726. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Clark, A.G. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 2005, 307, 108–110. [Google Scholar] [CrossRef]
- Bailey, J.A.; Gu, Z.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef]
- Coulombe-Huntington, J.; Majewski, J. Intron loss and gain in Drosophila. Mol. Biol. Evol. 2007, 24, 2842–2850. [Google Scholar] [CrossRef]
- Finet, C.; Kassner, V.A.; Carvalho, A.B.; Chung, H.; Day, J.P.; Day, S.; Delaney, E.K.; De Re, F.C.; Dufour, H.D.; Dupim, E.; et al. DrosoPhyla: Resources for Drosophilid Phylogeny and Systematics. Genome Biol. Evol. 2021, 13, evab179. [Google Scholar] [CrossRef]
- Ayala, F.J.; Tracey, M.L. Enzyme variability in the Drosophila willistoni group. VIII. Genetic differentiation and reproductive isolation between subespecies. J. Hered. 1973, 64, 120–124. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res 2021, 49, D899–D907. [Google Scholar] [CrossRef] [PubMed]
- Elgin, S.C.; Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef] [PubMed]
- Zanini, R.; Muller, M.J.; Vieira, G.C.; Valiati, V.H.; Depra, M.; Valente, V. Combining morphology and molecular data to improve Drosophila paulistorum (Diptera, Drosophilidae) taxonomic status. Fly 2018, 12, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Krsticevic, F.J.; Schrago, C.G.; Carvalho, A.B. Long-read single molecule sequencing to resolve tandem gene copies: The Mst77Y region on the Drosophila melanogaster Y chromosome. G3 2015, 5, 1145–1150. [Google Scholar] [CrossRef]
- Chapple, C.E.; Guigo, R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS ONE 2008, 3, e2968. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. “Patterns of speciation in Drosophila”. Evolution 1997, 51, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Winge, H.; Cordeiro, A.R. Experimental hybrids between Drosophila willistoni Sturtevant and Drosophila paulistorum Dobzhansky and Pavan from southern marginal populations. Heredity 1963, 18, 215–222. [Google Scholar] [CrossRef]
- Masly, J.P.; Jones, D.; Noor, M.A.; Locke, J.; Orr, H.A. Gene transposition as a cause of hybrid sterility in Drosophila. Science 2006, 313, 1448–1450. [Google Scholar] [CrossRef]
- Gvozdev, V.A.; Kogan, G.L.; Usakin, L.A. The Y chromosome as a target for acquired and amplified genetic material in evolution. Bioessays 2005, 27, 1256–1262. [Google Scholar] [CrossRef]
- Chen, P.; Kotov, A.A.; Godneeva, B.K.; Bazylev, S.S.; Olenina, L.V.; Aravin, A.A. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes. Dev. 2021, 35, 914–935. [Google Scholar] [CrossRef]
- Brown, E.J.; Nguyen, A.H.; Bachtrog, D. The Y chromosome may contribute to sex-specific ageing in Drosophila. Nat. Ecol. Evol. 2020, 4, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Lemos, B.; Araripe, L.O.; Hartl, D.L. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 2008, 319, 91–93. [Google Scholar] [CrossRef]
- Lamnissou, K.; Loukas, M.; Zouros, E. Incompatibilities between Y chromosome and autosomes are responsible for male hybrid sterility in crosses between Drosophila virilis and Drosophila texana. Heredity 1996, 76, 603–609. [Google Scholar] [CrossRef]
- Marais, G.A.; Nicolas, M.; Bergero, R.; Chambrier, P.; Kejnovsky, E.; Moneger, F.; Hobza, R.; Widmer, A.; Charlesworth, D. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 2008, 18, 545–549. [Google Scholar] [CrossRef]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; University Press: Cambridge, UK, 1973; 961p. [Google Scholar]
- Steinemann, M.; Steinemann, S. Degenerating Y-chromosome of Drosophila miranda-a trap for retrotransposons. Proc. Natl. Acad. Sci. USA 1992, 89, 7591–7595. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Wei, K.H.; Nalley, M.J.; Gibilisco, L.; Bachtrog, D. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 2018, 16, e2006348. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Hackstein, J.H.P.; Hochstenbach, R.; Hauschteck-Jungen, E.; Beukeboom, L.W. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? Bioessays 1996, 18, 317–323. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B-chromosome evolution. Philos. Trans. R. Soc. Lond. Ser. B-Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef]
- Nokkala, S.; Nokkala, C. Interaction of B chromosomes with A or B chromosomes in segregation in insects. Cytogenet. Genome Res. 2004, 106, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Grozeva, S.; Kuznetsova, V.; Maryanska-Nadachowska, A. The origin of the achiasmatic XY sex chromosome system in Cacopsylla peregrina (Frst.)(Psylloidea, Homoptera). Genetica 2003, 119, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Nokkala, S.; Kuznetsova, V.; Maryanska-Nadachowska, A. Achiasmate segregation of a B chromosome from the X chromosome in two species of psyllids (Psylloidea, Homoptera). Genetica 2000, 108, 181–189. [Google Scholar] [CrossRef]
- Fraisse, C.; Picard, M.A.L.; Vicoso, B. The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat. Commun. 2017, 8, 1486. [Google Scholar] [CrossRef] [PubMed]
- Steinemann, M.; Pinsker, W.; Sperlich, D. Chromosome homologies within the Drosophila obscura group probed by insitu hybridization. Chromosoma 1984, 91, 46–53. [Google Scholar] [CrossRef]
- Bachtrog, D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 2008, 179, 1513–1525. [Google Scholar] [CrossRef]
- Betran, E.; Thornton, K.; Long, M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar] [CrossRef]

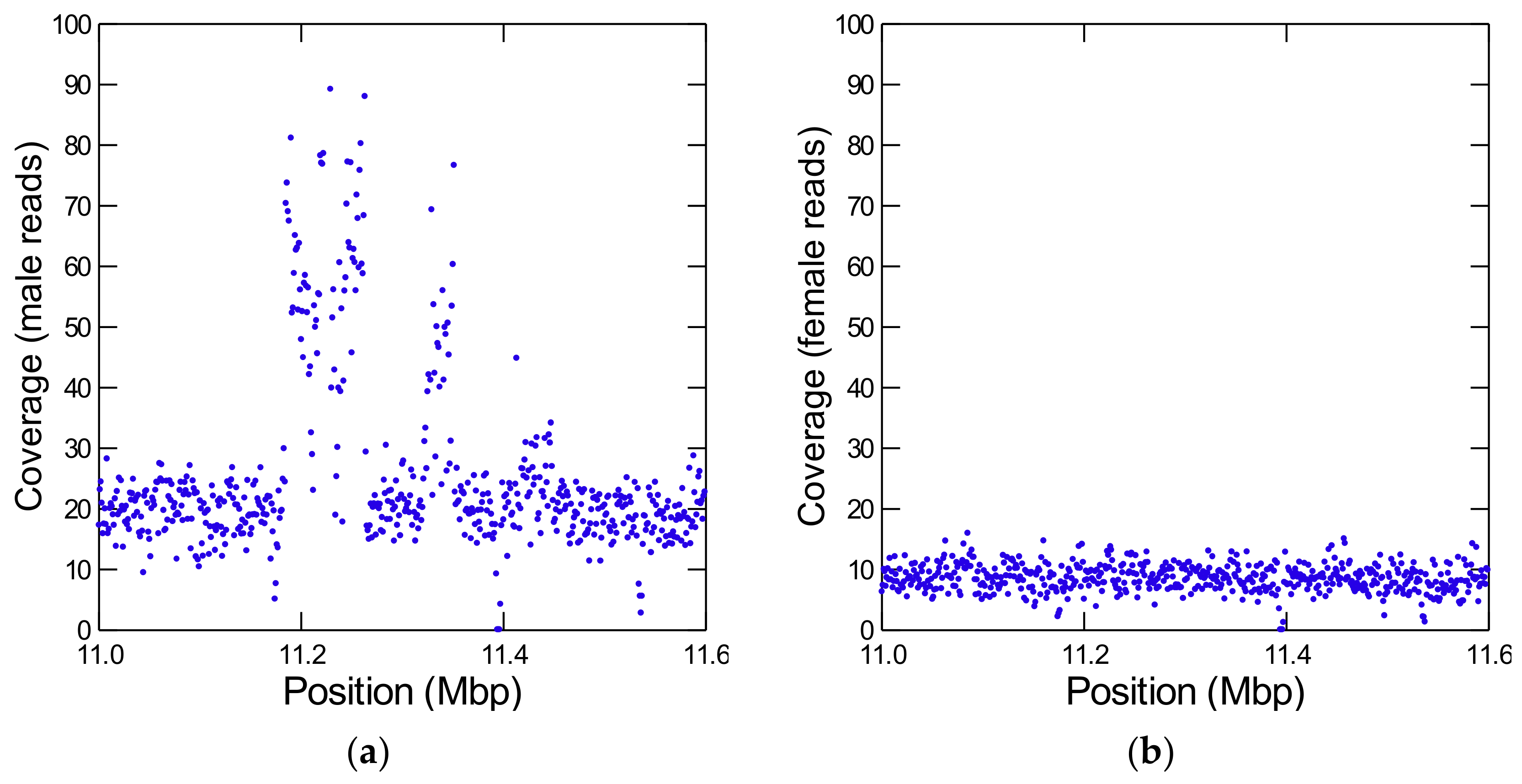

| D. willistoni Gene | D. melanogaster Ortholog | ||||
|---|---|---|---|---|---|
| Name | Copies 1 | Name | Loc. | Expression 2 | Predicted function/domains |
| GK21041 | 1 Y | CG18155 | X | testis-specific | fatty acid biosynthetic process; mitochondrion location |
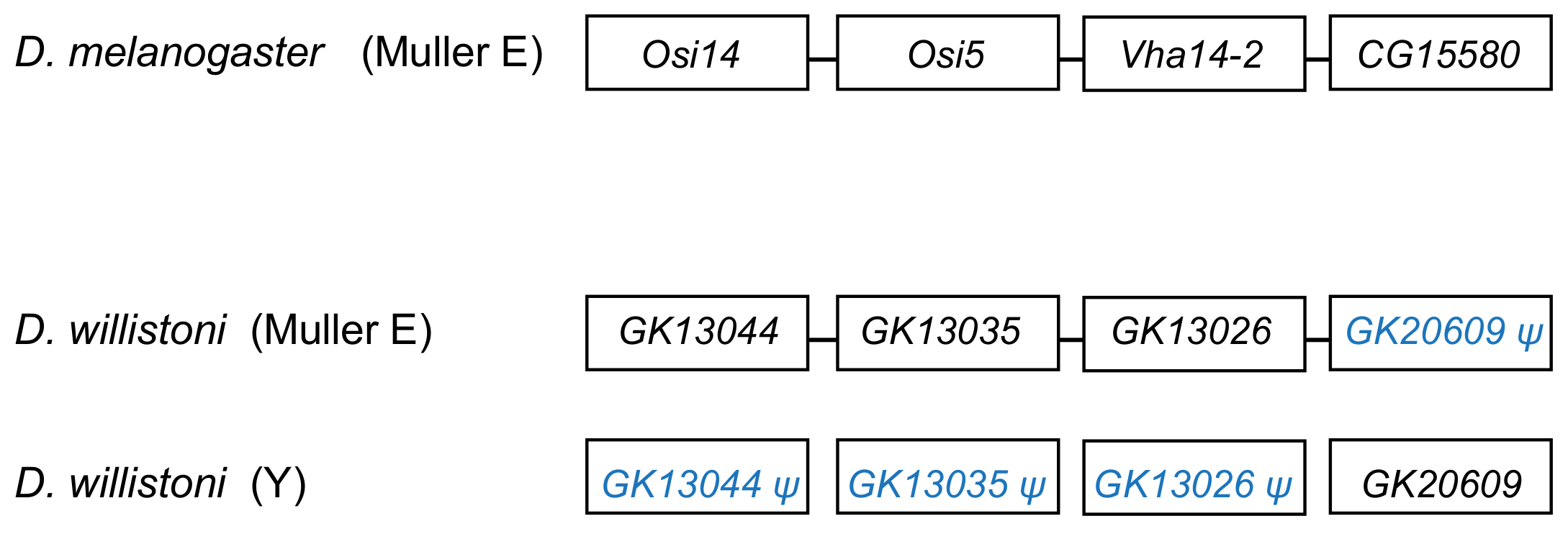
| GK20609 | 1 Y | CG15580 | 3R | testis-specific | leucine-rich repeat domain superfamily |
| GK13929 | 1 Y | CG10588 | 3L | testis + accessory gland | M16 metallo-endopeptidase protein present in spermatozoon |
| GK28041 | 1 Y | CG32650 | X | testis-specific | protein of unknown function DUF4763 |
| GK27472 | 1 Y | CG13539 | 2R | testis + accessory gland | protein of unknown function DUF1487 |
| YOgnWI030283 | 1 Y | CG34277 | 3R | testis-specific | ? |
| GK21220 | 1 Y | CG6052 | 3L | testis-specific | ATPase-coupled transmembrane transporter and lipid transporter. protein present in spermatozoon |
| YOgnWI000172 | 1 Y | CG14339 | 2L | testis-specific | cell division; regulation of mitotic sister chromatid separation. |
| GK27406 | 1 Y | Piezo-like | 3Rhet | testis-specific | mechanosensitive ion channel |
| GK28211 | 1 Y | Ran-like | 3L | testis-specific | GTP-binding protein involved in nucleocytoplasmic transport. |
| GK18510 | 10 Y, 1 A | ProtA | 2L | testis-specific | protamine protein: DNA packing in sperm |
| GK20591 | 1 Y, 1 A | CG6888 | 3L | testis-specific | thioredoxin peroxidase; cell redox homeostasis; protein present in spermatozoon. |
| YOgnWI018045 | 1 Y, 1 A | CG34175 | 2L | testis-specific | ? |
| GK20618, GK20619 | 4 Y, 1 A | Prosβ6 | 3L | widespread 3 | component of the proteasome |
| Autosomal Source | Y-Chromosome | |||||
|---|---|---|---|---|---|---|
| Chr. | Scaffold | Location (kb) | Size (kb) | Genes | Functional Genes | ψ Genes |
| E | CH964272 | 11,183–11,448 | 265 | 25 | 1 (GK20609) | 12 |
| C | CH963850 | 1340–1402 | 62 | 10 | 0 | 10 |
| B | CH963913 | 3149–3452 | 303 | 14 | 2 (GK18510, GK20618) | 10 |
| B | CH963857 | 10,827–10,901 | 74 | 9 | 1 (YOgnWI018045) | 4 |
| total | - | - | 704 | 58 | 4 | 36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricchio, J.; Uno, F.; Carvalho, A.B. New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni. Genes 2021, 12, 1815. https://doi.org/10.3390/genes12111815
Ricchio J, Uno F, Carvalho AB. New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni. Genes. 2021; 12(11):1815. https://doi.org/10.3390/genes12111815
Chicago/Turabian StyleRicchio, João, Fabiana Uno, and A. Bernardo Carvalho. 2021. "New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni" Genes 12, no. 11: 1815. https://doi.org/10.3390/genes12111815
APA StyleRicchio, J., Uno, F., & Carvalho, A. B. (2021). New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni. Genes, 12(11), 1815. https://doi.org/10.3390/genes12111815






