Zoo: Selecting Transcriptomic and Methylomic Biomarkers by Ensembling Animal-Inspired Swarm Intelligence Feature Selection Algorithms
Abstract
1. Introduction
2. Materials and Methods
2.1. Summary of Datasets
2.2. Performance Metrics
2.3. Stratified k-Fold Cross Validation Strategy
2.4. Nine Swarm Intelligence Feature Selection Algorithms
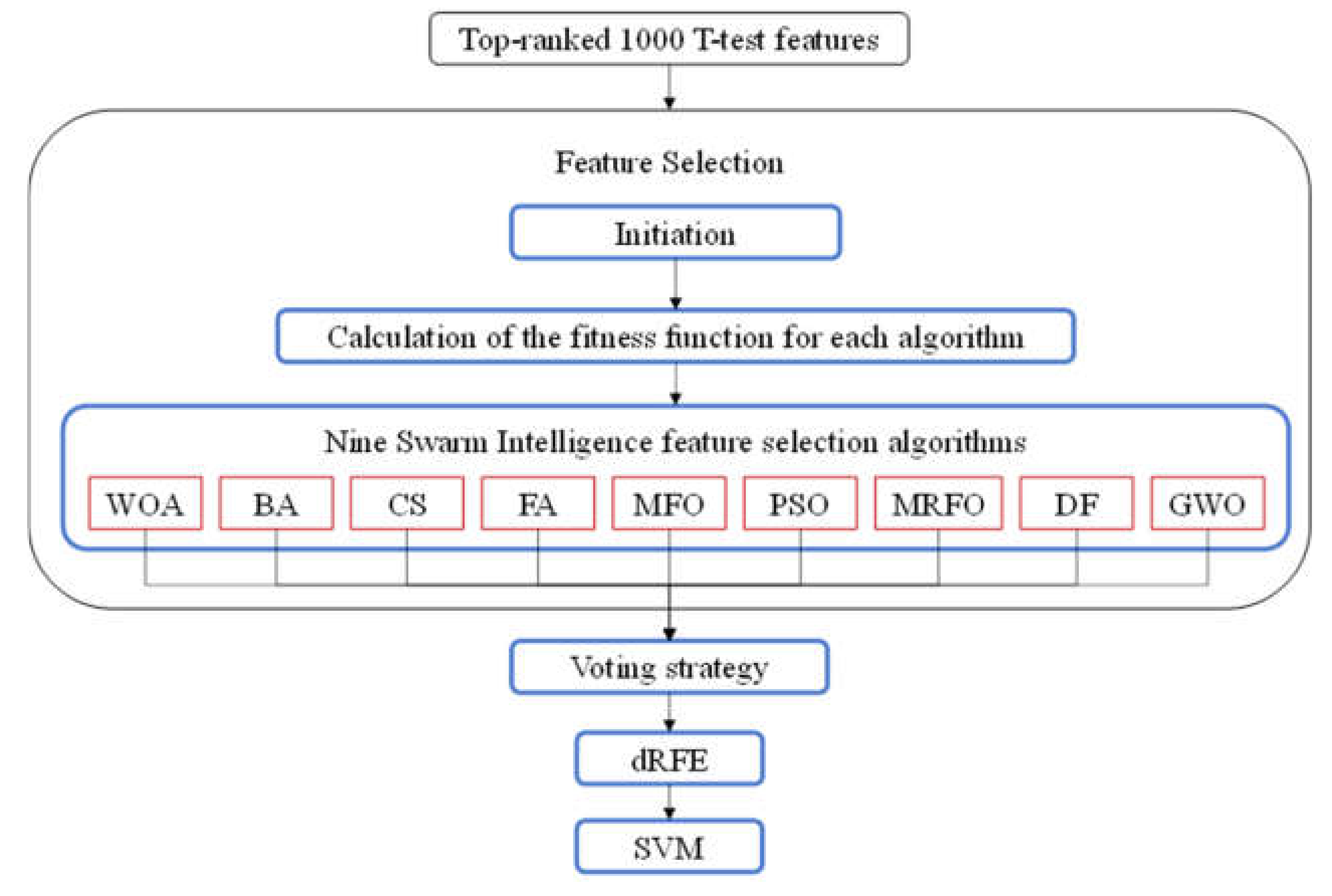
2.5. The Ensemble SI-Based Feature Selection Algorithm Zoo
2.6. Binary Animal-Inspired SI-Based Feature Selection Algorithms
2.7. The Existing Feature Selection Algorithms
3. Results
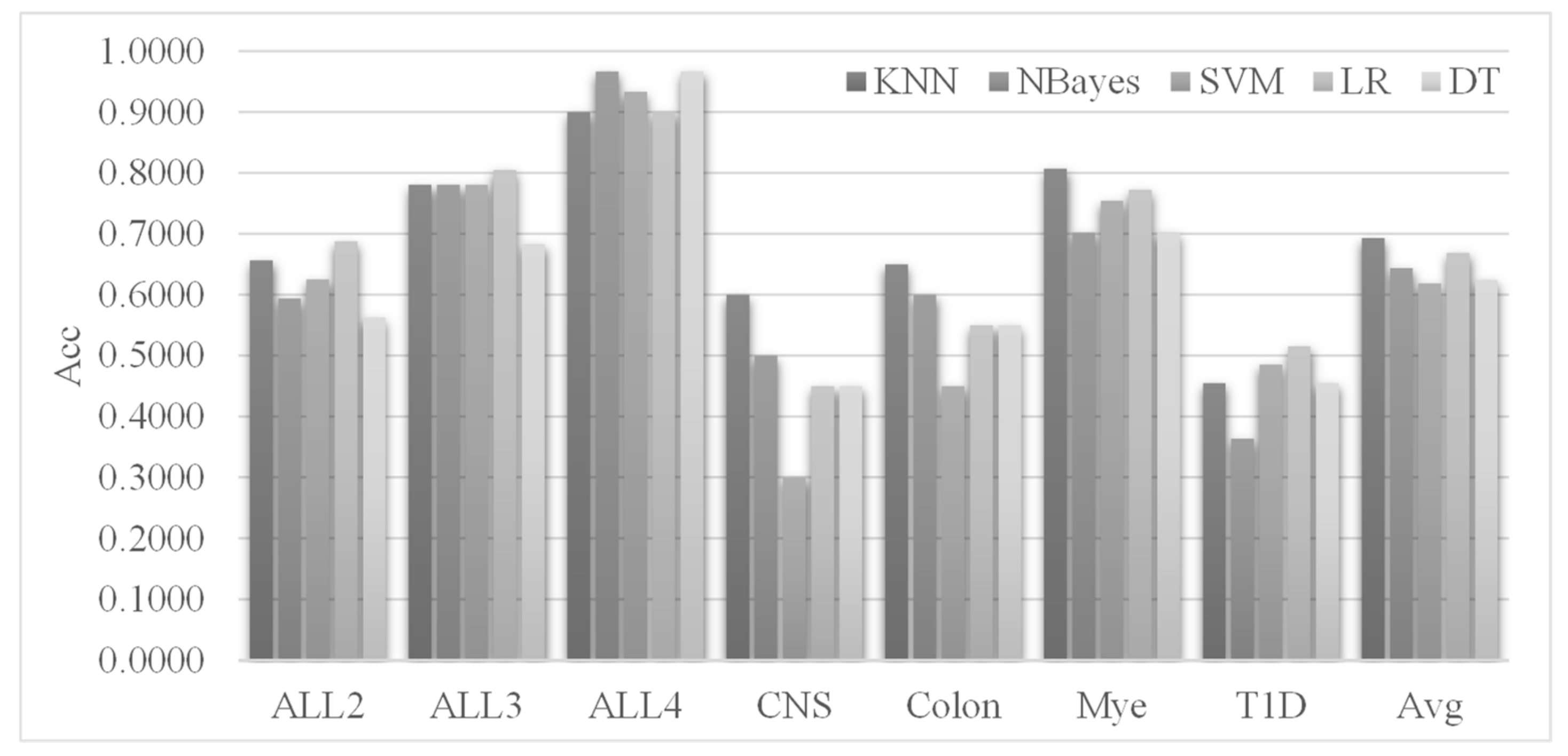
3.1. Evaluating the Classifiers for the Selected Features
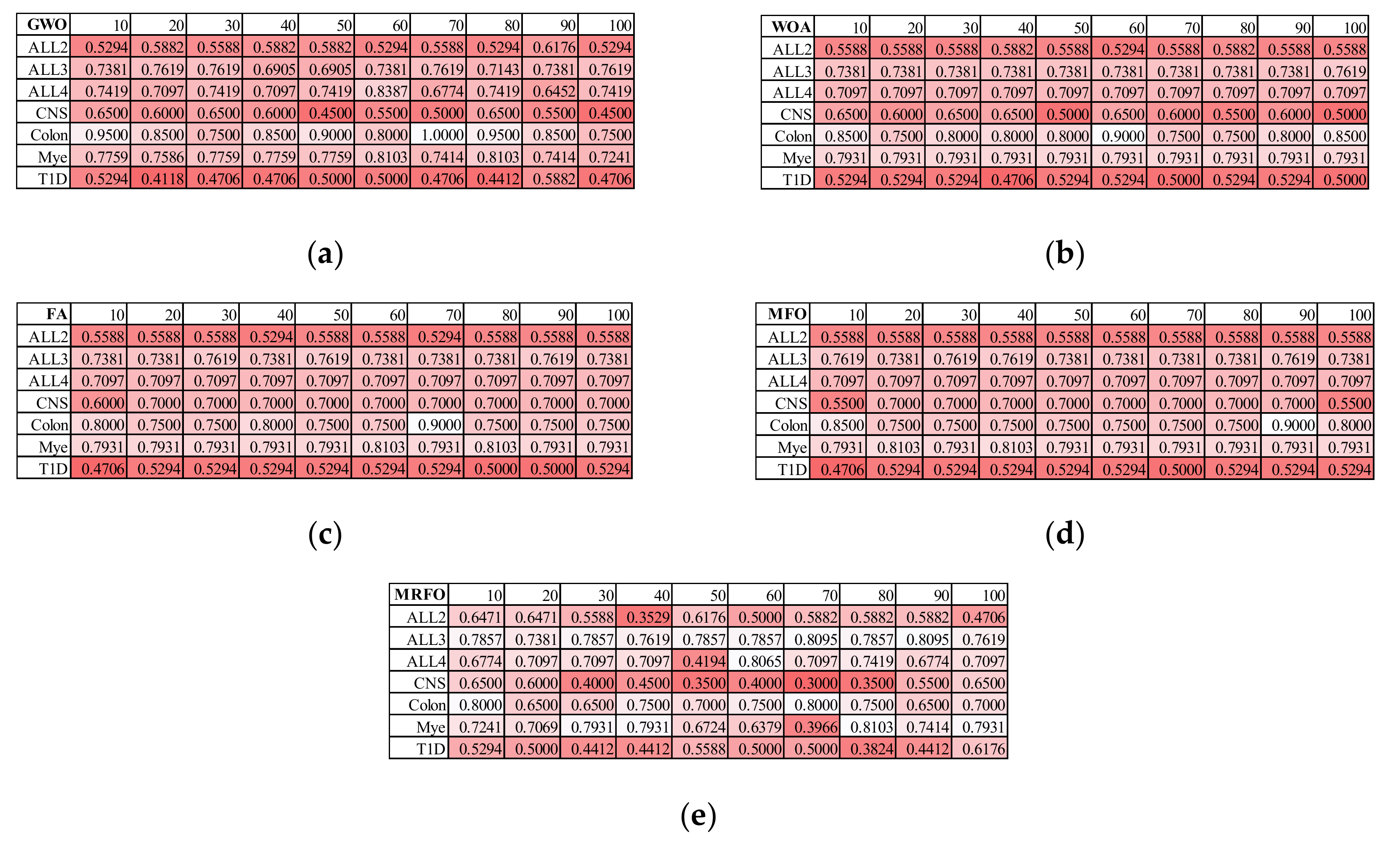
3.2. Finding the Best Population Size for Five SI Algorithms
3.3. Parameter Tunings of the Other Four SI Algorithms
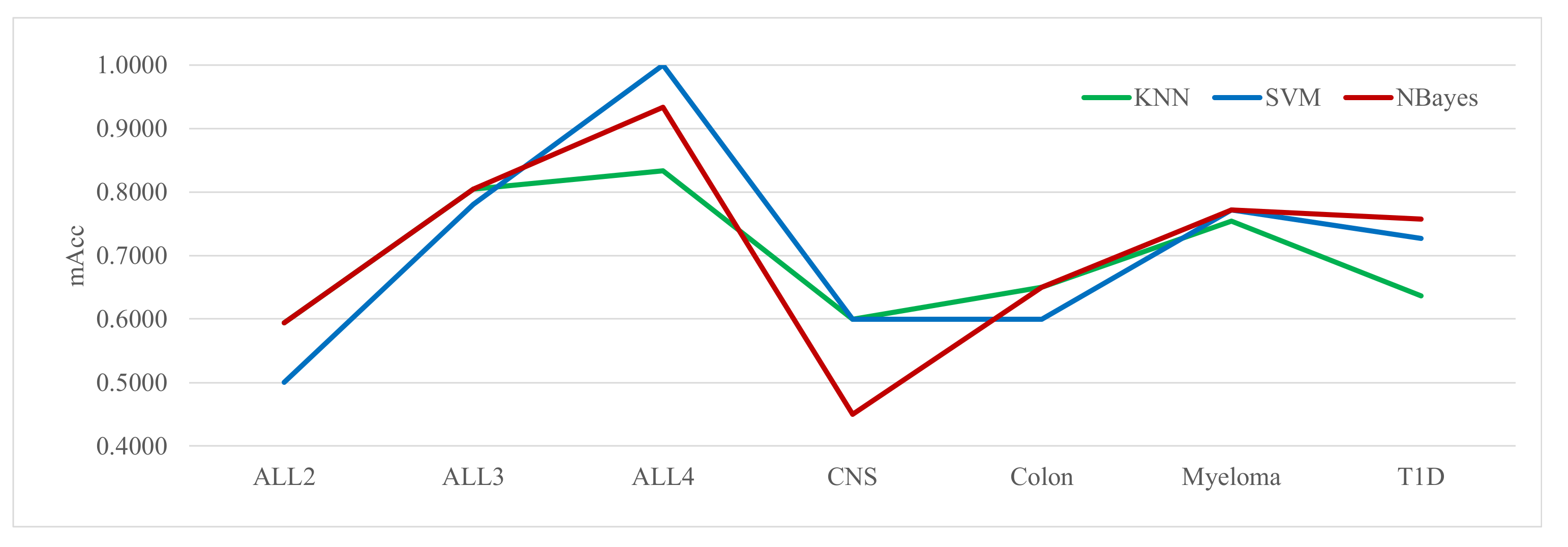
3.4. Finding the Best Classifier for Zoo
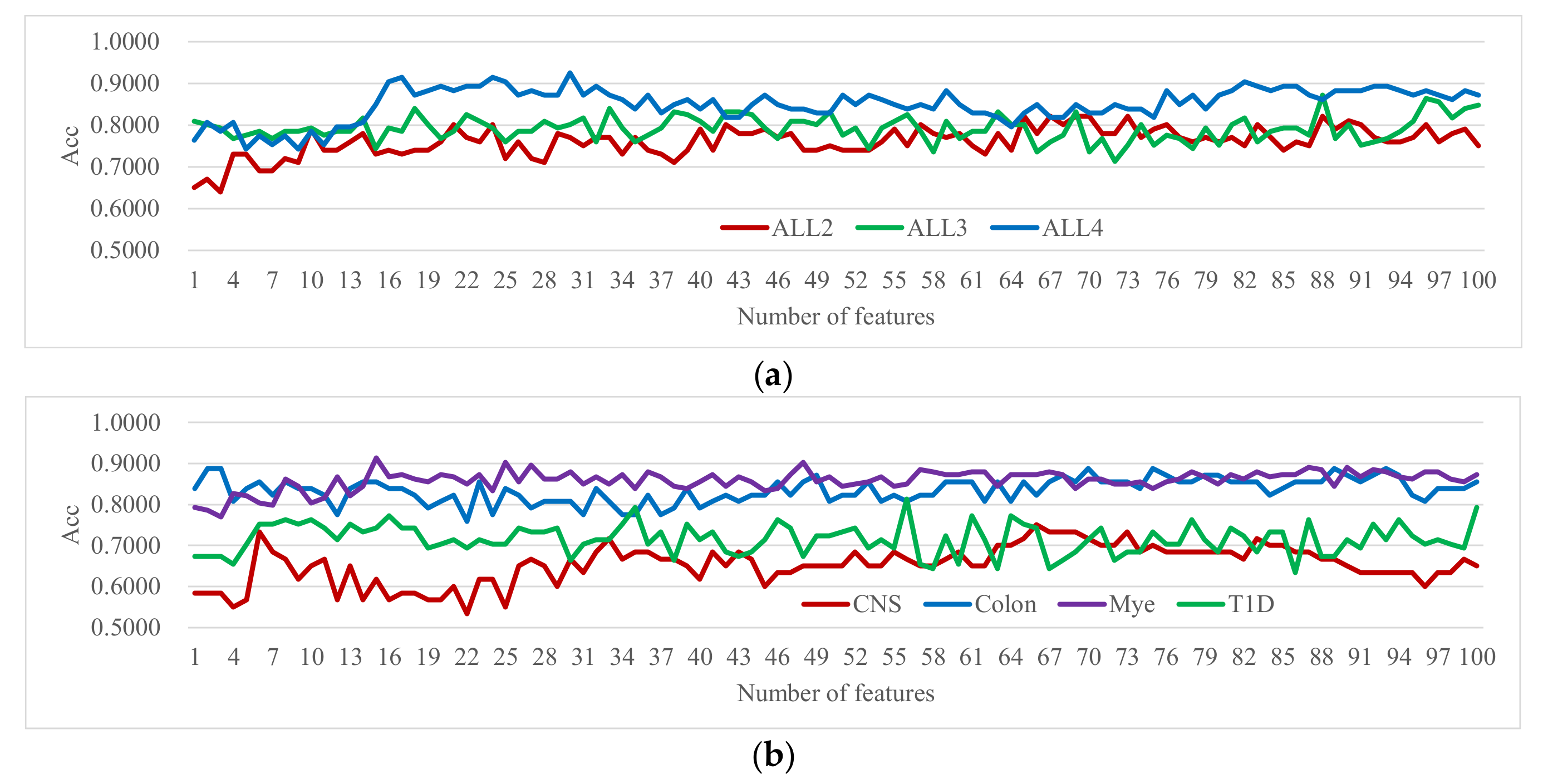
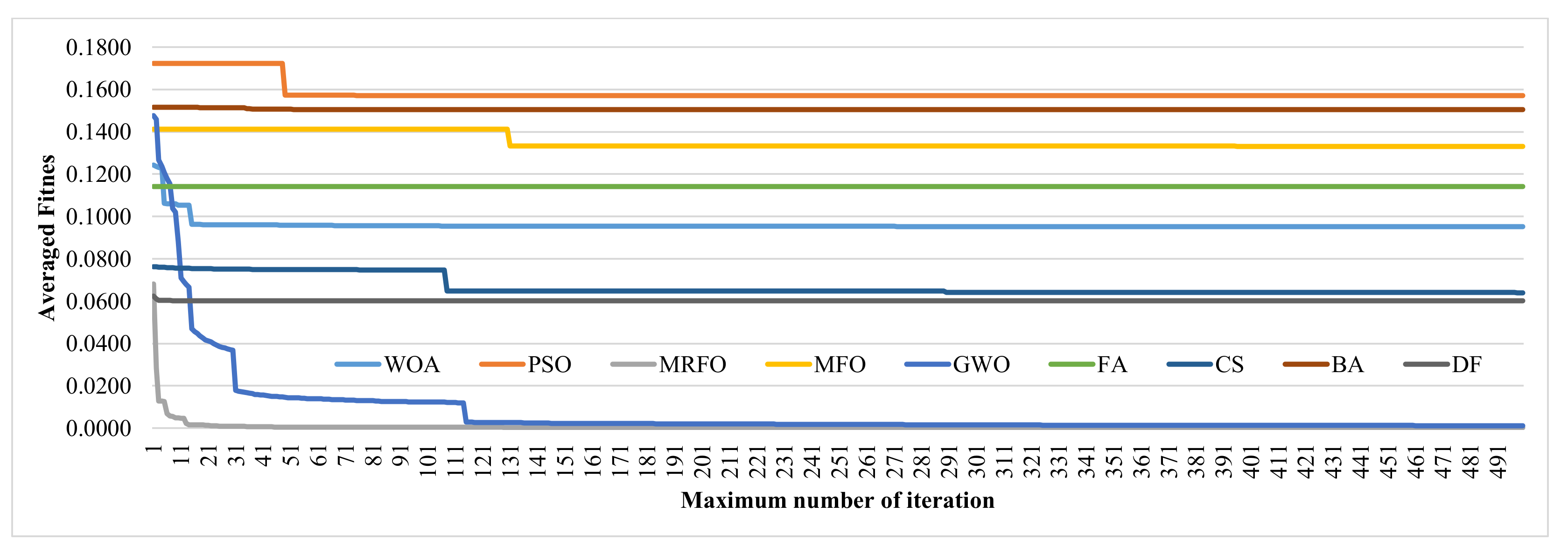
3.5. Choosing the Maximum Number of Iterations
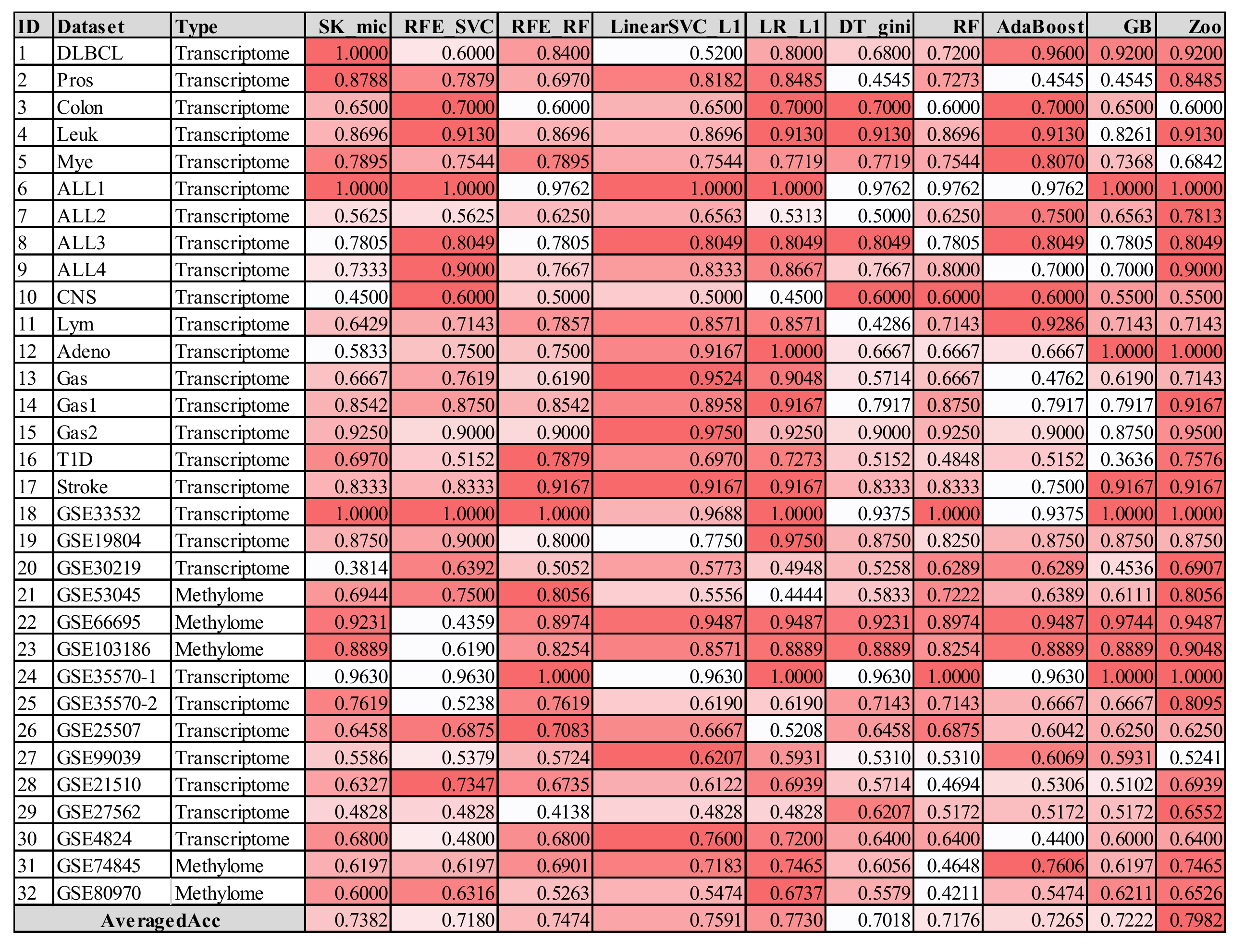
3.6. Comparison with Other Feature Selection Algorithms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Amelio, S.; Lombardo, F.; Pizzarelli, A.; Bellini, I.; Cavallero, S. Advances in Omic Studies Drive Discoveries in the Biology of Anisakid Nematodes. Genes 2020, 11, 801. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.G.; Chin, K.-V. Logistic regression for disease classification using microarray data: Model selection in a large p and small n case. Bioinformatics 2007, 23, 1945–1951. [Google Scholar] [CrossRef]
- Wu, W.; Ma, X. Joint learning dimension reduction and clustering of single-cell RNA-sequencing data. Bioinformatics 2020, 36, 3825–3832. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, P.; Feng, Y.; Xie, X.; Duan, M.; Fan, Y.; Liu, S.; Huang, L.; Zhou, F. RIFS2D: A two-dimensional version of a randomly restarted incremental feature selection algorithm with an application for detecting low-ranked biomarkers. Comput. Biol. Med. 2021, 133, 104405. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Ding, S.; Duan, M.; Liu, S.; Huang, L.; Zhou, F. FeSTwo, a two-step feature selection algorithm based on feature engineering and sampling for the chronological age regression problem. Comput. Biol. Med. 2020, 125, 104008. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, Y.; Zhang, X.; Hahn, J. Region of Interest Selection for Functional Features. Neurocomputing 2021, 422, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, M.; Maioli, G.; Colombo, G.I.; Piacentini, L. GARS: Genetic Algorithm for the identification of a Robust Subset of features in high-dimensional datasets. BMC Bioinform. 2020, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Fu, C.; Zhang, R.; Zhou, F. EnRank: An Ensemble Method to Detect Pulmonary Hypertension Biomarkers Based on Feature Selection and Machine Learning Models. Front. Genet. 2021, 12, 636429. [Google Scholar] [CrossRef]
- Li, F.; Yang, M.; Li, Y.; Zhang, M.; Wang, W.; Yuan, D.; Tang, D. An improved clear cell renal cell carcinoma stage prediction model based on gene sets. BMC Bioinform. 2020, 21, 232. [Google Scholar] [CrossRef] [PubMed]
- Sreejith, S.; Khanna Nehemiah, H.; Kannan, A. Clinical data classification using an enhanced SMOTE and chaotic evolutionary feature selection. Comput. Biol. Med. 2020, 126, 103991. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, G.; Movahedi, P.; Ebrahimi, M.; Pahikkala, T.; Plosila, J.; Tenhunen, H. GeFeS: A generalized wrapper feature selection approach for optimizing classification performance. Comput. Biol. Med. 2020, 125, 103974. [Google Scholar] [CrossRef]
- Ding, X.; Yang, F.; Jin, S.; Cao, J. An efficient alpha seeding method for optimized extreme learning machine-based feature selection algorithm. Comput. Biol. Med. 2021, 134, 104505. [Google Scholar] [CrossRef] [PubMed]
- Chalakkal, R.; Hafiz, F.; Abdulla, W.; Swain, A. An efficient framework for automated screening of Clinically Significant Macular Edema. Comput. Biol. Med. 2021, 130, 104128. [Google Scholar] [CrossRef]
- Phadikar, S.; Sinha, N.; Ghosh, R. Automatic Eyeblink Artifact Removal From EEG Signal Using Wavelet Transform With Heuristically Optimized Threshold. IEEE J. Biomed. Health Inform. 2021, 25, 475–484. [Google Scholar] [CrossRef]
- Abu Khurmaa, R.; Aljarah, I.; Sharieh, A. An intelligent feature selection approach based on moth flame optimization for medical diagnosis. Neural Comput. Appl. 2020, 33, 7165–7204. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, M.; Luo, Z.; Cai, Y. An ensemble deep learning method for vehicle type classification on visual traffic surveillance sensors. IEEE Access 2017, 5, 24417–24425. [Google Scholar] [CrossRef]
- Abd El Aziz, M.; Hassanien, A.E. Modified cuckoo search algorithm with rough sets for feature selection. Neural Comput. Appl. 2018, 29, 925–934. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The Whale Optimization Algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Yang, X.-S. A New metaheuristic bat-inspired algorithm. In Nicso 2010: Nature Inspired Cooperative Strategies for Optimization; Gonzalez, J.R., Pelta, D.A., Cruz, C., Terrazas, G., Krasnogor, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 284, pp. 65–74. [Google Scholar]
- Yang, X.-S. Firefly algorithm, stochastic test functions and design optimisation. Int. J. Bio-Inspired Comput. 2010, 2, 78–84. [Google Scholar] [CrossRef]
- Mirjalili, S. Moth-flame optimization algorithm: A novel nature-inspired heuristic paradigm. Knowl. -Based Syst. 2015, 89, 228–249. [Google Scholar] [CrossRef]
- Binh, T.; Zhang, M.; Xue, B. A PSO based hybrid feature selection algorithm for high-dimensional classification. In 2016 IEEE Congress on Evolutionary Computation; IEEE: Manhattan, NY, USA, 2016; pp. 3801–3808. [Google Scholar]
- Eberhart, R.; Kennedy, J. A new optimizer using particle swarm theory. In Proceedings of the MHS’95, 6th International Symposium on Micro Machine and Human Science, Nagoya, Japan, 4–6 October 1995; pp. 39–43. [Google Scholar]
- Zhao, W.; Zhang, Z.; Wang, L. Manta ray foraging optimization: An effective bio-inspired optimizer for engineering applications. Eng. Appl. Artif. Intell. 2020, 87, 103300. [Google Scholar] [CrossRef]
- Mirjalili, S. Dragonfly algorithm: A new meta-heuristic optimization technique for solving single-objective, discrete, and multi-objective problems. Neural Comput. Appl. 2016, 27, 1053–1073. [Google Scholar] [CrossRef]
- Rostami, M.; Berahmand, K.; Nasiri, E.; Forouzandeh, S. Review of swarm intelligence-based feature selection methods. Eng. Appl. Artif. Intell. 2021, 100, 104210. [Google Scholar] [CrossRef]
- Brezocnik, L.; Fister, I., Jr.; Podgorelec, V. Swarm Intelligence Algorithms for Feature Selection: A Review. Appl. Sci. 2018, 8, 1521. [Google Scholar] [CrossRef]
- Han, Y.; Huang, L.; Zhou, F. A dynamic recursive feature elimination framework (dRFE) to further refine a set of OMIC biomarkers. Bioinformatics 2021, 37, 2183–2189. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Zhou, M.; Luo, Y.; Meng, Q.; Mai, G.; Ma, D.; Wang, G.; Zhou, F. McTwo: A two-step feature selection algorithm based on maximal information coefficient. BMC Bioinform. 2016, 17, 142. [Google Scholar] [CrossRef]
- Tian, E.; Zhan, F.H.; Walker, R.; Rasmussen, E.; Ma, Y.P.; Barlogie, B.; Shaughnessy, J.D. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 2003, 349, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Grabsch, H.; Ivanova, T.; Tan, I.B.; Murray, J.; Ooi, C.H.; Wright, A.I.; West, N.P.; Hutchins, G.G.A.; Wu, J.; et al. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut 2013, 62, 1100. [Google Scholar] [CrossRef]
- Wang, G.; Hu, N.; Yang, H.H.; Wang, L.; Su, H.; Wang, C.; Clifford, R.; Dawsey, E.M.; Li, J.-M.; Ding, T.; et al. Comparison of Global Gene Expression of Gastric Cardia and Noncardia Cancers from a High-Risk Population in China. PLoS ONE 2013, 8, e63826. [Google Scholar] [CrossRef]
- Levy, H.; Wang, X.; Kaldunski, M.; Jia, S.; Kramer, J.; Pavletich, S.J.; Reske, M.; Gessel, T.; Yassai, M.; Quasney, M.W.; et al. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012, 13, 593–604. [Google Scholar] [CrossRef]
- Krug, T.; Gabriel, J.P.; Taipa, R.; Fonseca, B.V.; Domingues-Montanari, S.; Fernandez-Cadenas, I.; Manso, H.; Gouveia, L.O.; Sobral, J.; Albergaria, I.; et al. TTC7B emerges as a novel risk factor for ischemic stroke through the convergence of several genome-wide approaches. J. Cereb. Blood Flow Metab. 2012, 32, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Shipp, M.A.; Ross, K.N.; Tamayo, P.; Weng, A.P.; Kutok, J.L.; Aguiar, R.C.T.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.S.; et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Febbo, P.G.; Ross, K.; Jackson, D.G.; Manola, J.; Ladd, C.; Tamayo, P.; Renshaw, A.A.; D’Amico, A.V.; Richie, J.P.; et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002, 1, 203–209. [Google Scholar] [CrossRef]
- Chiaretti, S.; Li, X.C.; Gentleman, R.; Vitale, A.; Vignetti, M.; Mandelli, F.; Ritz, J.; Foa, R. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood 2004, 103, 2771–2778. [Google Scholar] [CrossRef]
- Pomeroy, S.L.; Tamayo, P.; Gaasenbeek, M.; Sturla, L.M.; Angelo, M.; McLaughlin, M.E.; Kim, J.Y.H.; Goumnerova, L.C.; Black, P.M.; Lau, C.; et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002, 415, 436–442. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.G.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Notterman, D.A.; Alon, U.; Sierk, A.J.; Levine, A.J. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001, 61, 3124–3130. [Google Scholar]
- Alon, U.; Barkai, N.; Notterman, D.A.; Gish, K.; Ybarra, S.; Mack, D.; Levine, A.J. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1999, 96, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef]
- Alter, M.D.; Kharkar, R.; Ramsey, K.E.; Craig, D.W.; Melmed, R.D.; Grebe, T.A.; Bay, R.C.; Ober-Reynolds, S.; Kirwan, J.; Jones, J.J.; et al. Autism and Increased Paternal Age Related Changes in Global Levels of Gene Expression Regulation. PLoS ONE 2011, 6, e16715. [Google Scholar] [CrossRef] [PubMed]
- Shamir, R.; Klein, C.; Amar, D.; Vollstedt, E.-J.; Bonin, M.; Usenovic, M.; Wong, Y.C.; Maver, A.; Poths, S.; Safer, H.; et al. Analysis of blood-based gene expression in idiopathic Parkinson disease. Neurology 2017, 89, 1676–1683. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Ishikawa, T.; Iida, S.; Ishiguro, M.; Mogushi, K.; Mizushima, H.; Uetake, H.; Tanaka, H.; Sugihara, K. Clinical Significance of Osteoprotegerin Expression in Human Colorectal Cancer. Clin. Cancer Res. 2011, 17, 2444–2450. [Google Scholar] [CrossRef] [PubMed]
- Michael Meister, A.B.; Xu, E.C.; Schnabel, P.; Warth, A.; Hoffmann, H.; Dienemann, H.; Riedlinger, J.; Bodenmueller, H.; Zolg, W.; Herth, F.J.F.; et al. Intra-tumor Heterogeneity of Gene Expression Profiles in Early Stage Non-Small Cell Lung Cancer. J. Bioinform. Res. Stud. 2014, 1, 1. [Google Scholar]
- Lu, T.-P.; Tsai, M.-H.; Lee, J.-M.; Hsu, C.-P.; Chen, P.-C.; Lin, C.-W.; Shih, J.-Y.; Yang, P.-C.; Hsiao, C.K.; Lai, L.-C.; et al. Identification of a Novel Biomarker, SEMA5A, for Non-Small Cell Lung Carcinoma in Nonsmoking Women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2590–2597. [Google Scholar] [CrossRef] [PubMed]
- LaBreche, H.G.; Nevins, J.R.; Huang, E. Integrating Factor Analysis and a Transgenic Mouse Model to Reveal a Peripheral Blood Predictor of Breast Tumors. BMC Med. Genom. 2011, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Debernardi, A.; Jacquiau, B.; Vitte, A.-L.; Vesin, A.; Nagy-Mignotte, H.; Moro-Sibilot, D.; Brichon, P.-Y.; Lantuejoul, S.; Hainaut, P.; et al. Ectopic Activation of Germline and Placental Genes Identifies Aggressive Metastasis-Prone Lung Cancers. Sci. Transl. Med. 2013, 5, 186ra66. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, W.W.; Chari, R.; Coe, B.P.; Girard, L.; MacAulay, C.; Lam, S.; Gazdar, A.F.; Minna, J.D.; Lam, W.L. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene 2008, 27, 4615–4624. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dogan, M.V.; Shields, B.; Cutrona, C.; Gao, L.; Gibbons, F.X.; Simons, R.; Monick, M.; Brody, G.H.; Tan, K.; Beach, S.R.; et al. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genom. 2014, 15, 151. [Google Scholar] [CrossRef]
- Jones, L.R.; Young, W.; Divine, G.; Datta, I.; Chen, K.M.; Ozog, D.; Worsham, M.J. Genome-Wide Scan for Methylation Profiles in Keloids. Dis Markers 2015, 2015, 943176. [Google Scholar] [CrossRef]
- Bartlett, T.E.; Chindera, K.; McDermott, J.; Breeze, C.E.; Cooke, W.R.; Jones, A.; Reisel, D.; Karegodar, S.T.; Arora, R.; Beck, S.; et al. Epigenetic reprogramming of fallopian tube fimbriae in BRCA mutation carriers defines early ovarian cancer evolution. Nat. Commun. 2016, 7, 11620. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.G.; Hannon, E.; de Jager, P.L.; Chibnik, L.; Lott, S.J.; Condliffe, D.; Smith, A.R.; Haroutunian, V.; Troakes, C.; Al-Sarraj, S.; et al. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement. 2018, 14, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.K.; Ramnarayanan, K.; Zhu, F.; Srivastava, S.; Xu, C.; Tan, A.L.K.; Lee, M.; Tay, S.; Das, K.; Xing, M.; et al. Genomic and Epigenomic Profiling of High-Risk Intestinal Metaplasia Reveals Molecular Determinants of Progression to Gastric Cancer. Cancer Cell 2018, 33, 137. [Google Scholar] [CrossRef]
- Feng, X.; Li, J.; Li, H.; Chen, H.; Li, F.; Liu, Q.; You, Z.-H.; Zhou, F. Age Is Important for the Early-Stage Detection of Breast Cancer on Both Transcriptomic and Methylomic Biomarkers. Front. Genet. 2019, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, E.; Kurz, C.; Marschner, S.; Avanzo, M.; Gagliardi, V.; Fanetti, G.; Franchin, G.; Stancanello, J.; Corradini, S.; Niyazi, M.; et al. Distant metastasis time to event analysis with CNNs in independent head and neck cancer cohorts. Sci. Rep. 2021, 11, 6418. [Google Scholar] [CrossRef] [PubMed]
- Hichem, H.; Elkamel, M.; Rafik, M.; Mesaaoud, M.T.; Ouahiba, C. A new binary grasshopper optimization algorithm for feature selection problem. J. King Saud. Univ. 2019. [Google Scholar] [CrossRef]
- Zamani, H.; Nadimi-Shahraki, M.-H. Feature selection based on whale optimization algorithm for diseases diagnosis. Int. J. Comput. Sci. Inf. Secur. 2016, 14, 1243. [Google Scholar]
- Nakamura, R.Y.; Pereira, L.A.; Costa, K.A.; Rodrigues, D.; Papa, J.P.; Yang, X.-S. BBA: A binary bat algorithm for feature selection. In Proceedings of the 2012 25th SIBGRAPI Conference on Graphics, Patterns and Images, Ouro Preto, Brazil, 22–25 August 2012; pp. 291–297. [Google Scholar]
- Yang, X.-S.; Deb, S. Cuckoo search via Lévy flights. In Proceedings of the 2009 World Congress on Nature & Biologically Inspired Computing (NaBIC), Coimbatore, India, 9–11 December 2009; pp. 210–214. [Google Scholar]
- Rodrigues, D.; Pereira, L.A.; Almeida, T.; Papa, J.P.; Souza, A.; Ramos, C.C.; Yang, X.-S. BCS: A binary cuckoo search algorithm for feature selection. In Proceedings of the 2013 IEEE International Symposium on Circuits and Systems (ISCAS), Beijing, China, 19–23 May 2013; pp. 465–468. [Google Scholar]
- Yang, X.-S. Nature-Inspired Metaheuristic Algorithms; Luniver Press: København, Denmark, 2008. [Google Scholar]
- Yang, X.-S. Firefly algorithms for multimodal optimization. In Proceedings of the International Symposium on Stochastic Algorithms, Sapporo, Japan, 26–28 October 2009; pp. 169–178. [Google Scholar]
- Zawbaa, H.M.; Emary, E.; Parv, B.; Sharawi, M. Feature selection approach based on moth-flame optimization algorithm. In Proceedings of the 2016 IEEE Congress on Evolutionary Computation (CEC), Vancouver, BC, Canada, 24–29 July 2016; pp. 4612–4617. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; pp. 1942–1948. [Google Scholar]
- Sharkawy, R.; Ibrahim, K.; Salama, M.; Bartnikas, R. Particle swarm optimization feature selection for the classification of conducting particles in transformer oil. IEEE Trans. Dielectr. Electr. Insul. 2011, 18, 1897–1907. [Google Scholar] [CrossRef]
- Ghosh, K.K.; Guha, R.; Bera, S.K.; Kumar, N.; Sarkar, R. S-shaped versus V-shaped transfer functions for binary Manta ray foraging optimization in feature selection problem. Neural Comput. Appl. 2021, 33, 11027–11041. [Google Scholar] [CrossRef]
- Mafarja, M.M.; Eleyan, D.; Jaber, I.; Hammouri, A.; Mirjalili, S. Binary dragonfly algorithm for feature selection. In Proceedings of the 2017 International Conference on New Trends in Computing Sciences (ICTCS), Amman, Jordan, 11–13 October 2017; pp. 12–17. [Google Scholar]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary grey wolf optimization approaches for feature selection. Neurocomputing 2016, 172, 371–381. [Google Scholar] [CrossRef]
- Yang, X.-S. Nature-inspired optimization algorithms: Challenges and open problems. J. Comput. Sci. 2020, 46, 101104. [Google Scholar] [CrossRef]
- Khurma, R.A.; Aljarah, I.; Sharieh, A.; Mirjalili, S. EvoloPy-FS: An open-source nature-inspired optimization framework in python for feature selection. In Evolutionary Machine Learning Techniques: Algorithms and Applications; Mirjalili, S., Faris, H., Aljarah, I., Eds.; Springer: Singapore, 2020; pp. 131–173. [Google Scholar]
- Faris, H.; Aljarah, I.; Mirjalili, S.; Castillo, P.A.; Merelo, J.J. EvoloPy: An Open-Source Nature-Inspired Optimization Framework in Python; SCITEPRESS—Science and Technology Publications: Porto, Portugal, 2016; pp. 171–177. [Google Scholar]
- Abdel-Basset, M.; El-Shahat, D.; El-henawy, I.; de Albuquerque, V.H.C.; Mirjalili, S. A new fusion of grey wolf optimizer algorithm with a two-phase mutation for feature selection. Expert Syst. Appl. 2020, 139, 112824. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, R.; Zheng, W.; Liu, S.; Zhou, F. RIFS: A randomly restarted incremental feature selection algorithm. Sci. Rep. 2017, 7, 13013. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Huang, L.; Zhou, F. Zoo: Selecting Transcriptomic and Methylomic Biomarkers by Ensembling Animal-Inspired Swarm Intelligence Feature Selection Algorithms. Genes 2021, 12, 1814. https://doi.org/10.3390/genes12111814
Han Y, Huang L, Zhou F. Zoo: Selecting Transcriptomic and Methylomic Biomarkers by Ensembling Animal-Inspired Swarm Intelligence Feature Selection Algorithms. Genes. 2021; 12(11):1814. https://doi.org/10.3390/genes12111814
Chicago/Turabian StyleHan, Yuanyuan, Lan Huang, and Fengfeng Zhou. 2021. "Zoo: Selecting Transcriptomic and Methylomic Biomarkers by Ensembling Animal-Inspired Swarm Intelligence Feature Selection Algorithms" Genes 12, no. 11: 1814. https://doi.org/10.3390/genes12111814
APA StyleHan, Y., Huang, L., & Zhou, F. (2021). Zoo: Selecting Transcriptomic and Methylomic Biomarkers by Ensembling Animal-Inspired Swarm Intelligence Feature Selection Algorithms. Genes, 12(11), 1814. https://doi.org/10.3390/genes12111814





