Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Plant Growth
2.2. Y1H Assay
2.3. Construction of proCRC:GUS Reportersystem and GUS Assays
2.4. Expression Analysis
2.5. In Silico Analysis of Genomic Loci, GO Enrichment and Co-Expression Analysis
3. Results
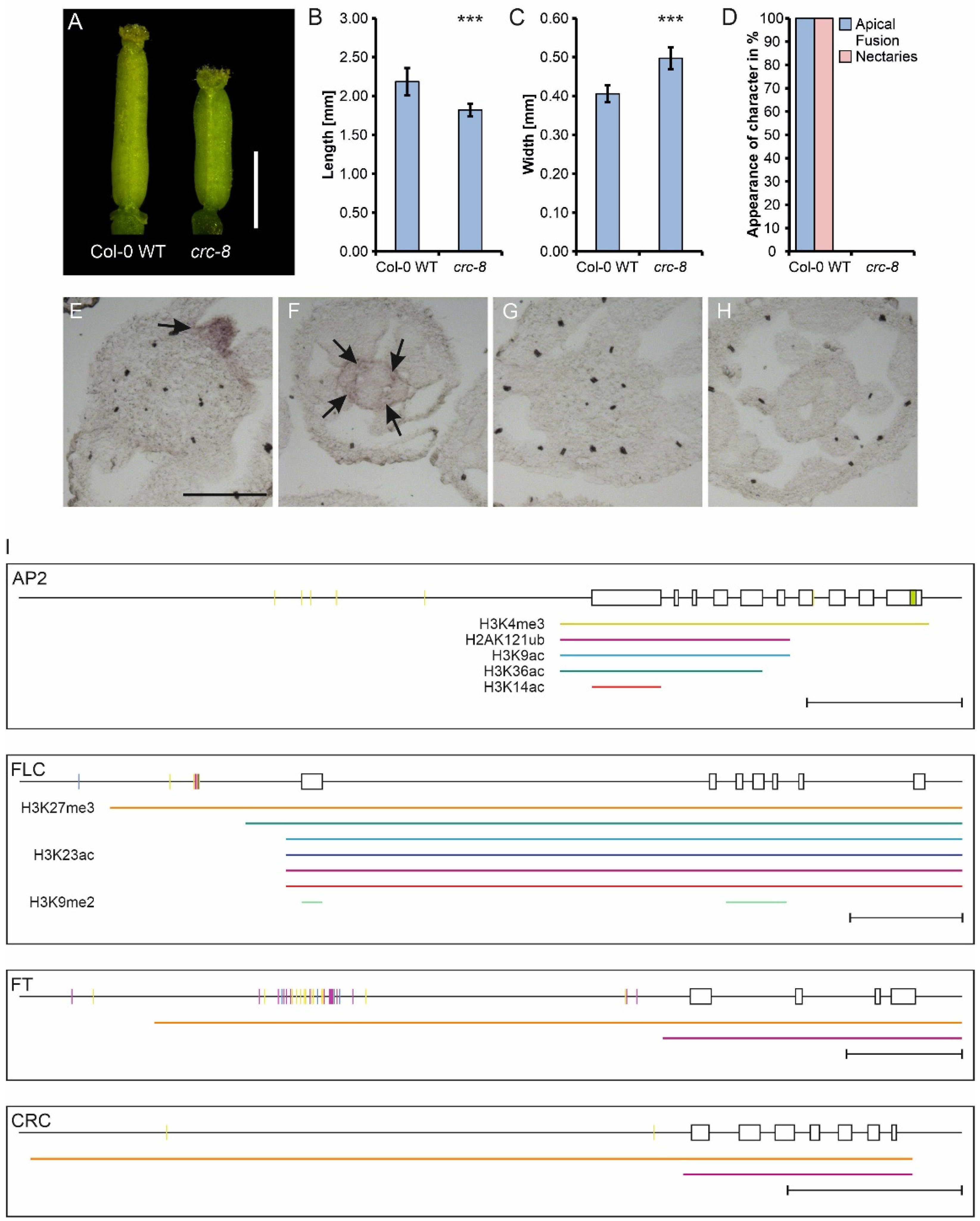
3.1. Regulation by DNA Methylation, Chromatin Modifications or miRNAs Plays Only a Minor Role in CRC Expression
3.2. Diverse Transcription Factors Bind to the CRC Promoter
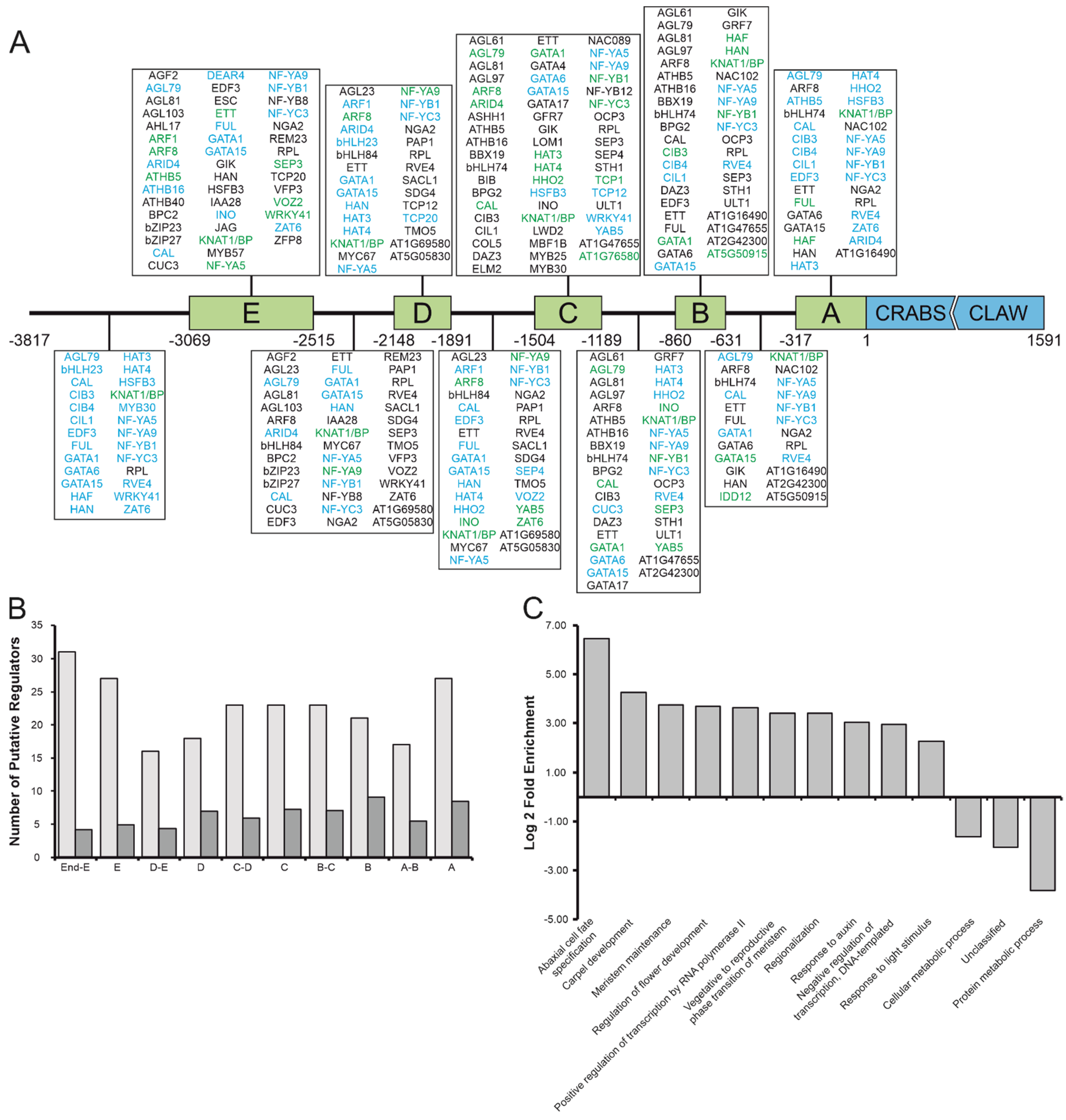
3.3. Relevant Promoter Fragments Are Enriched in TFBS and CRC Regulators Are Functionally Related
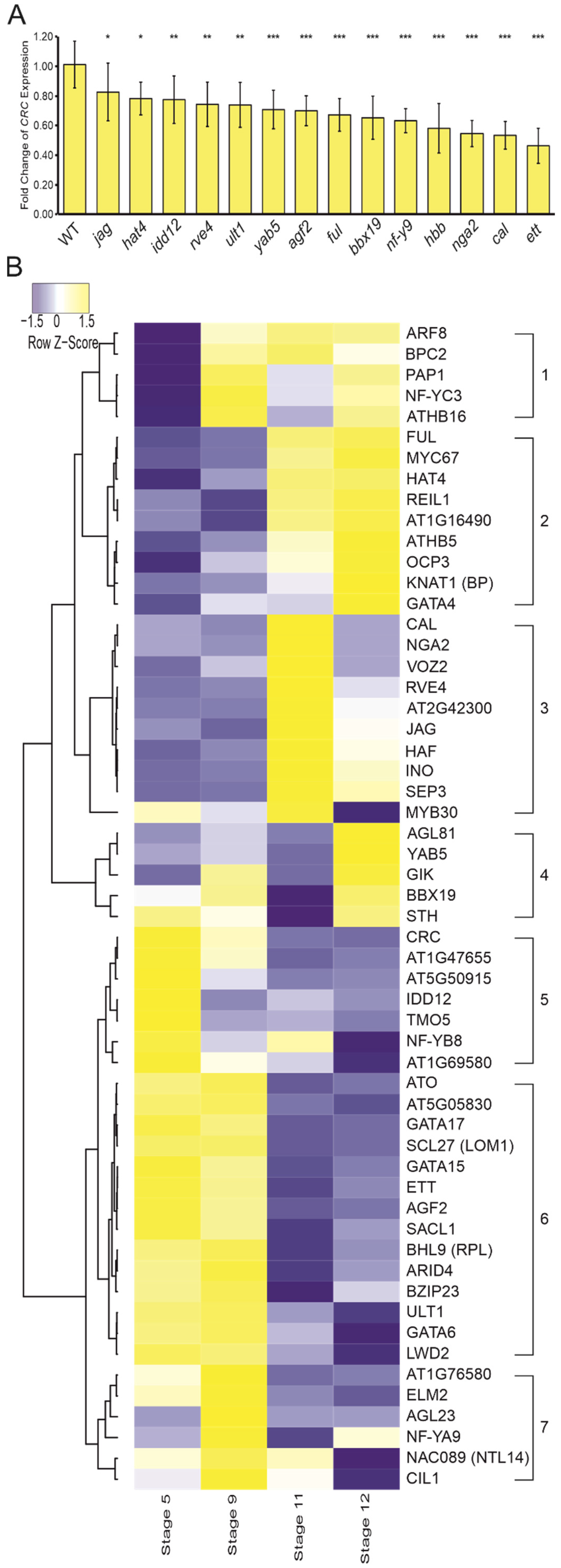
3.4. CRC Expression Is Activated by Diverse Developmental Regulators
3.5. Regulators of CRC Are Partially Co-Expressed during Flower Development
4. Discussion
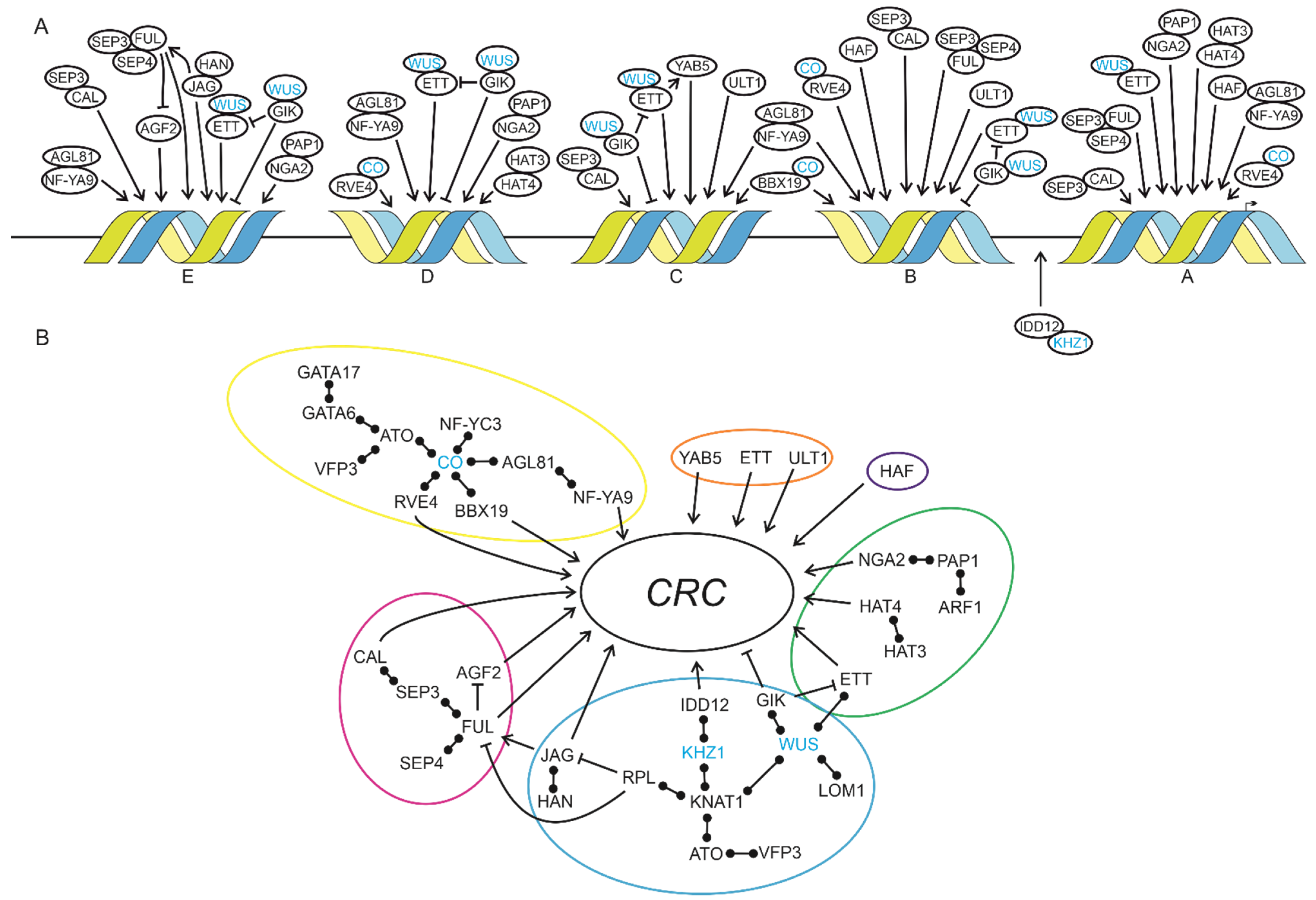
4.1. A Complex Interplay of Transcription Factors Regulates CRC Expression
4.2. Regulation of Complex Expression Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riethoven, J.-J.M. Regulatory Regions in DNA: Promoters, Enhancers, Silencers, and Insulators. Methods Mol. Biol. 2010, 674, 33–42. [Google Scholar] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta (BBA)-Bioenergy 2015, 1849, 1116–1131. [Google Scholar] [CrossRef]
- Porto, M.S.; Pinheiro, M.P.N.; Batista, V.G.L.; dos Santos, R.C.; Filho, P.D.A.M.; de Lima, L.M. Plant Promoters: An Approach of Structure and Function. Mol. Biotechnol. 2014, 56, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Sun, K.; Mackaluso, J.D.; Seddon, A.E.; Jin, R.; Thomashow, M.F.; Shiu, S.-H. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 14992–14997. [Google Scholar] [CrossRef]
- Thomas, M.C.; Chiang, C.-M. The General Transcription Machinery and General Cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Deal, R.B.; Henikoff, S. Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 2011, 14, 116–122. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nat. Cell Biol. 2004, 427, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Zicola, J.; Liu, L.; Tänzler, P.; Turck, F. Targeted DNA methylation represses two enhancers of Flowering Locus T in Arabidopsis thaliana. Nat. Plants 2019, 5, 300–307. [Google Scholar] [CrossRef]
- Chen, X. A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Miyashima, S.; Koi, S.; Hashimoto, T.; Nakajima, K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 2011, 138, 2303–2313. [Google Scholar] [CrossRef]
- Lieb, J.; Liu, X.; Botstein, D.; Brown, P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet. 2001, 28, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Moses, A.M.; Chiang, D.Y.; Pollard, D.A.; Iyer, V.N.; Eisen, M.B. MONKEY: Identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004, 5, R98. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Muino, J.M.; Østerås, M.; Farinelli, L.; Krajewski, P.; Angenent, G.C. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 2010, 5, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-S.; Wang, Z.; Miao, F.; Ma, H.; Kao, C.-T.; Hsu, T.-S.; Yu, J.-H.; Hung, E.-T.; Lin, C.-C.; Kuan, C.-Y.; et al. A novel synthetic-genetic-array—based yeast one-hybrid system for high discovery rate and short processing time. Genome Res. 2019, 29, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Moses, A.M.; Chiang, D.Y.; Kellis, M.; Lander, E.S.; Eisen, M.B. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 2003, 3, 19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bow-man, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Bowman, J.L. Distinct Mechanisms Promote Polarity Establishment in Carpels of Arabidopsis. Cell 1999, 99, 199–209. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Liu, T.; Newell, N.R.; Magnani, E.; Huang, T.; Kerstetter, R.; Michaels, S.; Barton, M.K. Establishing a Framework for the Ad/Abaxial Regulatory Network of Arabidopsis: Ascertaining Targets of Class III HOMEODOMAIN LEUCINE ZIPPER and KANADI Regulation. Plant Cell 2013, 25, 3228–3249. [Google Scholar] [CrossRef]
- Tatematsu, K.; Toyokura, K.; Miyashima, S.; Nakajima, K.; Okada, K. A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. 2015, 82, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW Acts as a Bifunctional Transcription Factor in Flower Development. Front. Plant Sci. 2018, 9, 835. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLA Win the Nectaries and Carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mena, C.; de Folter, S.; Costa, M.M.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.; Wuest, S.; Rae, L.; Raganelli, A.; Ryan, P.; Kwasniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of Reproductive Floral Organ Identity Specification in Arabidopsis by the C Function Regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- Wuest, S.; Ó’Maoiléidigh, D.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Huang, J.; Xu, Y.; Tanoi, K.; Ito, T. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef]
- Mitsuda, N.; Ikeda, M.; Takada, S.; Takiguchi, Y.; Kondou, Y.; Yoshizumi, T.; Fujita, M.; Shinozaki, K.; Matsui, M.; Ohme-Takagi, M. Efficient Yeast One-/Two-Hybrid Screening Using a Library Composed Only of Transcription Factors in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 2145–2151. [Google Scholar] [CrossRef] [PubMed]
- Hemsley, A.; Arnheim, N.; Toney, M.D.; Cortopassi, G.; Galas, D.J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989, 17, 6545–6551. [Google Scholar] [CrossRef] [PubMed]
- Lampropoulos, A.; Sutikovic, Z.; Wenzl, C.; Maegele, I.; Lohmann, J.U.; Forner, J. GreenGate—A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. PLoS ONE 2013, 8, e83043. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.M.; Hall, A.; Millar, A.J.; Darrah, C.; Davis, S.J. Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 2009, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Glazebrook, J. Arabidopsis: Alaboratorymanual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2002; ISBN 0-87969-573-0. [Google Scholar]
- Brewer, P.B.; Heisler, M.; Hejatko, J.; Friml, J.; Benková, E. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat. Protoc. 2006, 1, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Huang, S.-s.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Zheng, H.-Q.; Wu, N.-Y.; Chien, C.-H.; Huang, H.-D.; Lee, T.-Y.; Chiang-Hsieh, Y.-F.; Hou, P.-F.; Yang, T.-Y.; Chang, W.-C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012, 41, D377–D386. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kivivirta, K.; Herbert, D.; Lange, M.; Beuerlein, K.; Altmüller, J.; Becker, A. A protocol for laser microdissection (LMD) followed by transcriptome analysis of plant reproductive tissue in phylogenetically distant angiosperms. Plant Methods 2019, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.; Kasianov, A.S.; Gerasimov, E.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Ratner, B. The correlation coefficient: Its values range between +1/−1, or do they? J. Target. Meas. Anal. Mark. 2009, 17, 139–142. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Schölkopf, B.; Weigel, D.; Lohmann, J. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef]
- Li, B.; Gao, Z.; Liu, X.; Sun, D.; Tang, W. Transcriptional Profiling Reveals a Time-of-Day-Specific Role of REVEILLE 4/8 in Regulating the First Wave of Heat Shock–Induced Gene Expression in Arabidopsis. Plant Cell 2019, 31, 2353–2369. [Google Scholar] [CrossRef]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The REVEILLE Clock Genes Inhibit Growth of Juvenile and Adult Plants by Control of Cell Size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef]
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef]
- Sessions, A.; Nemhauser, J.L.; McColl, A.; Roe, J.L.; Feldmann, K.A.; Zambryski, P.C. EETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 1997, 124, 4481–4491. [Google Scholar] [CrossRef]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [CrossRef]
- Crawford, B.C.W.; Yanofsky, M.F. HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 2011, 138, 2999–3009. [Google Scholar] [CrossRef]
- Ohno, C.K.; Reddy, G.V.; Heisler, M.; Meyerowitz, E.M. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 2004, 131, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.P.; Goldshmidt, A.; Efroni, I.; Bowman, J.L.; Eshed, Y. The NGATHA Distal Organ Development Genes Are Essential for Style Specification in Arabidopsis. Plant Cell 2009, 21, 1373–1393. [Google Scholar] [CrossRef] [PubMed]
- Prunet, N.; Morel, P.; Thierry, A.-M.; Eshed, Y.; Bowman, J.L.; Negrutiu, I.; Trehin, C. REBELOTE, SQUINT, and ULTRAPETALA1Function Redundantly in the Temporal Regulation of Floral Meristem Termination in Arabidopsis thaliana. Plant Cell 2008, 20, 901–919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carles, C.C.; Fletcher, J.C. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009, 23, 2723–2728. [Google Scholar] [CrossRef] [PubMed]
- Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y. Signals Derived from YABBY Gene Activities in Organ Primordia Regulate Growth and Partitioning of Arabidopsis Shoot Apical Meristems. Plant Cell 2008, 20, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Kempin, S.A.; Savidge, B.; Yanofsky, M.F. Molecular basis of the cauliflower phenotype in Arabidopsis. Science 1995, 267, 522–525. [Google Scholar] [CrossRef]
- Kivivirta, K.I.; Herbert, D.; Roessner, C.; de Folter, S.; Marsch-Martinez, N.; Becker, A. Transcriptome analysis of gynoecium morphogenesis uncovers the chronology of gene regulatory network activity. Plant Physiol. 2021, 185, 1076–1090. [Google Scholar] [CrossRef]
- Margolin, A.A.; Wang, K.; Lim, W.K.; Kustagi, M.; Nemenman, I.; Califano, A. Reverse engineering cellular networks. Nat. Protoc. 2006, 1, 662–671. [Google Scholar] [CrossRef]
- Ng, K.-H.; Yu, H.; Ito, T. AGAMOUS Controls GIANT KILLER, a Multifunctional Chromatin Modifier in Reproductive Organ Patterning and Differentiation. PLoS Biol. 2009, 7, e1000251. [Google Scholar] [CrossRef]
- Sun, B.; Ito, T. Regulation of floral stem cell termination in Arabidopsis. Front. Plant Sci. 2015, 6, 17. [Google Scholar] [CrossRef]
- Obayashi, T.; Mutwil, M.; Giorgi, F.; Bassel, G.; Tanimoto, M.; Chow, A.; Steinhauser, D.; Persson, S.; Provart, N.J. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009, 32, 1633–1651. [Google Scholar] [CrossRef]
- Bottomley, M.J.; Collard, M.W.; Huggenvik, J.I.; Liu, Z.; Gibson, T.J.; Sattler, M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Genet. 2001, 8, 626–633. [Google Scholar] [CrossRef]
- Garcia, D.; Collier, S.A.; Byrne, M.; Martienssen, R.A. Specification of Leaf Polarity in Arabidopsis via the trans-Acting siRNA Pathway. Curr. Biol. 2006, 16, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Crawford, B.C.; Ditta, G.; Yanofsky, M.F. The NTT Gene Is Required for Transmitting-Tract Development in Carpels of Arabidopsis thaliana. Curr. Biol. 2007, 17, 1101–1108. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative Regulation of the SHATTERPROOF Genes by FRUITFULL during Arabidopsis Fruit Development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef]
- Bemer, M.; Van Mourik, H.; Muino, J.M.; Ferrándiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef] [PubMed]
- Trigueros, M.; Navarrete-Gómez, M.; Sato, S.; Christensen, S.K.; Pelaz, S.; Weigel, D.; Yanofsky, M.F.; Ferrándiz, C. TheNGATHAGenes Direct Style Development in theArabidopsisGynoecium. Plant Cell 2009, 21, 1394–1409. [Google Scholar] [CrossRef] [PubMed]
- Melzer, R.; Theißen, G. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 2009, 37, 2723–2736. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Wellmer, F. Molecular Regulation of Flower Development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar]
- Vercruysse, J.; Baekelandt, A.; Gonzalez, N.; Inzé, D. Molecular networks regulating cell division during Arabidopsis leaf growth. J. Exp. Bot. 2020, 71, 2365–2378. [Google Scholar] [CrossRef]
- Ballester, P.; Ferrándiz, C. Shattering fruits: Variations on a dehiscent theme. Curr. Opin. Plant Biol. 2017, 35, 68–75. [Google Scholar] [CrossRef]
- Hackett, S.R.; Baltz, E.A.; Coram, M.; Wranik, B.J.; Kim, G.; Baker, A.; Fan, M.; Hendrickson, D.G.; Berndl, M.; McIsaac, R.S. Learning causal networks using inducible transcription factors and transcriptome-wide time series. Mol. Syst. Biol. 2020, 16, e9174. [Google Scholar] [CrossRef]
- Hofhuis, H.F.; Heidstra, R. Transcription factor dosage: More or less sufficient for growth. Curr. Opin. Plant Biol. 2018, 45, 50–58. [Google Scholar] [CrossRef]
- Chen, D.; Yan, W.; Fu, L.-Y.; Kaufmann, K. Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Simonini, S.; Bencivenga, S.; Trick, M.; Østergaard, L. Auxin-Induced Modulation of ETTIN Activity Orchestrates Gene Expression in Arabidopsis. Plant Cell 2017, 29, 1864–1882. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.; Arreola, A.; Gallagher, T.L.; Gasser, C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Trigg, S.; Garza, R.M.; MacWilliams, A.; Nery, J.R.; Bartlett, A.; Castanon, R.; Goubil, A.; Feeney, J.; O’Malley, R.; Huang, S.-S.C.; et al. CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 2017, 14, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-P.; Lin, J.-J.; Li, W.-H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6, 25164. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Yin, Y.; Nitta, K.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nat. Cell Biol. 2015, 527, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Inukai, S.; Kock, K.H.; Bulyk, M.L. Transcription factor—DNA binding: Beyond binding site motifs. Curr. Opin. Genet. Dev. 2017, 43, 110–119. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gross, T.; Becker, A. Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis. Genes 2021, 12, 1663. https://doi.org/10.3390/genes12111663
Gross T, Becker A. Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis. Genes. 2021; 12(11):1663. https://doi.org/10.3390/genes12111663
Chicago/Turabian StyleGross, Thomas, and Annette Becker. 2021. "Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis" Genes 12, no. 11: 1663. https://doi.org/10.3390/genes12111663
APA StyleGross, T., & Becker, A. (2021). Transcription Factor Action Orchestrates the Complex Expression Pattern of CRABS CLAW in Arabidopsis. Genes, 12(11), 1663. https://doi.org/10.3390/genes12111663






