Abstract
Midbrain raphe nuclei (MRNs) contain a large number of serotonergic neurons associated with the regulation of numerous types of psychoemotional states and physiological processes. The aim of this work was to study alterations of the MRN transcriptome in mice with prolonged positive or negative fighting experience and to identify key gene networks associated with the regulation of serotonergic system functioning. Numerous genes underwent alterations of transcription in the MRNs of male mice that either manifested aggression or experienced social defeat in daily agonistic interactions. The expression of the Tph2 gene encoding the rate-limiting enzyme of the serotonin synthesis pathway correlated with the expression of many genes, 31 of which were common between aggressive and defeated mice and were downregulated in the MRNs of mice of both experimental groups. Among these common differentially expressed genes (DEGs), there were genes associated with behavior, learning, memory, and synaptic signaling. These results suggested that, in the MRNs of the mice, the transcriptome changes associated with serotonergic regulation of various processes are similar between the two groups (aggressive and defeated). In the MRNs, more DEGs correlating with Tph2 expression were found in defeated mice than in the winners, which is probably a consequence of deeper Tph2 downregulation in the losers. It was shown for the first time that, in both groups of experimental mice, the changes in the transcription of genes controlling the synthesis and transport of serotonin directly correlate with the expression of genes Crh and Trh, which control the synthesis of corticotrophin- and thyrotropin-releasing hormones. Our findings indicate that CRH and TRH locally produced in MRNs are related to serotonergic regulation of brain processes during a chronic social conflict.
1. Introduction
The serotonergic system is an important neurotransmitter system that is involved in the brain-driven regulation of various behaviors and physiological, metabolic, hormonal, and psychoemotional processes [1,2,3,4] as well as in the development of affective and neurological disorders (reviewed in refs. [5,6,7,8,9,10]). Nonetheless, due to the multifunctionality of serotonin, the question of the specificity of its role has always been raised, because alterations in the activity of the serotonergic system may be either a consequence of serotonin-driven regulation of neurobiological processes or a symptom of a developing disease.
The effect of serotonin neurotransmission on social cognition and emotion management can be modulated by both genotype [11,12] and other factors, such as gender [13], age [14], as well as environmental factors [15,16], which makes it difficult to study the basic molecular genetic mechanisms that determine the synthesis of serotonin and its role in the formation of the phenotype on the human population. Despite efforts to study genetic control of serotonergic regulation of various processes in the brain, this field remains largely unexplored.
Midbrain raphe nuclei (MRNs) are some of the most relevant brain structures for research on the mechanisms underlying serotonergic regulation of behavior because this brain area contains a large number of serotonergic neurons [1,17] and has interconnections with multiple brain regions. Thus, this brain area has been chosen to investigate the neurogenomic pathways of regulation of serotonergic activity in MRNs of mice in the sensory contact model described in [18,19]. The essence of this model is that, under the conditions of a chronic social conflict, aggressive mice with prolonged positive fighting experience (winners) and defeated mice with negative fighting experience (losers) develop various pathological forms of behavior. Male mice exhibiting repeated aggression in daily agonistic interactions enter a pathological psychotic-like state characterized, e.g., by symptoms of increased aggressiveness, hyperactivity, stereotypical behavior, and disturbances in social recognition (reviewed in [20,21]). Mice with chronic social defeat experience develop a mixed anxiety/depression-like state with symptoms of hypoactivity, behavioral deficits, anhedonia, and other problems [22,23,24,25,26]. We have previously demonstrated that, in the MRNs of both groups of animals with the opposite social experiences, there is underexpression of several key genes responsible for the synthesis, transport, and binding of serotonin [27,28,29].
Recently, next-generation sequencing showed high efficiency in the elucidation (in cells and tissues) of the molecular genetic mechanisms underlying the development of various pathologies. Earlier, using high-throughput RNA sequencing (RNA-Seq), we have found that the induction of aggressive or depressive behavior in mice in the model of sensory contact is associated with changes in the expression of many genes in the ventral tegmental area (VTA) [30]. In that study, a comprehensive analysis of these shifts in transcriptomes made it possible to identify the genes most likely associated with the specific features of VTA activity during the development of opposite types of behavior in experimental animals [30].
The aim of the present investigation was to study in detail the features of transcriptome changes in the MRNs of mice that had repeated experiences of either victories or defeats (winners and losers) in daily agonistic interactions. As mentioned above, this area of the brain is suitable for research on expression alterations of genes participating in the regulation of the serotonergic system and other systems functionally related to it under the influence of various external and/or endogenous factors that affect behavior. Therefore, most attention in this article is given to genes associated with the functioning of the serotonin system in the MRNs of mice under the conditions of chronic social conflict.
2. Materials and Methods
2.1. Animals
The experiments were carried out using 10–12 week-old C57BL/6J male mice. The mice were kept in the Conventional Vivarium (federal research center Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia) under standard conditions at 22 ± 2 °C on a 12/12 h light–dark cycle (lights on at 8:00 AM) with dry laboratory feed and water available ad libitum. All procedures were conducted in compliance with European Communities Council Directive 210/63/EU of 22 September 2010. The study protocol was approved by Scientific Council No. 9 of the Institute of Cytology and Genetics SB RAS of 24 March 2010, No. 613 (Novosibirsk, Russia).
2.2. Induction of Opposite Types of Social Behavior in Mice by Agonistic Interactions
The opposite types of social behavior were induced by means of daily agonistic interactions (intermale confrontations) of mice as described previously [19,22]. Pairs of weight-matched mice were each placed in a cage (28 × 14 × 10 cm) bisected by a perforated transparent partition allowing the animals to see, hear, and smell each other but preventing physical contact. The animals were left undisturbed for 2 or 3 days to adapt to the unfamiliar housing conditions before they were exposed to encounters. Every day at 14:00–17:00 PM (Russian local time), the cage lid was replaced by a transparent one, and after 5 min (the period necessary to activate agonistic interactions), the partition was removed for 10 min for the intermale confrontation. The mouse that attacked, bit, and chased the opponent was considered the winner. The superiority of the winner was established according to the outcome of two or three encounters with the same opponent. A mouse that showed only defensive behavior (sideways postures, upright postures, withdrawal, lying on the back, or freezing) was defined as the loser. If the aggressive attacks were very active and long, then the interactions between the mice were stopped after 3 min (or even earlier) by re-insertion of the partition to prevent injuries in the defeated mouse (only males were studied). This means that the painful effects of agonistic interactions are absent in defeated mice in this model.
Each defeated mouse (loser) was exposed to the same winner for 3 days, and then, to continue the agonistic interactions, the defeated mouse was placed in an unfamiliar cage with an unfamiliar winner behind the partition. Each winner remained in its original cage. The intermale confrontation procedure was performed once a day for 21 days and yielded equal numbers of winners and losers.
Three groups of animals were set up: (1) controls, i.e., mice without a daily experience of agonistic interactions; (2) winners, i.e., a group of aggressive mice chronically winning during 21 days in the daily agonistic interactions (intermale confrontations); and (3) losers, i.e., mice with chronic experience of defeats during 21 days in the daily agonistic interactions. These animals with the opposite types of social experience developed various pathological behaviors. Mice with a prolonged experience of aggression and victories (winners) are known to exhibit increased aggressiveness, hyperactivity, stereotypical behaviors, anxiety, impaired social recognition, irritability, autistic spectrum symptoms, a condition similar to drug addiction, and other problems (reviewed in [20]). The losers manifest mixed anxiety/depression-like behaviors accompanied by full immobility, avoidance of any social interactions, helplessness, indifference, and other aberrations [25,31].
Twenty-one pairs of C57BL/6J mice were employed to induce the opposite types of social behavior. Winners and losers with the most pronounced behavioral phenotypes were chosen for transcriptome analysis. The following criteria were used. To be designated as losers, during the activation period (5 min before a fight), the mice had to demonstrate all symptoms of depressive behavior: to not approach the partition, to sit in the cage corner opposite to the partition or with the nose into a corner or litter; the mice had to exhibit immobility, freezing during a winner’s attack, or indifference in all experimental situations (without behavioral reactions); no inversions of behavior to the opposite one after a change of aggressors; and avoidance and passive defense when attacked by the aggressor. To be designated as winners, during the activation period, the mice had to demonstrate strong aggressive motivation, and every day, they had to attack the opponent immediately after partition removal, stopping only for rest and to display manic motivation to bite the opponent in spite of full submission.
The control animals and all experimental mice were decapitated simultaneously. Experimental mice were decapitated 24 h after the last agonistic interaction. The brain regions were dissected by the same experimenter according to a relevant map in the Allen Mouse Brain Atlas [32]. All biological samples were placed in the RNAlater solution (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −70 °C until sequencing.
2.3. RNA-Seq
The frozen MRN samples from the mice (winners (n = 3), losers (n = 3), and controls (n = 3)) were sent to JSC Genoanalytica (Moscow, Russia), which specializes in RNA-Seq. For transcriptome profiling, mRNA was extracted by means of the Dynabeads mRNA Purification Kit (Ambion, Thermo Fisher Scientific, Waltham, MA, USA). cDNA libraries were constructed using the NEBNext mRNA Library Prep Reagent Set for Illumina (NEB, Ipswich, MA, USA). Single-end sequencing of cDNA libraries was performed on the Illumina HiSeq 1500 platform (Illumina Sequencing, San Diego, CA, USA) with a read length of 50 bases. All samples were analyzed as three biological replicates. Quality metrics of the mapped data (Supplementary Table S1) were determined in the Spliced Transcripts Alignment to a Reference (STAR) software [33]. The sequencing data were mapped to the mouse reference genome (GRCm38.p3) available in GenBank using the TopHat2 aligner (Center for Bioinformatics and Computational Biology, University of Maryland, College Park, MD, USA) [34]. The RNA-Seq datasets are available in the European Nucleotide Archive (Accession No. PRJEB47635).
Cufflinks/Cuffdiff programs were utilized to estimate gene expression levels in fragments per kilobase of transcript per million mapped reads (FPKM) and to identify differentially expressed genes (DEGs) in the experimental groups relative to the control. Genes were considered differentially expressed at a false discovery rate (q value) <5% [35]. Only annotated gene sequences were included in the analysis.
2.4. Validation of RNA-Seq Data
We have previously conducted studies on gene expression in males in similar experiments by the real-time PCR method with a larger number of samples in each experimental group, i.e., winners and losers (>10 animals). In comparison with the control, the direction and extent of changes in the expression of Tph2, Slc6a4, and several other genes in the MRNs of males in these experimental groups are generally consistent between the two utilized methods: real-time PCR [27,28] and RNA-Seq [29]. To cross-validate our results, we also employed a unique resource from Stanford University, USA [36], and noted significant concordance with our RNA-Seq data pool [37]. These findings suggested that the transcriptome analyses of the data provided by JSC Genoanalytica were valid, and that the method reflects the actual processes that occur in the brain under our experimental paradigm.
2.5. Functional Annotation of DEGs
This procedure was performed using the DAVID (database for annotation, visualization, and integrated discovery) gene annotation tool [38]. The Mus musculus genome served as the background list for over-representation analysis. The gene ontology (GO) option in DAVID as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database [39] were used to identify significantly (p < 0.05) enriched biological processes and metabolic pathways. To determine the association of DEGs with a behavior/neurological phenotype, the Neurological Disease Portal (Phenotypes, Mouse) in the rat genome database [40] was employed. An atlas of combinatorial transcriptional regulation in mice and humans [41] was used to identify DEGs encoding transcription factor genes.
2.6. Statistical Methods
The acquired RNA-Seq data (in FPKM values) were log-transformed, centered, and normalized. The normalized RNA-Seq data were subjected to correlation analysis. A 99% and 99.9% (df = 4; p < 0.01 (0.917); p < 0.001 (0.974)) two-tailed confidence interval was considered significant. Software packages Statistica 6.0 (StatSoft, Tulsa, OK, USA) and JACOBI4 [42] were used for data analysis and presentation.
3. Results
3.1. DEGs in the MRNs of Winners vs. Control Mice
A total of 14474 expressed genes were detected. Among them, 348 genes were found to be differentially expressed in the MRNs of aggressive mice (winners) compared to controls (Supplementary Table S2). Of these, 276 genes (79.3%) had a lower transcription level in the winners than in controls. Functional annotation of the DEGs revealed 94 genes associated with a behavior/neurological phenotype, among which there were genes associated with abnormal emotion/affect behavior, abnormal aggression-related behavior, increased aggression towards mice, abnormal depression-related behavior, and abnormal fear/anxiety-related behavior (Supplementary Table S2).
Among the DEGs, there were 34 genes encoding transcription factors, 11 of which are associated with a behavior/neurological phenotype (Supplementary Table S2). Functional annotation in the KEGG database allowed us to identify the metabolic pathways most significantly enriched in the DEG set (Table 1). A detailed description of the DEGs related to these pathways is given in Supplementary Table S3. Most of the DEGs assigned to the listed metabolic pathways had a lower transcription level in the winners than in the control animals.

Table 1.
KEGG terms most significantly related to the DEGs in the MRNs of the winning mice compared to controls.
3.2. DEGs in the MRNs of Losers vs. Controls
A total of 14445 expressed genes were detected, among which 214 genes were found to be differentially expressed in the MRNs of the defeated mice (losers) compared to the controls (Supplementary Table S4). Most of these DEGs, namely 185 genes (86.4%), had a lower level of transcription in the MRNs of the defeated mice as compared to the controls. Altogether, 76 DEGs proved to be associated with a behavior/neurological phenotype, among which there were genes related to abnormal emotion/affect behavior, abnormal aggression-related behavior, increased aggression towards mice, abnormal depression-related behavior, and abnormal fear/anxiety-related behavior (Supplementary Table S4). Twenty-two DEGs encode transcription factors, and among them, 10 genes are associated with a behavior/neurological phenotype. Table 2 presents the metabolic pathways most significantly enriched in this DEG set. A detailed description of the DEGs related to these pathways is given in Supplementary Table S5. Most of the DEGs assigned to the listed metabolic pathways have a lower transcription level in the defeated mice than in the control animals.

Table 2.
KEGG terms most significantly related to the DEGs in the MRNs of the losers (defeated mice) compared to controls.
Most of the metabolic pathways thus identified in the MRNs of the defeated mice were similar to those in the aggressive animals. The difference is that, in the MRNs of the defeated mice, DEGs dealing with the cholinergic synapse were found. On the other hand, the terms specific to MRNs of the aggressive mice were ribosome, cardiac muscle contraction, oxytocin signaling pathway, GABAergic synapse, fatty acid biosynthesis, amphetamine addiction, tyrosine metabolism, ECM-receptor interaction, butanoate metabolism, morphine addiction, dopaminergic synapse, and long-term depression.
3.3. DEGs Shared by the Winners and Losers (Common DEGs)
A comparison of the DEG lists revealed 158 common genes that manifested significantly altered expression in the MRNs of both the winners and losers as compared to the controls (Supplementary Table S6). For all the common DEGs in the MRNs, the transcription level changed unidirectionally between the winners and losers with respect to the controls. One hundred forty-two (89.9%) out of 158 common DEGs were downregulated by the social confrontation. Fifty-five common DEGs are associated with a behavior/neurological phenotype (Supplementary Table S6). KEGG analysis indicated that the terms most significantly enriched in the set of common DEGs are MAPK signaling pathway, serotonergic synapse, focal adhesion, calcium signaling pathway, and type II diabetes mellitus (Supplementary Table S7). All genes assigned to these metabolic pathways turned out to be significantly downregulated in the MRNs of mice of both experimental groups.
3.4. Expression of DEGs Encoding Proteins Responsible for the Synthesis and Transport of Serotonin and Hormones
In MRNs, many neurons are serotonergic. Our data indicated that expression of genes encoding key proteins that control the synthesis (TPH2 and DDC) and transport (reuptake) of serotonin (SLC6A4) was low in the MRNs of mice of both experimental groups (Table 3).

Table 3.
Expression of DEGs dealing with the synthesis and transport of serotonin and DEGs encoding neuropeptides CRH and TRH.
It is known that, when the body is exposed to endogenous or exogenous factors, functional changes in tissues and organs are primarily determined by neurotransmitter systems and through the action of biologically active substances (hormones). According to our data, in the MRNs of both winners and defeated mice that experienced chronic social confrontation for 21 days, there was a highly significant decrease in the expression of genes Crh and Trh encoding corticotropin-releasing hormone (CRH) and thyrotropin-releasing hormone (TRH) (Table 3).
3.5. Expression of Neurotransmitter and Hormone Receptors
Our findings revealed shifts in the expression of several genes encoding neurotransmitter and hormone receptors (Table 4). Readers can see that, under the conditions of social confrontation, the effects of various neurotransmitter and hormonal signals can modulate the functioning of MRNs in mice. The expression of six of the genes presented in Table 4 (Chrm1, Gabra4, Hcrtr1, Htr5b, Irs4, and Nr2f2) significantly changed in the MRNs of both winners and defeated animals under the conditions of chronic social confrontations. The most significant alterations in transcription levels were documented for Htr5b, which encodes 5-hydroxytryptamine (serotonin) receptor 5B.

Table 4.
Expression of DEGs coding for neurotransmitter and hormone receptors.
3.6. Correlation between mRNA Levels of Genes Encoding Proteins Responsible for the Synthesis and Transport of Serotonin and the Expression of Genes Coding for Hormones or Neurotransmitter or Hormone Receptors
In both experimental groups of mice, changes in the transcription level of genes controlling the synthesis (Tph2 and Ddc) and transport (Slc6a4) of serotonin directly and most significantly correlated with the expression of genes encoding serotonin (Htr5b) and insulin (Irs4) receptors as well as with transcription levels of genes Crh and Trh encoding corticotropin-releasing hormone and thyrotropin-releasing hormone, respectively (Table 5). There was an inverse correlation with the expression of the Nr2f2 gene. Expression of DEGs encoding cholinergic receptors and hypocretin (orexin) receptor 1 did not correlate with mRNA levels of genes encoding proteins that control serotonin synthesis and transport. Regarding GABA receptors, in both experimental groups, the Ddc transcription level positively correlated with Gabra4 expression (Table 5).

Table 5.
Correlation between the mRNA level of genes encoding proteins participating in the synthesis and transport of serotonin and the expression of genes coding for hormones or receptors of neurotransmitters or hormones.
3.7. A Comparison of Molecular Mechanisms Correlating with mRNA Expression of Genes Encoding Proteins That Govern Serotonin Synthesis in the MRNs of Mice with the Opposite Social Experiences in Daily Confrontations
To identify the main features associated with decreased expression of genes involved in serotonin synthesis in the MRNs of aggressive and defeated mice, we analyzed correlations between DEG expression and the expression of Tph2, which encodes the rate-limiting enzyme of serotonin biosynthesis.
The results of the correlation analysis showed that the expression of 33 DEGs found in the MRNs of winning mice correlated with Tph2 expression (Supplementary Table S8), whereas in the MRNs of defeated mice, the expression of 98 DEGs correlated with Tph2 expression (Supplementary Table S9). It is important to note that 31 genes are common between these lists. Accordingly, we can say that decreased expression of genes involved in the synthesis and transport of serotonin in the MRNs of mice is a nonspecific mechanism behind the functional response of this brain region to social confrontation. Furthermore, the observed more significant Tph2 underexpression in the MRNs of the defeated mice probably involves a larger number of genes related to the formation of phenotypic features of the serotonergic-system activity in the MRNs of losers. Biological processes associated with behavior, learning, memory, and synaptic signaling were identified in the analysis of DEGs correlating with Tph2 expression in the MRNs of both experimental groups of mice. In contrast to the winners, in the MRNs of the losers, changes in processes associated with neurogenesis were present too (Figure 1 and Figure 2).
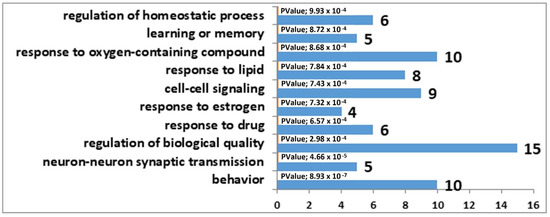
Figure 1.
GO terms linked with the DEGs that correlated with Tph2 expression in the MRN transcriptome analysis of the winners. The horizontal axis represents the number of genes.
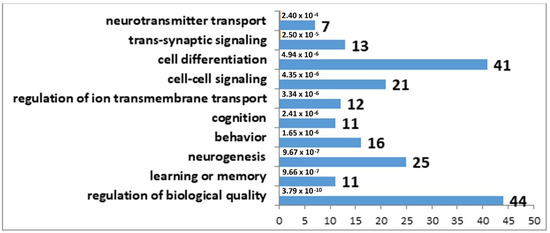
Figure 2.
GO terms found to be enriched in the set of DEGs that correlated with Tph2 expression in the MRN transcriptome analysis of the losers. The horizontal axis denotes the number of genes.
In the set of the 33 DEGs whose expression correlated with Tph2 transcription in the MRNs of the winners, the largest number of correlations (33) was found for three genes, Gch1, Slc6a4, and Trh; their expression correlated with all DEGs in this set of genes (Supplementary Table S8). In the set of 98 DEGs whose expression correlated with Tph2 transcription in the MRNs of losers, the largest number of correlations was found for Tac2, A2m, and Crh; their expression levels correlated with almost all DEGs (97, 96, and 96, respectively) in this gene set (Supplementary Table S9).
4. Discussion
We analyzed the impact of chronic social confrontations on levels of gene transcription in the MRNs of male C57BL/6J mice. It is reported here that, in the MRNs of experimental mice, significant changes in the expression of a large number of genes occur both in winners and in defeated males. During the manifestation of the opposite types of social behavior, transcription levels of some genes are expected to change in opposite directions, as shown earlier in the analysis of the VTA transcriptome of the same groups of animals [30]; however, in the MRNs of winners and defeated mice, no genes were found that changed the level of transcription in a direction opposite to that in the opponent group. Furthermore, in the MRNs, numerous genes (158 DEGs) underwent transcription alterations that were unidirectional between the winners and defeated animals. The MAPK signaling pathway is linked with the regulation of a wide range of cellular processes, including proliferation, differentiation, apoptosis, and stress responses in both normal and pathological conditions [43]; this signaling cascade was identified here as a common and most severely altered metabolic pathway (highly enriched in DEG sets). This finding suggests that mice in both experimental groups were stressed.
MRNs are known as a brain region characterized by high density of serotonergic neurons, the functioning of which is associated with the regulation of behavior and psychoemotional states [44]. According to our functional annotation in KEGG, there was significant downregulation of several genes related to the serotonergic synapse metabolic pathway in the MRNs of mice of both experimental groups; these can be considered key genes in this work because their products control the serotonin synthesis (Tph2 and Ddc), transport (Slc6a4), and binding (Htr5b). In earlier reports from our group, the decrease in mRNA levels of genes encoding the proteins that manage the synthesis and transport of serotonin in the MRNs of mice that had opposite social experiences has already been discussed in detail, and the reduced transcription has been confirmed by real-time PCR [27,28]. The objectives of the present project were to identify additional most significant molecular determinants of serotonergic regulation of aggressive and submissive behaviors. We assumed that the genes whose expression would significantly correlate with the mRNA levels of genes encoding the proteins controlling the synthesis and transport of serotonin would be highly likely associated with serotonergic signaling.
Several serotonin receptors are expressed in MRNs; however, here, significant changes in the transcription level were documented only for two of them: Htr3a and Htr5b. The expression of Htr3a in the MRNs was statistically significantly lower only in defeated mice, and diminished Htr5b transcription in the MRNs was registered in both experimental groups of mice.
Recently, it was shown that Htr3a (encoding 5-hydroxytryptamine (serotonin) receptor 3A) is implicated in the regulation of goal-oriented behavior via the serotonergic projection from the median raphe nucleus to ventral hippocampus [45]. By contrast, in our study, Htr3a expression did not correlate with the expression of genes encoding the proteins responsible for serotonin synthesis.
Htr5b codes for 5-hydroxytryptamine (serotonin) receptor 5B, which is expressed in serotonergic neurons both in the dorsal raphe nucleus (DRN) and median raphe nucleus [46], where it functions as an autoreceptor [47], i.e., is localized on the presynaptic membrane of the neuron and binds a specific ligand released by that same neuron, thereby implementing a feedback mechanism for monitoring the neurotransmitter synthesis and/or release.
Htr5b expression highly positively correlated here with mRNA levels of genes encoding the proteins that manage serotonin synthesis (Tph2 and Ddc) and transport (Slc6a4). Additionally, it was shown for the first time that the expression of Htr5b, Tph2, Ddc, and Slc6a4 highly positively correlates with the expression of genes Crh and Trh. According to results of single-cell RNA-Seq analysis of serotonergic neurons in the murine DRN and median raphe nucleus, the genes encoding both neuropeptides (CRH and TRH) are expressed predominantly in neurons of the DRN [46].
The contribution of CRH concentration changes in raphe nuclei to the regulation of behavior and psychoemotional states is poorly understood at present. Some experimental evidence that alterations in CRH levels in the DRN may mediate stress-related and emotional/affective phenomena has been obtained and reviewed [48]. There are two known genes (Crhr1 and Crhr2) coding for CRH receptors, and the interaction of CRH with either receptor can trigger sensitization of DRN neurons thereby leading to a subsequent greater release of serotonin in response to CRH input; this phenomenon may, at least in part, be responsible for the behavioral aberrations associated with depression or anxiety [48]. Microinjection of CRH into the DRN has revealed that the interaction of CRH with these receptors can modulate behavioral consequences of uncontrolled stress in a dose-dependent manner. It has been suggested that low doses of injected CRH preferentially bind to CRHR1 and inhibit DRN 5-HT activity, whereas higher doses of CRH are expected to bind to both receptor subtypes and to no longer inhibit DRN serotonin activity. CRHR2 is thought to mediate excitation of DRN 5-HT neurons [49]. Even though CRH is presumably expressed predominantly in DRN cells [46], a number of studies also point to an important role of the median raphe nucleus in CRH-related signaling, which contributes to behavior regulation. It has been reported that CRH injection into the median raphe nucleus significantly elevates memory-dependent fear expression in rats [50]. The results of another experiment showed that the increased medial-prefrontal-cortex serotonin release caused by the infusion of CRH into the DRN can be abrogated by inactivation of the median raphe nucleus [51]. It was concluded in that research article that neurons of both the DRN and median raphe nucleus are involved in complex CRH-driven modulation of the serotonergic activity in the medial prefrontal cortex [51].
In our study, Crhr1 expression did not change under the influence of social confrontations, whereas Crhr2 expression was not detectable in the MRNs of mice of both experimental groups. We did not find a significant correlation between expression levels of Crh and Crhr1; however, we did detect a significant positive correspondence of expression levels between Crh and Crhbp (encoding CRH-binding protein (Table 6), which regulates CRH bioavailability) [52,53]. These findings suggest that, in our experiment, CRH signaling and its possible effect on behavior are primarily associated not with receptors CRHR1 and CRHR2 but rather with the level of Crhbp expression. In support of this hypothesis, it is possible to cite a number of studies indicating an important function of CRHBP in the regulation of behavior. Genotypic analysis suggests that a single-nucleotide substitution in the CRHBP gene may be related to a decrease in aggressive behavior [54]. In another study, genotype analysis revealed a positive correlation between the CRHBP gene and a suicidal tendency [55]. Our results support a recently advanced hypothesis that not only the CRH synthesized in the hypothalamus but also the CRH locally produced in CRH-containing raphe nuclei in the brainstem is important for stress adaptation [56].

Table 6.
The expression of genes encoding receptors for CRH or TRH or encoding the proteins that implement their inactivation as well as correlations.
It has been repeatedly demonstrated that thyroid hormones can serve as an effective adjunctive treatment of affective disorders [57]. A review article on a modulatory impact of exogenous thyroid hormones on the serotonin system of the brain in affective disorders indicates that raphe nuclei participate in these processes [58]. As for TRH, which is endogenously synthesized in MRNs [46], its role in the regulation of serotonin synthesis and behavior has hardly been studied.
Table 6 shows that, in our work, Trh expression did not correlate with the expression of its receptors (Trhr and Trhr2) and thyrotropin-releasing-hormone-degrading enzyme (Trhde) in the MRNs. Nevertheless, Trh mRNA expression corresponded to mRNA levels of genes encoding the proteins that control the synthesis and transport of serotonin as well as to the expression of the Irs4 gene encoding insulin receptor substrate 4, which is known as an adapter molecule involved in the signal transduction of both insulin [59] and leptin [60]. Mutations in IRS4 are associated with central hypothyroidism [61], and the TSH response to the TRH test is blunted in male IRS4 mutation carriers having central hypothyroidism [62]. This evidence suggests that IRS4 is required for proper TRH signaling.
The expression of the genes discussed above positively correlated here with the expression of genes encoding proteins that control the synthesis and transport of serotonin and negatively correlated with Nr2f2 expression in the MRNs of mice of both experimental groups. Nuclear receptor subfamily 2 group F member 2 gene (Nr2f2, also known as COUP-TFII) codes for a member of the steroid/thyroid hormone superfamily of nuclear receptors. NR2F2 is a ligand-activated transcription factor exerting complex pleiotropic effects on glucose homeostasis, insulin sensitivity, and lipid metabolism in various tissues [63,64]. It is reported that insulin represses NR2F2 gene expression in pancreatic β-cells [65], and that the regulation of NR2F2 promoter activity is cell type specific [66,67]. Nevertheless, on the basis of our data, we can assume that insulin can play an important part in the modulation of biological processes in the MRNs of mice of both experimental groups, and that its influence on the level of Nr2f2 transcription can be regulated through IRS4. Moreover, it is known that Nr2f2 belongs to the group of genes whose transcriptional activation is associated with all-trans-retinoic acid [66,68]. In early studies, it was reported that COUP-TF dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of vitamin D3, thyroid hormone, and retinoic acid receptors [69]. Therefore, even though specific features of the regulation of NR2F2 activity in raphe nuclei have not yet been studied, our data are in good agreement with what was described above about this regulation in other tissues.
Our results indicate mRNA underexpression of TPH2 (the rate-limiting enzyme of brain serotonin synthesis) and of DDC and SLC6A4 (proteins controlling serotonin synthesis and transport) in the MRNs of both winners and defeated mice. Additionally, it turned out that the expression of a large number of DEGs in the MRNs in both experimental groups correlates with Tph2 expression. Moreover, 31 genes were common between these lists and the changes in their transcription were unidirectional between the lists. Accordingly, our data imply that a decrease in the synthesis and transport of serotonin in murine MRNs is a nonspecific mechanism behind the cellular response to chronic social confrontation.
5. Conclusions
Our results mean that chronic social confrontations significantly affect transcription levels of numerous genes in the MRNs of both aggressive and defeated mice. mRNA underexpression of key genes encoding the proteins that control the synthesis of serotonin (TPH2 and DDC) and transport (reuptake) of serotonin (SLC6A4) is demonstrated in the MRNs of mice of both experimental groups. Data analysis suggests that the reduction in serotonin synthesis in the MRNs of the winners and losers as a consequence of chronic social confrontation is nonspecific, because Tph2 expression correlated with the expression of many common genes whose MRN transcription levels shifted in the same direction between the experimental groups. Among these DEGs, genes associated with behavior, learning, memory, and synaptic signaling were identified. A larger number of DEGs whose expression correlated with the Tph2 expression level was found in the MRNs of the defeated mice, which can probably be explained by a more substantial decrease in Tph2 transcription in the MRNs of the losers.
Here, it was shown for the first time that, in both groups of experimental mice, alterations in expression of genes encoding the proteins that govern the synthesis and transport of serotonin directly correlated with expression of genes Htr5b, Crh, Trh, and Irs4. An inverse correlation was observed with the expression of Nr2f2. We can hypothesize that, along with genes encoding the proteins that manage the synthesis and transport of serotonin, these genes play a key role in the regulation of processes associated with the functioning of serotonergic neurons in MRNs of mice under the conditions of a chronic social conflict. Our findings support the recently advanced hypothesis that not only the CRH synthesized in the hypothalamus but also the CRH locally produced in CRH-containing raphe nuclei in the brainstem is important for stress adaptation. In addition, our results indicate that TRH locally produced in MRNs is also linked with serotonergic regulation of brain processes responsible for social behavior accompanying chronic positive or negative experiences under the conditions of chronic agonistic interactions. This may also be true in other cases of organismal adaptation to the effects of other stressors.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12111811/s1, Supplementary Table S1: Summary statistics for the sequenced libraries; Supplementary Table S2: DEGs in the MRNs of winners versus control mice; Supplementary Table S3: Description of the DEGs related to the metabolic pathways most significantly altered in winning mice compared to controls; Supplementary Table S4: DEGs in the MRNs of losers versus control mice; Supplementary Table S5: Description of DEGs related to the metabolic pathways most significantly affected in losers (defeated mice) compared to controls; Supplementary Table S6: DEGs identified in the MRNs of both winners and losers (common DEGs); Supplementary Table S7: KEGG pathways for the common DEGs; Supplementary Table S8: DEGs the expression of which correlates with Tph2 expression in the MRNs of winners; Supplementary Table S9: DEGs the expression of which correlates with Tph2 expression in the MRNs of losers.
Author Contributions
Conceptualization, N.N.K.; Data curation, V.N.B., D.A.S., I.L.K. and A.G.G.; Formal analysis, O.E.R., V.N.B., D.A.S., I.L.K. and A.G.G.; Funding acquisition, N.N.K.; Investigation, O.E.R. and V.N.B.; Methodology, N.N.K.; Supervision, N.N.K.; Writing—original draft, O.E.R.; Writing—review and editing, O.E.R., V.N.B., D.A.S., I.L.K., A.G.G. and N.N.K. All authors have read and agreed to the published version of the manuscript.
Funding
The current study was supported by the Russian Science Foundation (grant No. 19-15-00026 to N.N.K.). The preparation and maintenance of the experimental animals were carried out in the Conventional Vivarium at the Institute of Cytology and Genetics SB RAS and were supported by publicly funded project No. 0259-2021-0016. The funding body had no role in the design of the study, in the collection, analysis, and interpretation of the data, and in the writing of the manuscript.
Institutional Review Board Statement
All procedures were conducted in compliance with European Communities Council Directive 210/63/EU of September 22, 2010. The study protocol was approved by Scientific Council No. 9 of the Institute of Cytology and Genetics SB RAS of 24 March 2010, No. 613 (Novosibirsk, Russia).
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-Seq datasets are available in the European Nucleotide Archive (Accession No. PRJEB47635).
Acknowledgments
The authors are grateful to JSC Genoanalytica (Moscow, Russia) for conducting the technological part of the experiment and the primary statistical analysis. The authors are grateful to Nikolai A. Shevchuk for help in correcting the English version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CRH | corticotropin-releasing hormone (corticotropin-releasing factor) |
| DEGs | differentially expressed genes |
| FPKM | fragments per kilobase of transcript per million mapped reads |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes pathway database |
| MRN | midbrain raphe nuclei |
| TRH | thyrotropin-releasing hormone |
References
- Jacobs, B.L.; Azmitia, E.C. Structure and function of the brain serotonin system. Physiol. Rev. 1992, 72, 165–229. [Google Scholar] [CrossRef] [PubMed]
- Popova, N.K. From gene to aggressive behavior: The role of brain serotonin. Neurosci. Behav. Physiol. 2008, 38, 471–475. [Google Scholar] [CrossRef]
- Berger, M.; Gray, J.A.; Roth, B.L. The expanded biology of serotonin. Annu. Rev. Med. 2009, 60, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Yabut, J.M.; Crane, J.D.; Green, A.E.; Keating, D.J.; Khan, W.I.; Steinberg, G.R. Emerging Roles for Serotonin in Regulating Metabolism: New Implications for an Ancient Molecule. Endocr. Rev. 2019, 40, 1092–1107. [Google Scholar] [CrossRef]
- Yildirim, B.O.; Derksen, J.J. Systematic review, structural analysis, and new theoretical perspectives on the role of serotonin and associated genes in the etiology of psychopathy and sociopathy. Neurosci. Biobehav. Rev. 2013, 37, 1254–1296. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Lee, L.T.; Yang, Y.K. Serotonin and mental disorders: A concise review on molecular neuroimaging evidence. Clin. Psychopharmacol. Neurosci. 2014, 12, 196–202. [Google Scholar] [CrossRef]
- Oathes, D.J.; Hilt, L.M.; Nitschke, J.B. Affective neural responses modulated by serotonin transporter genotype in clinical anxiety and depression. PLoS ONE 2015, 10, e0115820. [Google Scholar] [CrossRef]
- Dorszewska, J.; Florczak-Wyspianska, J.; Kowalska, M.; Stanski, M.; Kowalewska, A.; Kozubski, W. Serotonin in Neurological Diseases. In Serotonin—A Chemical Messenger between All Types of living Cells; Kaneez, F.S., Ed.; IntechOpen: London, UK, 2017; pp. 219–239. [Google Scholar]
- Michely, J.; Eldar, E.; Martin, I.M.; Dolan, R.J. A mechanistic account of serotonin’s impact on mood. Nat. Commun. 2020, 11, 2335. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Xu, Q.; Jiang, M.; Feng, R.; Jiang, S.; Yuan, B.; Xu, S.; Wu, T.; Wang, F.; Huang, J.H. Emotion Induced Monoamine Neuromodulator Release Affects Functional Neurological Disorders. Front. Cell Dev. Biol. 2021, 9, 633048. [Google Scholar] [CrossRef]
- Lin, C.H.; Tseng, Y.L.; Huang, C.L.; Chang, Y.C.; Tsai, G.E.; Lane, H.Y. Synergistic effects of COMT and TPH2 on social cognition. Psychiatry 2013, 76, 273–294. [Google Scholar] [CrossRef]
- Zill, P.; Baghai, T.C.; Zwanzger, P.; Schule, C.; Eser, D.; Rupprecht, R.; Moller, H.J.; Bondy, B.; Ackenheil, M. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol. Psychiatry 2004, 9, 1030–1036. [Google Scholar] [CrossRef]
- Dykens, E.M.; Roof, E.; Bittel, D.; Butler, M.G. TPH2 G/T polymorphism is associated with hyperphagia, IQ, and internalizing problems in Prader-Willi syndrome. J. Child Psychol. Psychiatry 2011, 52, 580–587. [Google Scholar] [CrossRef][Green Version]
- Kataja, E.L.; Leppanen, J.M.; Kantojarvi, K.; Pelto, J.; Haikio, T.; Korja, R.; Nolvi, S.; Karlsson, H.; Paunio, T.; Karlsson, L. The role of TPH2 variant rs4570625 in shaping infant attention to social signals. Infant Behav. Dev. 2020, 60, 101471. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Reynolds, G.P.; Yuan, Y.; Shi, Y.; Pu, M.; Zhang, Z. TPH-2 Polymorphisms Interact with Early Life Stress to Influence Response to Treatment with Antidepressant Drugs. Int. J. Neuropsychopharmacol. 2016, 19, pyw070. [Google Scholar] [CrossRef]
- Spagnolo, P.A.; Norato, G.; Maurer, C.W.; Goldman, D.; Hodgkinson, C.; Horovitz, S.; Hallett, M. Effects of TPH2 gene variation and childhood trauma on the clinical and circuit-level phenotype of functional movement disorders. J. Neurol. Neurosurg. Psychiatry 2020, 91, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Sos, K.E.; Mayer, M.I.; Cserep, C.; Takacs, F.S.; Szonyi, A.; Freund, T.F.; Nyiri, G. Cellular architecture and transmitter phenotypes of neurons of the mouse median raphe region. Brain Struct. Funct. 2017, 222, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, N.N. Sensory contact model: Protocol, control, applications. In Horizons in Neuroscience Research; Costa, A., Villalba, E., Eds.; NOVA Science Publishers Inc.: New York, NY, USA, 2011; Volume 3, pp. 81–100. [Google Scholar]
- Kudryavtseva, N.N.; Smagin, D.A.; Kovalenko, I.L.; Vishnivetskaya, G.B. Repeated positive fighting experience in male inbred mice. Nat. Protoc. 2014, 9, 2705–2717. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, N.N. The psychopathology of repeated aggression: A neurobiological aspect. In Perspectives on the Psychology of Aggression; Morgan, J.P., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2006; pp. 35–64. [Google Scholar]
- Kudryavtseva, N.N. Positive fighting experience, addiction-like state, and relapse: Retrospective analysis of experimental studies. Aggress. Viol. Behav. 2020, 52, 101403. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N. The sensory contact model for the study of aggressive and submissive behaviors in male mice. Aggress. Behav. 1991, 17, 285–291. [Google Scholar] [CrossRef]
- Avgustinovich, D.F.; Alekseenko, O.V.; Bakshtanovskaia, I.V.; Koriakina, L.A.; Lipina, T.V.; Tenditnik, M.V.; Bondar, N.P.; Kovalenko, I.L.; Kudriavtseva, N.N. Dynamic changes of brain serotonergic and dopaminergic activities during development of anxious depression: Experimental study. Usp. Fiziol. Nauk. 2004, 35, 19–40. [Google Scholar]
- Bondar, N.P.; Kovalenko, I.L.; Avgustinovich, D.F.; Smagin, D.A.; Kudryavtseva, N.N. Anhedonia in the shadow of chronic social defeat stress, or When the experimental context matters. Open Behav. Sci. J. 2009, 3, 17–27. [Google Scholar] [CrossRef]
- Galyamina, A.G.; Kovalenko, I.L.; Smagin, D.A.; Kudryavtseva, N.N. Interaction of depression and anxiety in the development of mixed anxiety/depression disorder: Experimental studies of the mechanisms of comorbidity (review). Neurosci. Behav. Physiol. 2017, 47, 699–713. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N. Development of Mixed Anxiety/Depression-Like State as a Consequences of Chronic Anxiety: Review of Experimental Data. In Neuroscience of Social Stress; Miczek, K.A., Sinha, R., Eds.; Current Topics in Behavioral Neurosciences Series; Springer Publishing: New York, NY, USA, 2021. [Google Scholar]
- Boyarskikh, U.A.; Bondar, N.P.; Filipenko, M.L.; Kudryavtseva, N.N. Downregulation of serotonergic gene expression in the Raphe nuclei of the midbrain under chronic social defeat stress in male mice. Mol. Neurobiol. 2013, 48, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Smagin, D.A.; Boyarskikh, U.A.; Bondar, N.P.; Filipenko, M.L.; Kudryavtseva, N.N. Reduction of serotonergic gene expression in the midbrain raphe nuclei under positive fighting experience. Adv Biosci Biotech. 2013, 4, 36–44. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Smagin, D.A.; Kovalenko, I.L.; Galyamina, A.G.; Vishnivetskaya, G.B.; Babenko, V.N.; Orlov, Y.L. Serotonergic genes in the development of anxiety/depression-like state and pathology of aggressive behavior in male mice: RNA-seq data. Mol. Biol. 2017, 51, 251–262. [Google Scholar] [CrossRef]
- Redina, O.; Babenko, V.; Smagin, D.; Kovalenko, I.; Galyamina, A.; Efimov, V.; Kudryavtseva, N. Gene Expression Changes in the Ventral Tegmental Area of Male Mice with Alternative Social Behavior Experience in Chronic Agonistic Interactions. Int. J. Mol. Sci. 2020, 21, 6599. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Bakshtanovskaya, I.V.; Koryakina, L.A. Social model of depression in mice of C57BL/6J strain. Pharmacol. Biochem. Behav. 1991, 38, 315–320. [Google Scholar] [CrossRef]
- Allen. Allen Institute for Brain Science. Allen Mouse Brain Atlas. 2004. Available online: http://mouse.brain-map.org/static/atlas (accessed on 24 April 2005).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- Babenko, V.N.; Smagin, D.A.; Kudryavtseva, N.N. RNA-Seq mouse brain regions expression data analysis: Focus on ApoE functional network. J. Integr. Bioinform. 2017, 14, 20170024. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Kanehisa, M. Post-Genome Informatics; Kyoto Encyclopedia of Genes and Genomes; Oxford University Press: Oxford, UK, 2000; p. 148. Available online: http://www.genome.jp/kegg (accessed on 16 July 2021).
- RGD. Neurological Disease Portal, Rat Genome Database Web Site; Medical College of Wisconsin: Milwaukee, WI, USA; Available online: http://rgd.mcw.edu/ (accessed on 7 January 2021).
- Ravasi, T.; Suzuki, H.; Cannistraci, C.V.; Katayama, S.; Bajic, V.B.; Tan, K.; Akalin, A.; Schmeier, S.; Kanamori-Katayama, M.; Bertin, N.; et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 2010, 140, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Polunin, D.; Shtaiger, I.; Efimov, V. JACOBI4 software for multivariate analysis of biological data. bioRxiv 2019, 803684. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Andrade, T.G.; Zangrossi, H., Jr.; Graeff, F.G. The median raphe nucleus in anxiety revisited. J. Psychopharmacol. 2013, 27, 1107–1115. [Google Scholar] [CrossRef]
- Yoshida, K.; Drew, M.R.; Mimura, M.; Tanaka, K.F. Serotonin-mediated inhibition of ventral hippocampus is required for sustained goal-directed behavior. Nat. Neurosci. 2019, 22, 770–777. [Google Scholar] [CrossRef]
- Ren, J.; Isakova, A.; Friedmann, D.; Zeng, J.; Grutzner, S.M.; Pun, A.; Zhao, G.Q.; Kolluru, S.S.; Wang, R.; Lin, R.; et al. Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. eLife 2019, 8, e49424. [Google Scholar] [CrossRef]
- Serrats, J.; Raurich, A.; Vilaro, M.T.; Mengod, G.; Cortes, R. 5-ht5B receptor mRNA in the raphe nuclei: Coexpression with serotonin transporter. Synapse 2004, 51, 102–111. [Google Scholar] [CrossRef]
- Maier, S.F.; Watkins, L.R. Stressor controllability and learned helplessness: The roles of the dorsal raphe nucleus, serotonin, and corticotropin-releasing factor. Neurosci. Biobehav. Rev. 2005, 29, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Hammack, S.E.; Pepin, J.L.; DesMarteau, J.S.; Watkins, L.R.; Maier, S.F. Low doses of corticotropin-releasing hormone injected into the dorsal raphe nucleus block the behavioral consequences of uncontrollable stress. Behav. Brain Res. 2003, 147, 55–64. [Google Scholar] [CrossRef]
- Ohmura, Y.; Yamaguchi, T.; Izumi, T.; Matsumoto, M.; Yoshioka, M. Corticotropin releasing factor in the median raphe nucleus is involved in the retrieval of fear memory in rats. Eur. J. Pharmacol. 2008, 584, 357–360. [Google Scholar] [CrossRef]
- Forster, G.L.; Pringle, R.B.; Mouw, N.J.; Vuong, S.M.; Watt, M.J.; Burke, A.R.; Lowry, C.A.; Summers, C.H.; Renner, K.J. Corticotropin-releasing factor in the dorsal raphe nucleus increases medial prefrontal cortical serotonin via type 2 receptors and median raphe nucleus activity. Eur. J. Neurosci. 2008, 28, 299–310. [Google Scholar] [CrossRef]
- Behan, D.P.; De Souza, E.B.; Lowry, P.J.; Potter, E.; Sawchenko, P.; Vale, W.W. Corticotropin releasing factor (CRF) binding protein: A novel regulator of CRF and related peptides. Front. Neuroendocrinol. 1995, 16, 362–382. [Google Scholar] [CrossRef]
- Westphal, N.J.; Seasholtz, A.F. CRH-BP: The regulation and function of a phylogenetically conserved binding protein. Front. Biosci. 2006, 11, 1878–1891. [Google Scholar] [CrossRef]
- Tang, L.; Chen, Y.; Xiang, Q.; Xiang, J.; Tang, Y.; Li, J. The GCAG Haplotype of the CRHBP Gene May Decrease the Risk for Robbery Behavior Among the Han Chinese. Genet. Test. Mol. Biomark. 2020, 24, 436–442. [Google Scholar] [CrossRef]
- De Luca, V.; Tharmalingam, S.; Zai, C.; Potapova, N.; Strauss, J.; Vincent, J.; Kennedy, J.L. Association of HPA axis genes with suicidal behaviour in schizophrenia. J. Psychopharmacol. 2010, 24, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Chaves, T.; Fazekas, C.L.; Horvath, K.; Correia, P.; Szabo, A.; Torok, B.; Banrevi, K.; Zelena, D. Stress Adaptation and the Brainstem with Focus on Corticotropin-Releasing Hormone. Int. J. Mol. Sci. 2021, 22, 9090. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.; Avni, J. Thyroid hormones in the treatment of affective disorders. Acta Psychiatr. Scand. 1988, 77, 623–636. [Google Scholar] [CrossRef]
- Bauer, M.; Heinz, A.; Whybrow, P.C. Thyroid hormones, serotonin and mood: Of synergy and significance in the adult brain. Mol. Psychiatry 2002, 7, 140–156. [Google Scholar] [CrossRef]
- Lavan, B.E.; Fantin, V.R.; Chang, E.T.; Lane, W.S.; Keller, S.R.; Lienhard, G.E. A novel 160-kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J. Biol. Chem. 1997, 272, 21403–21407. [Google Scholar] [CrossRef] [PubMed]
- Wauman, J.; De Smet, A.S.; Catteeuw, D.; Belsham, D.; Tavernier, J. Insulin receptor substrate 4 couples the leptin receptor to multiple signaling pathways. Mol. Endocrinol. 2008, 22, 965–977. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heinen, C.A.; de Vries, E.M.; Alders, M.; Bikker, H.; Zwaveling-Soonawala, N.; van den Akker, E.L.T.; Bakker, B.; Hoorweg-Nijman, G.; Roelfsema, F.; Hennekam, R.C.; et al. Mutations in IRS4 are associated with central hypothyroidism. J. Med. Genet. 2018, 55, 693–700. [Google Scholar] [CrossRef]
- Patyra, K.; Makkonen, K.; Haanpaa, M.; Karppinen, S.; Viikari, L.; Toppari, J.; Reeve, M.P.; Kero, J. Screening for Mutations in Isolated Central Hypothyroidism Reveals a Novel Mutation in Insulin Receptor Substrate 4. Front. Endocrinol 2021, 12, 658137. [Google Scholar] [CrossRef] [PubMed]
- Bardoux, P.; Zhang, P.; Flamez, D.; Perilhou, A.; Lavin, T.A.; Tanti, J.F.; Hellemans, K.; Gomas, E.; Godard, C.; Andreelli, F.; et al. Essential role of chicken ovalbumin upstream promoter-transcription factor II in insulin secretion and insulin sensitivity revealed by conditional gene knockout. Diabetes 2005, 54, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, X.; Qin, J.; Jeha, G.S.; Saha, P.K.; Yan, J.; Haueter, C.M.; Chan, L.; Tsai, S.Y.; Tsai, M.J. The nuclear orphan receptor COUP-TFII plays an essential role in adipogenesis, glucose homeostasis, and energy metabolism. Cell Metab. 2009, 9, 77–87. [Google Scholar] [CrossRef]
- Boutant, M.; Ramos, O.H.; Lecoeur, C.; Vaillant, E.; Philippe, J.; Zhang, P.; Perilhou, A.; Valcarcel, B.; Sebert, S.; Jarvelin, M.R.; et al. Glucose-dependent regulation of NR2F2 promoter and influence of SNP-rs3743462 on whole body insulin sensitivity. PLoS ONE 2012, 7, e35810. [Google Scholar] [CrossRef]
- Soosaar, A.; Neuman, K.; Nornes, H.O.; Neuman, T. Cell type specific regulation of COUP-TF II promoter activity. FEBS Lett. 1996, 391, 95–100. [Google Scholar] [CrossRef]
- Sabra-Makke, L.; Tourrel-Cuzin, C.; Denis, R.G.; Moldes, M.; Pegorier, J.P.; Luquet, S.; Vasseur-Cognet, M.; Bossard, P. The nutritional induction of COUP-TFII gene expression in ventromedial hypothalamic neurons is mediated by the melanocortin pathway. PLoS ONE 2010, 5, e13464. [Google Scholar] [CrossRef] [PubMed]
- Laursen, K.B.; Mongan, N.P.; Zhuang, Y.; Ng, M.M.; Benoit, Y.D.; Gudas, L.J. Polycomb recruitment attenuates retinoic acid-induced transcription of the bivalent NR2F1 gene. Nucleic Acids Res. 2013, 41, 6430–6443. [Google Scholar] [CrossRef] [PubMed]
- Cooney, A.J.; Tsai, S.Y.; O’Malley, B.W.; Tsai, M.J. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol. Cell. Biol. 1992, 12, 4153–4163. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).