1. Introduction
Avocado (
Persea americana Mill.) is an important fruit plant cultivated in tropical and subtropical climates. The fruit consumption is increasing worldwide, although much of the global production is in South and Mesoamerica [
1,
2].
P. americana is a polymorphic species with three botanical groups or horticultural races (the West Indian, Guatemalan, and Mexican) that are ecologically distinguishable [
3]. Individuals in each botanical group also have some common genetic characteristics that distinguish them from members of other groups [
4,
5].
Evaluating the genetic variation existing in a given germplasm is essential to understand its potential application in crop breeding. The knowledge is also important in estimating the loss of genetic diversity, in providing proofs of the evolutionary forces shaping the genotypic variations, and also in selecting genotypes to be prioritized in conservation strategies [
6]. Different markers have been used in avocado germplasm characterization, management, and conservation. Morphological markers were used to characterize avocado germplasm in California [
7], Florida [
8], Ghana [
9,
10], Mexico [
11], Indonesia [
12], and Tanzania [
13], among others. However, besides being labor intensive, morphological traits are associated with some shortcomings, such as low variability (polymorphism) and heritability, late expression, influence by environmental factors, and subjectivity [
14,
15]. Nowadays, avocado germplasm characterization has been improved by the use of genetic markers, which can even discriminate closely related individuals. Some genetic markers that have been applied are isozymes [
16], minisatellites [
17], variable number tandem repeats (VNTRs) [
18], randomly amplified polymorphic DNA (RAPD) [
19], and restriction fragment length polymorphisms (RFLP) [
20,
21]. Others are inter-simple sequence repeats (ISSR) [
22], simple sequence repeat (SSR) [
15,
23,
24,
25], and single nucleotide polymorphisms (SNPs) [
5,
26,
27]. Choosing which marker type to employ in a diversity study depends on the study objectives and available financial resources, expertise, and facilities [
28].
Population genetics has been used in describing the genetic composition of avocado populations and mechanisms affecting the composition [
15,
23,
24,
25]. Bayesian cluster analysis employed in the STRUCTURE and discriminant analysis of principal components has been widely utilized in studying the population structure of crops, including avocado [
5,
24,
25,
26,
29]. Bayesian cluster analysis generates genetic clusters (genetic populations) with individuals in each cluster having distinctive allele frequencies at the investigated loci [
30,
31,
32]. In avocado research, these genetic clusters have, sometimes, been shown to conform to the horticultural origin of the crop [
4,
5].
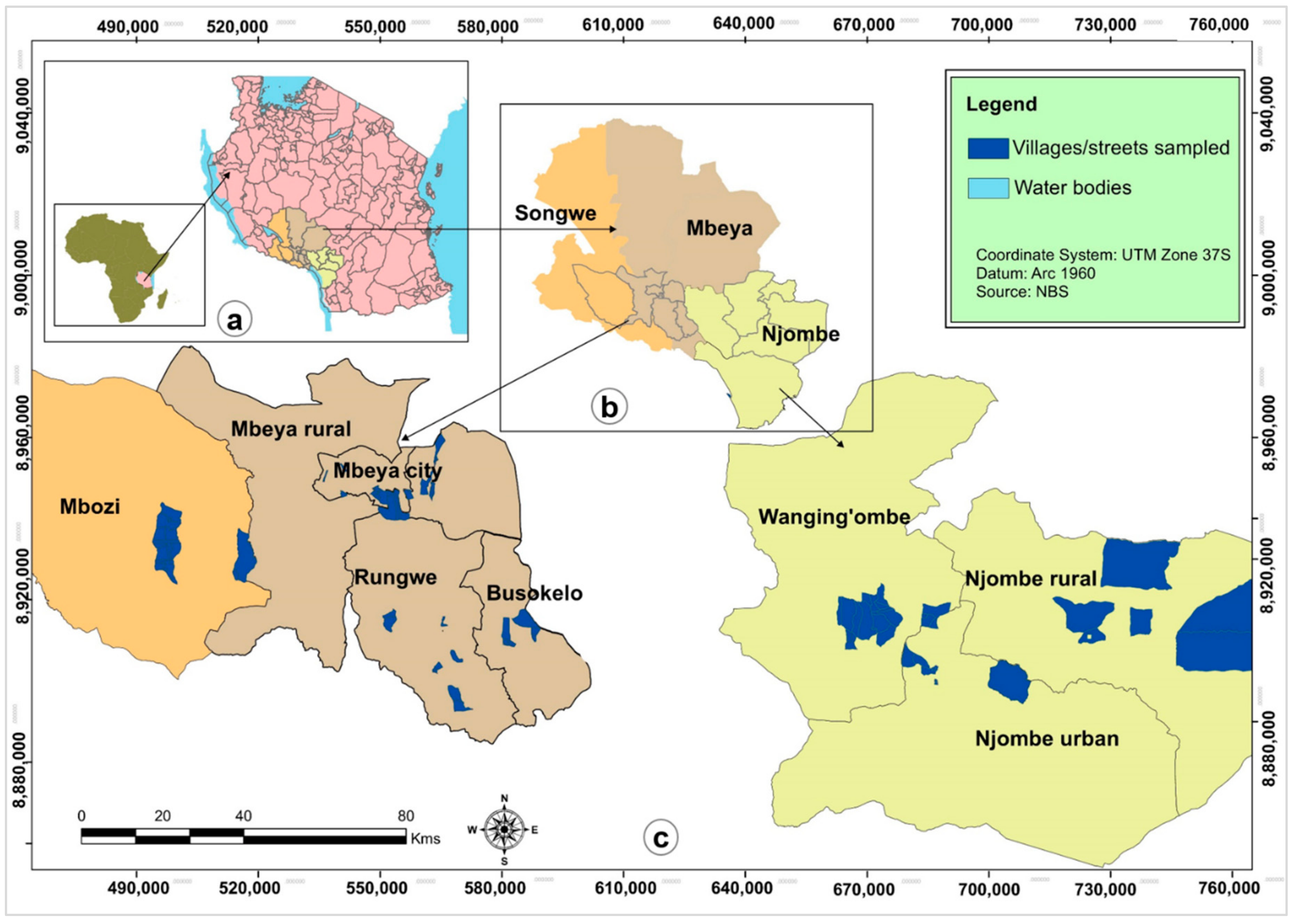
Tanzania rises from the sea level to more than 2900 m above sea level. The country has varying topographies, soils, and climates, which support the growth of different cultivars of avocados [
13,
25]. Although avocado is grown in several regions of Tanzania for the export and domestic markets [
33], only two studies have been executed to characterize this germplasm based on morphological traits [
13] and SSR markers [
25]. The present study aimed to compare morphological and genetic characteristics of this germplasm and uncover correlations existing among the morphological and genetic characteristics and the geographical sampling locations. Such insights can provide important information for plant breeders to plan breeding programs in the future. In addition, the insights can increase awareness about avocado genetic resources that could be exploited for management and utilization in Tanzania.
4. Discussion
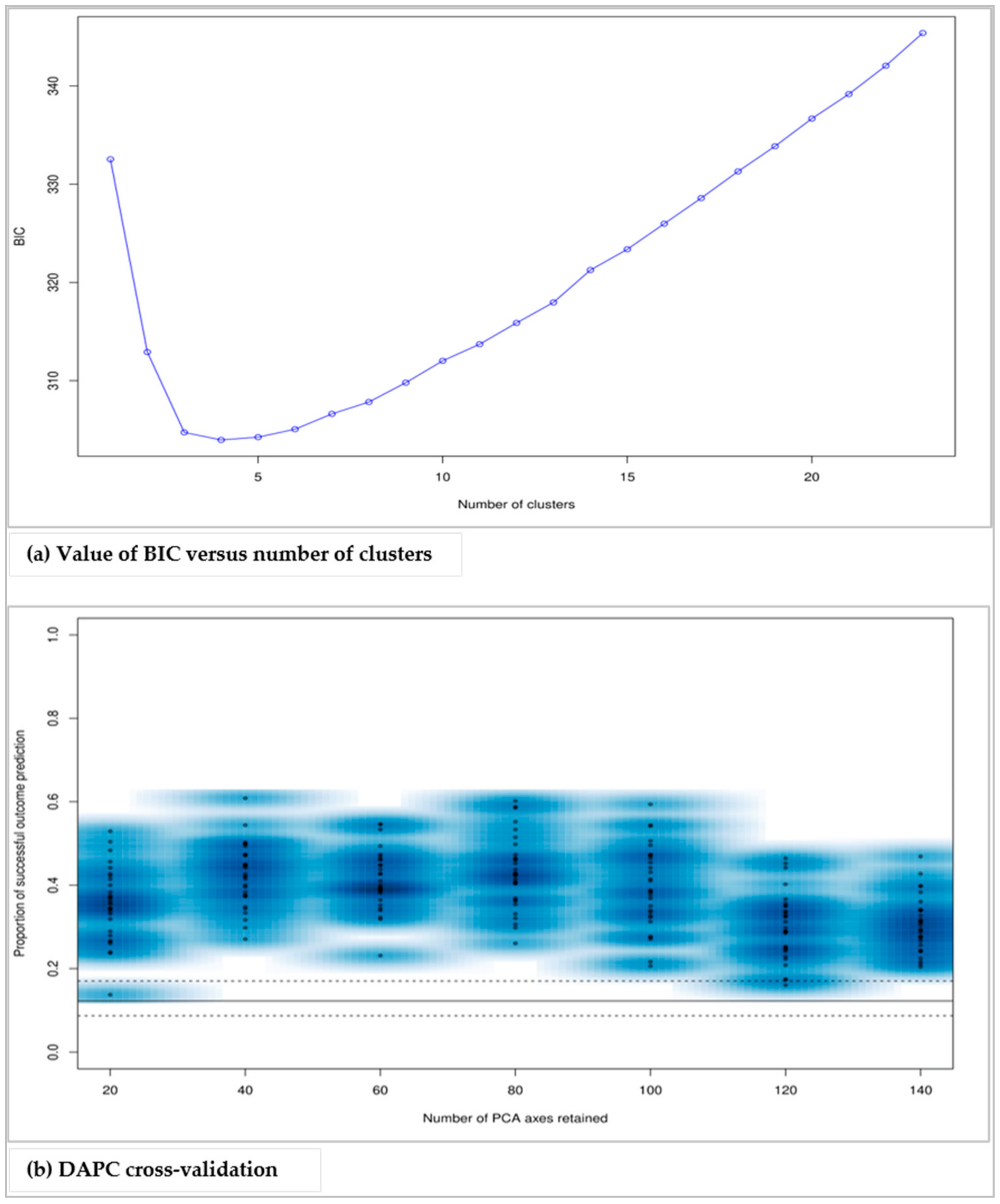
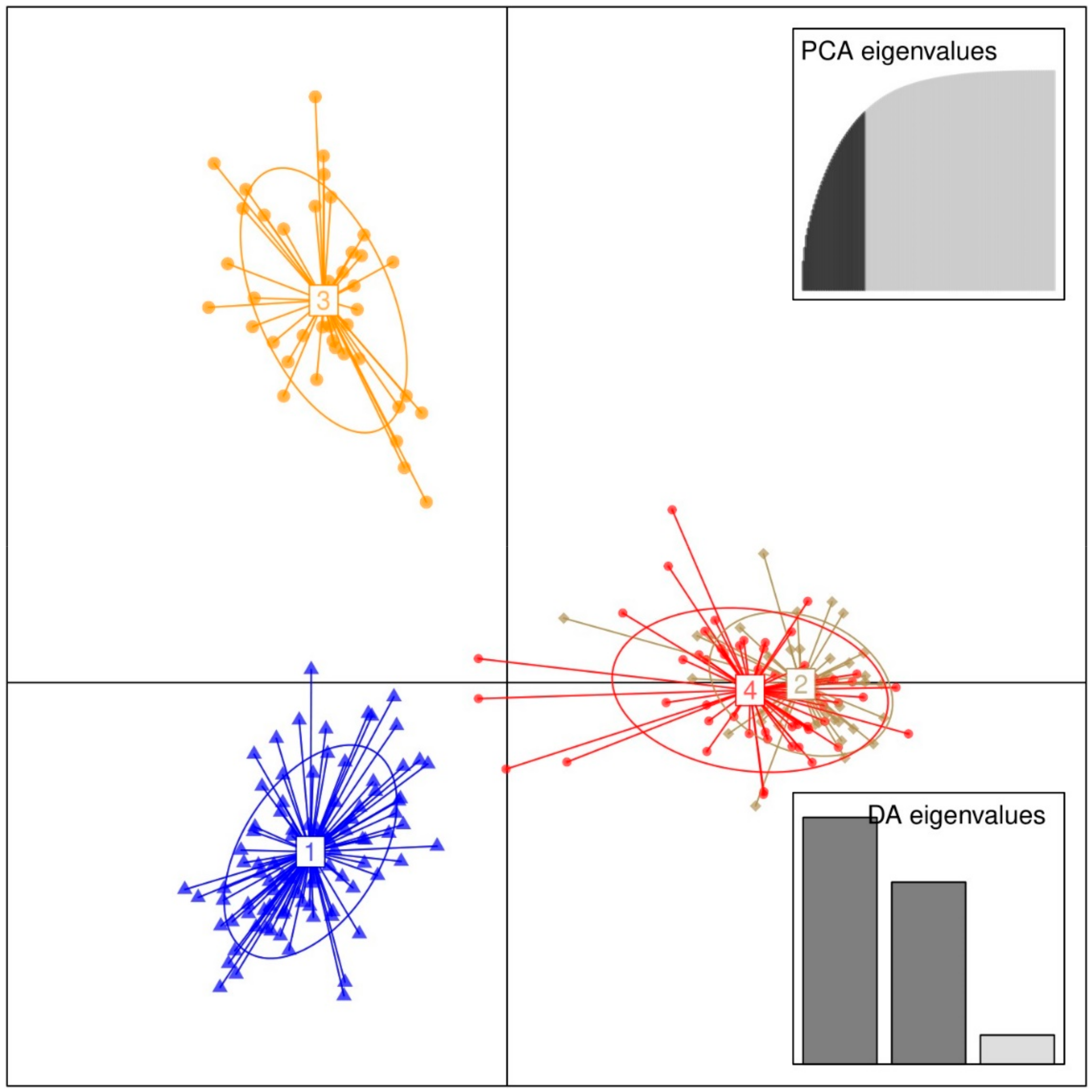
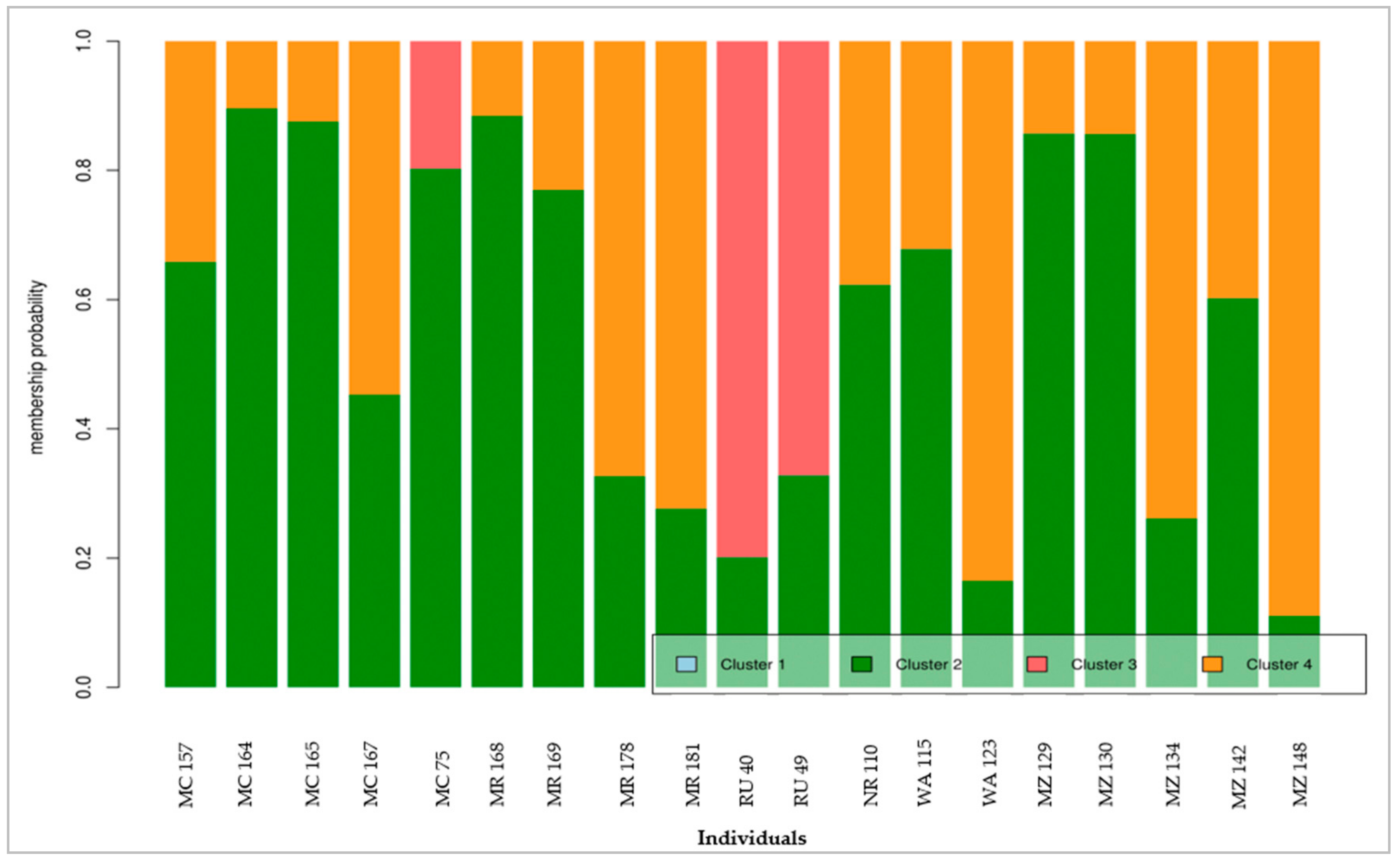
The present study has demonstrated the effectiveness of the genetic markers (microsatellite markers) over traditional morphological markers in characterizing avocado, exploring the diversity and the relationships among the individuals. Likewise, the study has shown the utility of DAPC in establishing the population structure of avocado crops and providing in-depth information on the individuals of the identified genetic clusters, which is an important step for practical plant breeding and conservation.
High diversity was noticed among the individuals of the four genetic clusters at the ten microsatellite loci. The mean number of different alleles per locus among the four clusters ranged from 6.60 (cluster 3) to 11.80 (cluster 4), with an average of 9.40 across the four clusters and loci (
Table 4). Gross-German and Viruel [
37] found a range of 3.7 (West Indian group) to 7.10 (hybrid group) with an average of 5.58 for the four populations they investigated, which consisted of a total of 41 avocado samples. Boza et al. [
4] reported a range of 7.93 (Mexican group) to 9.78 (Guatemalan group), among the three horticultural groups, with a much higher overall mean of 9.09. Similarly, Schnell et al. [
23] got a range of 6.00 (Mexican × West Indian group) to 13.35 (Mexican group) with an overall average of 10.26 for six populations of avocado comprising 221 samples. Cañas-Gutiérrez et al. [
49] reported a lower overall mean, 4.46 for 18 geographical populations. In the present work, allele richness was lowest in cluster 3 (6.00) and highest in cluster 4 (9.48) with an overall mean value of 7.69. This suggests that clusters 3 and 4 were the least and the most genetically diverse clusters, respectively. The most genetically diverse groups would be offered protection in conservation programs, and they may provide the best plant materials for breeding programs, whereas the least genetically diverse groups would deserve special conservation management [
50]. Guzmán et al. [
24] recorded a comparatively lower allelic richness, 5.95 (Mexican group) to 6.22 (West Indian group) with an overall average of 6.10, for the three avocado racial groups. While the current study’s private allele richness ranged from 1.00 (cluster 3) to 2.09 (cluster 1) with an overall average of 1.45, Guzmán et al. [
24] recorded a range of 0.63 (Mexican group) to 0.89 (Guatemalan group) with an overall mean of 0.74 for the three avocado populations. The average observed and expected heterozygosity for the four clusters was found to be 0.65 and 0.74, respectively. Lower values were reported by Boza et al. [
4],
Ho: 0.53 and
He: 0.64, for the three horticultural races included in their study. Higher values were estimated by Gross-German and Viruel [
37],
Ho: 0.66 and
He: 0.71 (four populations), and Schnell et al. [
23],
Ho: 0.71 and
He: 0.77 (six populations), indicating a comparatively higher diversity. While the overall average gene diversity in the present work was 0.59, Boza et al. [
4] obtained a higher value (0.63) for the three avocado races they investigated.
The number of private alleles per locus ranged from 0.50 (cluster 2) to 2.30 (cluster 1) with a grand mean of 1.23 across all populations and loci (
Table 4). Private alleles are a measure of population differentiation, thus the highest value for the number of private alleles per locus detected in cluster 1 indicates the greatest genetic differentiation of this cluster as was also revealed by its largest mean F
ST. Boza et al. [
4] reported the number of private allele per locus ranging from 0.65 (Mexican group) to 0.71 (West Indian group) among the three avocado races, and 0.02 to 0.07 among their six hybrid groups with a grand mean value of 0.23 for the nine populations, which is lower than the value obtained in our study. While, in the present study, the lowest and highest number of rare alleles per locus was 1.80 (cluster 3) and 5.80 (cluster 4), Boza et al. [
4] got a range of 3.31 (Mexican group) to 6.24 (West Indian group) among the three botanical groups, and 0.00 to 3.44 among their six hybrid groups. Rare alleles are significant in plant breeding as they may be associated with adaptations to biotic and abiotic stresses [
51]. In our study, the number of common alleles per locus varied from 4.40 (cluster 1 and cluster 3) to 6.00 (cluster 4), whereas Boza et al. [
4] got a range of 3.22 (West Indian group) to 4.67 (Guatemalan group) among the three botanical groups, and 3.80 to 4.64 among their six hybrid groups.
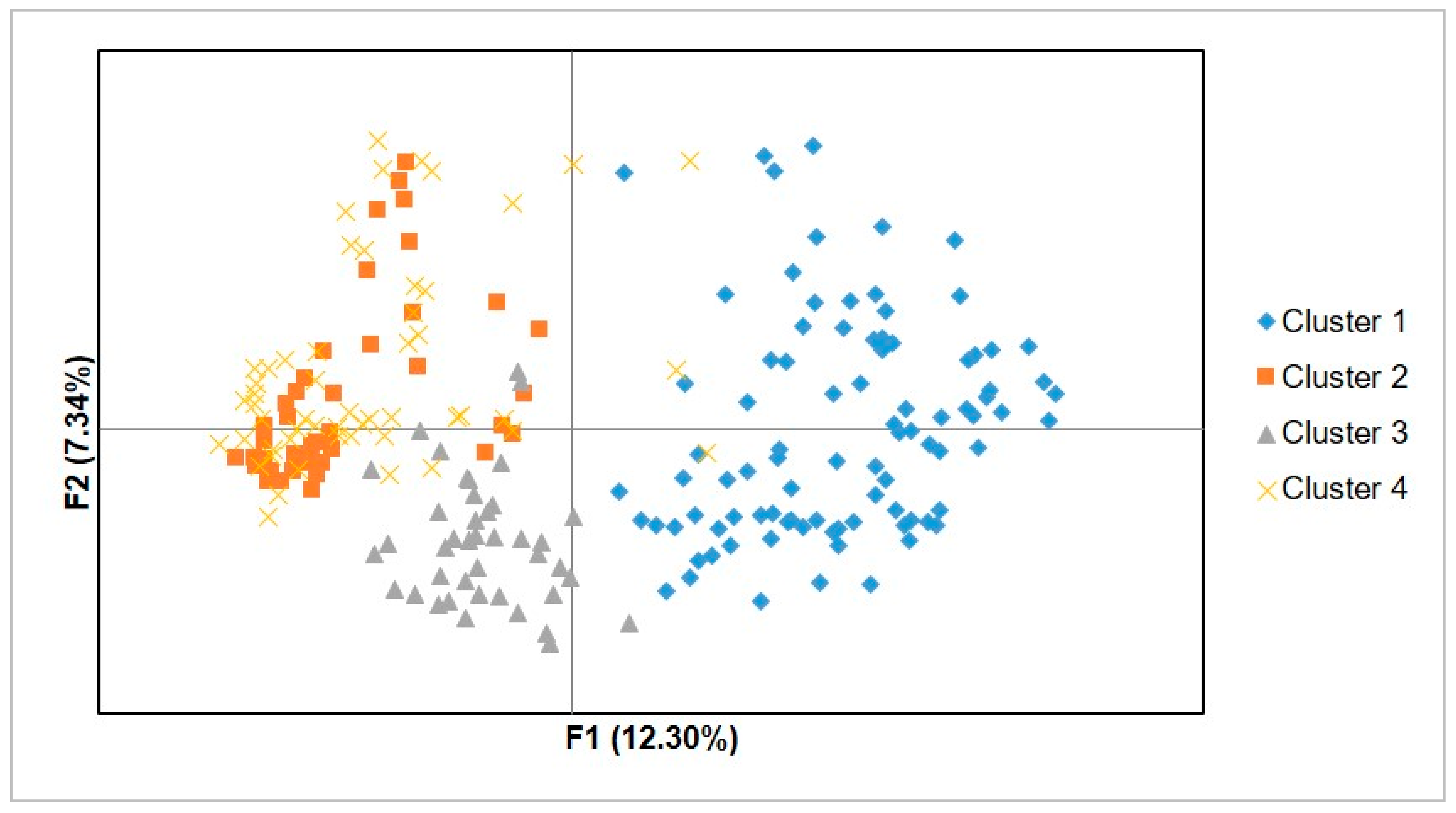
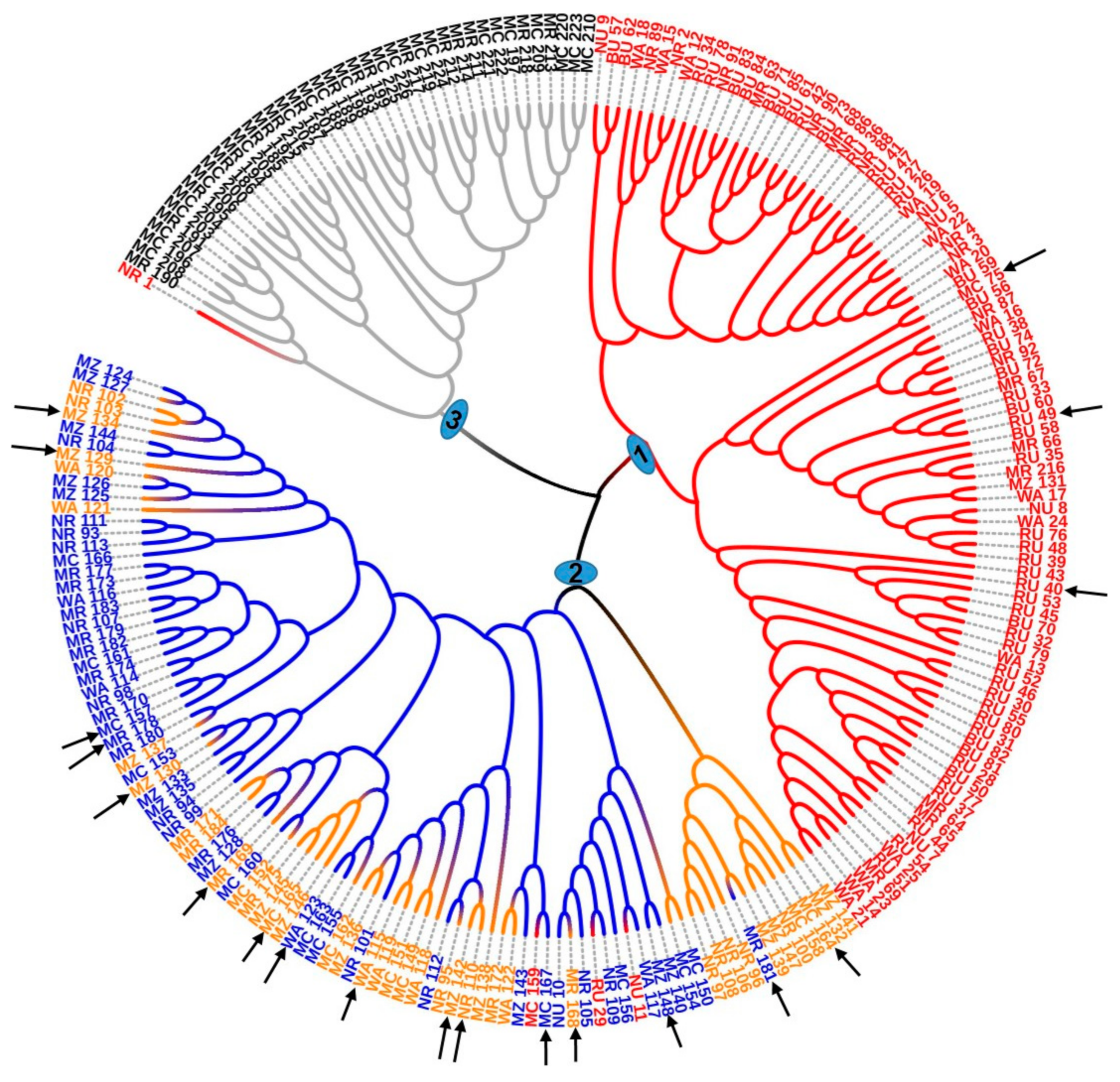
The PCoA (
Figure 6) and dendrogram (
Figure 7) obtained from microsatellite marker-based analyses resolved the studied trees into groups that were more or less similar to the four genetic clusters established by the DAPC analysis. Gross-German and Viruel [
37] observed that the model-based (STRUCTURE) genetic clustering, PCoA, and cluster analysis results were in line with the distribution of avocado into botanical races, i.e., the Mexican, West Indian, and interracial Guatemalan × Mexican. Similarly, Alcaraz and Hormaza [
15] observed that the UPGMA based dendrogram grouped 75 avocado accessions into three major groups that mainly corresponded to the botanical races. The four genetic clusters (groups) generated in the present study might represent the three avocado races and a hybrid group. This was also indicated by Juma et al. [
13], as Tanzanian avocado germplasm analyzed using different morphological traits was shown to contain material from all three races. Traits included were trunk surface and peel thickness. Smooth trunk surface was reported as an attribute of the Mexican and Guatemalan races, and the rough and very rough trunk surface is attributed to the West Indian race [
52]. Thin ripe peel (≤1 mm thick) is ascribed to the West Indian and Mexican races, and a thick ripe peel (2–3 mm thick) was ascribed to the Guatemalan race [
53]. Other traits were the doughy and buttery flesh textures ascribed to the Guatemalan and Mexican races and the watery flesh texture attributed to the West Indian group [
53]. However, in the present study, the examination of these characteristics showed that they appeared among individuals of all four clusters. More genetic studies need to be carried out on the Tanzanian avocado germplasm together with representative samples of the three avocado races to confirm the germplasm’s racial origin.
The AMOVA indicated that the overall genetic differentiation among the four avocado genetic clusters, F
ST, was 0.159 (
p < 0.0001). This implies a substantial amount of diversity harbored by the trees investigated and that the four genetic clusters were significantly distinct. The level of population differentiation (F
ST) observed in this study was higher than the values reported by Juma et al. [
25] for the same plant material when AMOVA was carried out on district-based populations (F
ST = 0.061,
p < 0.0001) and altitudinal groups (F
ST = 0.025,
p < 0.0001). Gross-German and Viruel [
37] and Boza et al. [
4] found an overall population differentiation of 0.25 and 0.193, respectively, which are comparatively higher than the value obtained in our study. In both studies, populations were based on the racial origin of avocado. Contrary to that, Cañas-Gutiérrez et al. [
49] noted an overall population differentiation of 0.054 among the municipality-based populations, which is about 69% less than the value observed in the present study. Considering the AMOVA-based findings from the mentioned studies, it can be concluded that the overall population differentiation among avocado groups is higher if the grouping is based on racial origin than if it is based on geographical origin.
Pairwise comparison of population differentiation (F
ST) and divergence (Nei’s genetic distance) revealed significant differentiation among all the clusters, with the lowest differentiation/genetic distance between clusters 2 and 4 (0.310;
Table 6). The comparatively low Nei’s genetic distance between clusters 2 and 4 explains why the two clusters were less resolved from one another on the DAPC and microsatellite-based PCoA and dendrogram.
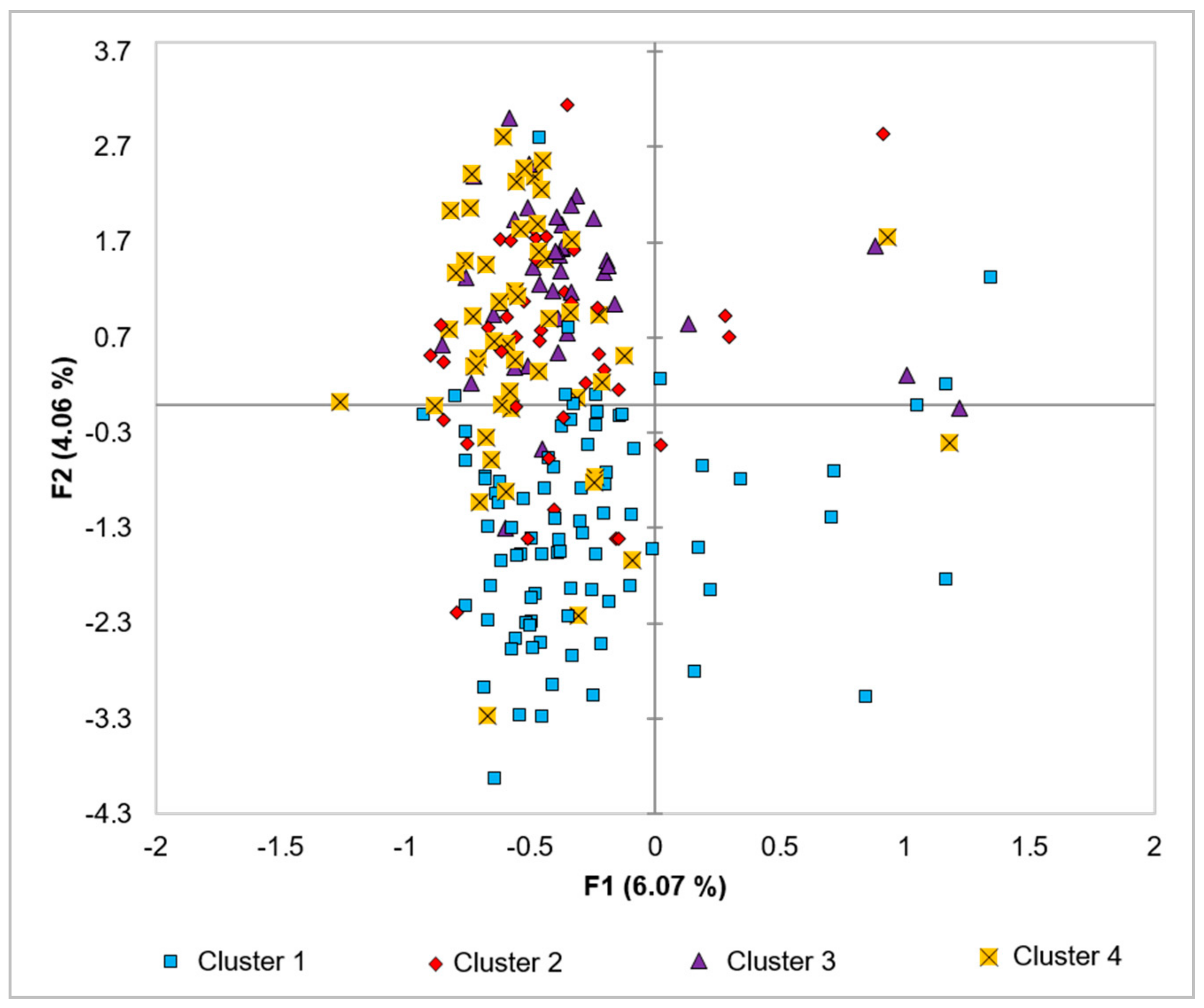
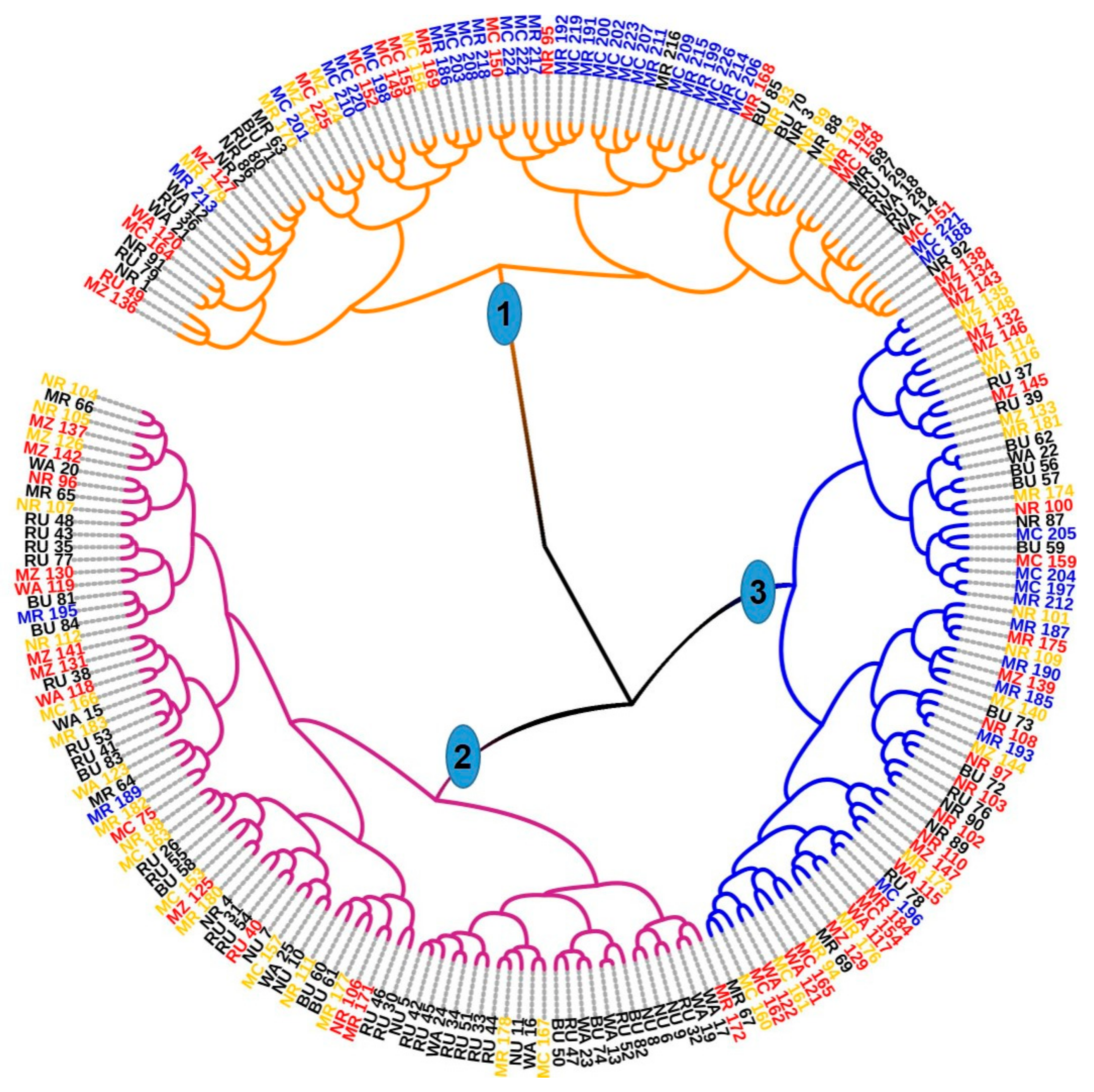
The morphology-based-PCAmix and dendrogram did not group the analyzed trees into their genetic clusters. The two analyses showed the intermingling of the individual trees from the four clusters. This finding suggests that the SSR loci investigated were not linked to the genes governing the investigated morphological traits. Another explanation is that the environment significantly influenced the phenotypes if linkage exists.
A weak positive correlation was revealed between the geographical distance of the sampling locations and the genetic distance (
r = 0.15,
p = 0.001) and between the geographical distance and the morphological dissimilarity matrix (
r = 0.08,
p = 0.001). Prohens et al. [
54] observed a lack of correlation between geographical distance and AFLP-based genetic distance (
r = 0.11,
p < 0.10) in their study of 28 Spanish eggplant accessions (
Solanum melongena L). Contrary to our study, they observed a comparatively higher correlation between the geographical and morphological distances (
r = 0.25,
p < 0.01). Sreekumar et al. [
55] reported a highly significant correlation between geographical distance and AFLP-based genetic distance (
r = 0.73,
p = 0.009), whereas no correlation could be found between geographical distance and morphological trait-based distance (
r = 0.44,
p = 0.07) in their study of 60 breadfruit samples (
Artocarpus altilis) in India. The weak correlation between geographical and genetic or morphological distances observed in the present study could be due to persistent movements and sharing of seeds between farmers of different areas [
13,
25,
33]. In the present study, a weak positive correlation was also noticed between the genetic and morphological distances (
r = 0.11,
p = 0.001). This suggests that there was no strong association between the studied morphological traits and the 10 SSR loci investigated. It also suggests that the morphological trait variation cannot fully display the pattern of genetic diversity in avocado. Working with 62 Ethiopian maize accessions, Beyene et al. [
28] noticed a moderate positive significant correlation between AFLP-based genetic and morphological distances (
r = 0.39,
p = 0.001), and also between SSR-based genetic and morphological distances (
r = 0.43,
p = 0.001). In a similar study on Vietnamese and Cambodian sesame accessions, Pham et al. [
56] reported a highly significant positive correlation (
r = 0.88,
p = 0.001) between agro-morphological and RAPD marker based distances between the accessions. Contrary to that, Roldan-Ruiz et al. [
57] observed an absence of correlation between AFLP-based genetic and morphological distances (
r = −0.06,
p < 0.375) and a weak correlation between the sequence tag sites (STS)-based genetic and morphological distances (
r = 0.18,
p < 0.12) in 16 ryegrass varieties. Similarly, Sreekumar et al. [
55] reported an absence of correlation between the AFLP-based genetic distance and the morphological distance (
r = 0.01,
p = 0.5) of breadfruit in India. Smith and Smith [
14] asserted that phenotypic variation sometimes does not follow genetic variation due to the influence of the environment on the phenotypic expression of the genotypes and potential multiple gene action on the traits.