Patterns of Epigenetic Diversity in Two Sympatric Fish Species: Genetic vs. Environmental Determinants
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Site Selection
2.1.1. Biological Models
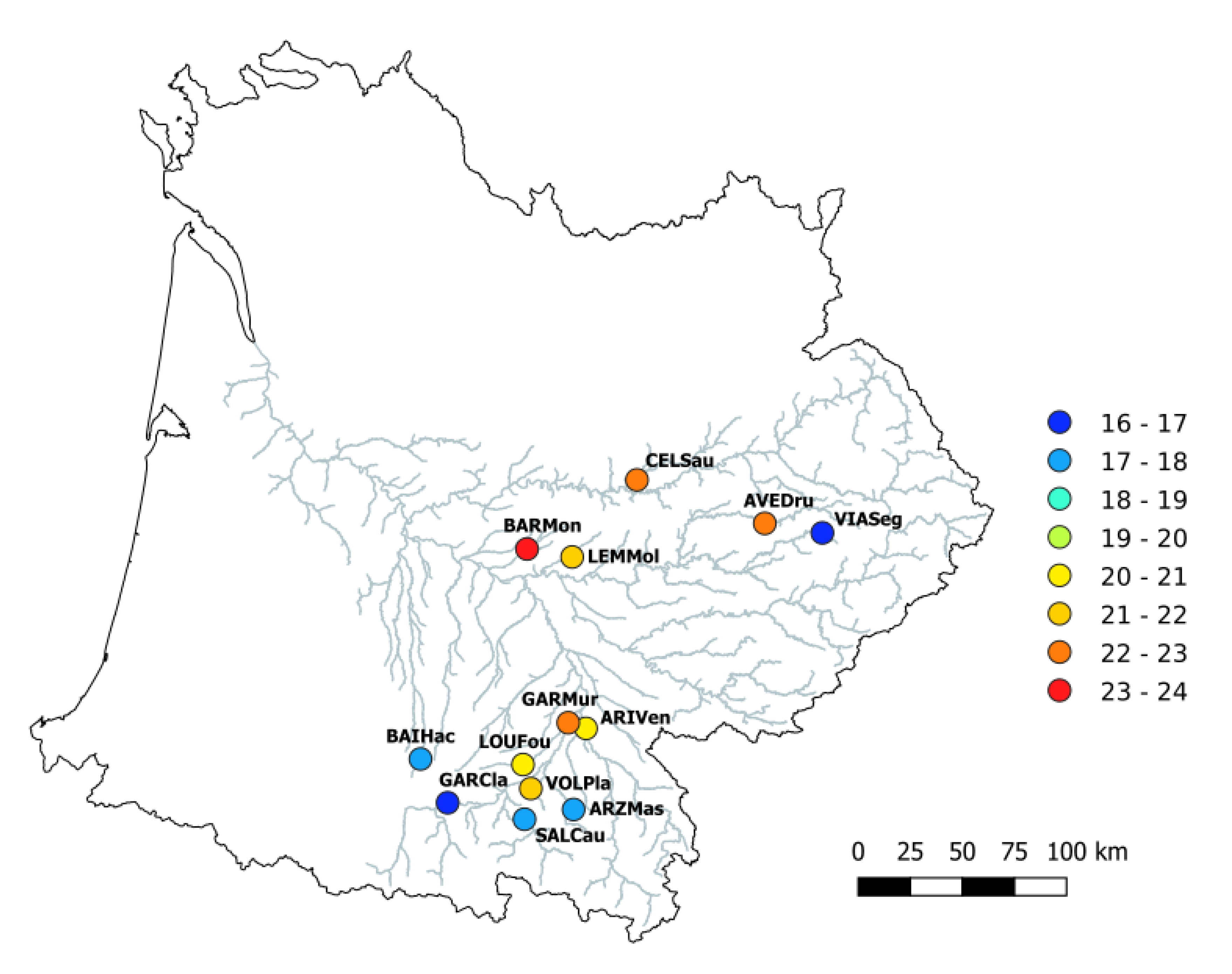
2.1.2. Study Area and Sampling
2.2. Environmental Data
2.3. Genetic and Epigenetic Data
2.3.1. Genetic Data
2.3.2. Epigenetic Data
2.3.3. MS-AFLP Protocol
2.4. Statistical Analyses
3. Results
3.1. Genetic and Epigenetic Discrimination
3.2. Simple Associations between Epigenetic, Genetic, and Environmental Distances
3.3. Multiple Associations between Epigenetic, Genetic, and Environmental Distances
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Site Code | River Name | Latitude | Longitude | River Flow (m3/s) | River Width (m) | River Slope (%) | Altitude (m) | Nitrates (mg·L−1) | Nitrites (mg·L−1) | Ortho-Phosphates (mg·L−1) | Oxygen (mg·L−1) | BOD (mg·L−1) | Water Conductivity (mS·cm−1) | pH | SM (mg·L−1) | Oxygen Saturation (%) | Temperature (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARIVen | Ariege | 573460.37 | 6261035.16 | 59 | 60.00 | 1 | 160 | 23.03 | 0.09 | 0.27 | 8.80 | 1.03 | 796.67 | 8.17 | 52.40 | 92.00 | 20.33 |
| ARZMas | Arize | 567474.38 | 6221932.39 | 4 | 6.35 | 2 | 284 | 3.70 | 0.02 | 0.03 | 9.37 | 0.67 | 343.00 | 8.17 | 4.63 | 100.33 | 17.23 |
| AVEDru | Aveyron | 659451.23 | 6359878.75 | 9 | 14.85 | 8 | 481 | 5.96 | 0.06 | 0.39 | 9.02 | 1.20 | 489.67 | 8.32 | 13.33 | 114.67 | 22.80 |
| BAIHac | Baise | 493842.07 | 6246291.53 | 0 | 6.00 | 6 | 279 | 2.67 | 0.02 | 0.04 | 9.20 | 0.60 | 157.67 | 8.00 | 39.33 | 99.00 | 17.67 |
| BARMon | Barguelonne | 545090.06 | 6347536.21 | 2 | 6.05 | 1 | 85 | 12.43 | 0.04 | 0.01 | 7.57 | 2.03 | 531.33 | 8.03 | 10.33 | 88.33 | 23.33 |
| CELSau | Cele | 597974.87 | 6380867.77 | 19 | 17.25 | 1 | 143 | 8.40 | 0.03 | 0.11 | 8.68 | 0.53 | 249.67 | 7.92 | 3.20 | 100.57 | 22.27 |
| GARCla | Garonne | 506893.70 | 6225179.17 | 53 | 67.00 | 4 | 388 | 1.73 | 0.01 | 0.02 | 10.33 | 1.17 | 165.33 | 8.07 | 18.37 | 109.67 | 16.63 |
| GARMur | Garonne | 564902.27 | 6263764.69 | 109 | 83.50 | 1 | 155 | 2.33 | 0.03 | 0.02 | 9.65 | 0.67 | 214.71 | 8.38 | 6.70 | 112.00 | 22.06 |
| LEMMol | Lemboulas | 566796.13 | 6343683.15 | 1 | 3.95 | 2 | 101 | 9.81 | 0.12 | 0.11 | 6.95 | 1.57 | 689.33 | 7.85 | 14.67 | 77.53 | 21.73 |
| LOUFou | Louge | 543137.54 | 6243604.74 | 1 | 9.25 | 1 | 249 | 4.90 | 0.03 | 0.05 | 8.83 | 0.80 | 264.50 | 8.08 | 34.70 | 99.50 | 20.08 |
| SALCau | Salat | 543868.20 | 6217247.36 | 34 | 32.50 | 5 | 355 | 1.33 | 0.01 | 0.03 | 10.30 | 1.13 | 165.83 | 8.33 | 3.23 | 107.50 | 17.48 |
| VIASeg | Viaur | 687141.24 | 6355312.09 | 1 | 5.70 | 6 | 750 | 10.32 | 0.02 | 0.03 | 9.11 | 0.93 | 140.00 | 7.74 | 22.67 | 100.33 | 16.40 |
| VOLPla | Volp | 546927.08 | 6232038.71 | 1 | 10.15 | 5 | 251 | 2.70 | 0.01 | 0.02 | 8.57 | 0.53 | 435.67 | 8.10 | 2.93 | 100.67 | 21.93 |
Appendix B
| Name | Sequence (5’-3’) | Dye | Number of loci |
|---|---|---|---|
| EcoRI adaptors | F: CTCGTAGACTGCGTACC R: AATTGGTACGCAGTCTAC | – | – |
| MspI/HpaII adaptors | F: GATCATGAGTCCTGCT R: CGAGCAGGACTCATGA | – | – |
| Preselective primers | F: GACTGCGTACCAATTC+A R: ATCATGAGTCCTGCTCGG+A | – | – |
| Selective primers G. occitaniae | F: GACTGCGTACCAATTC+AGA R: ATCATGAGTCCTGCTCGG+ATC | HEX | 66 |
| F: GACTGCGTACCAATTC+AAT R: ATCATGAGTCCTGCTCGG+AAT | AT550 | 69 | |
| F: GACTGCGTACCAATTC+AAT R: ATCATGAGTCCTGCTCGG+ATC | AT550 | 116 | |
| Selective primers P. phoxinus | F: GACTGCGTACCAATTC+AAC R: ATCATGAGTCCTGCTCGG+AAA | FAM | 71 |
| F: GACTGCGTACCAATTC+AAC R: ATCATGAGTCCTGCTCGG+AAT | FAM | 105 | |
| F: GACTGCGTACCAATTC+AGA R: ATCATGAGTCCTGCTCGG+ACT | HEX | 98 |
Appendix C.—PCR Conditions
References
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Holderegger, R.; Kamm, U.; Gugerli, F. Adaptive vs. neutral genetic diversity: Implications for landscape genetics. Landsc. Ecol. 2006, 21, 797–807. [Google Scholar] [CrossRef]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking epigenetics and biological conservation: Towards a conservation epigenetics perspective. Funct. Ecol. 2020, 34, 414–427. [Google Scholar] [CrossRef]
- Rey, O.; Danchin, E.; Mirouze, M.; Loot, C.; Blanchet, S. Adaptation to Global Change: A Transposable Element–Epigenetics Perspective. Trends Ecol. Evol. 2016, 31, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Alonso, C.; Becker, C.; Bossdorf, O.; Bucher, E.; Colomé-Tatché, M.; Durka, W.; Engelhardt, J.; Gaspar, B.; Gogol-Döring, A.; et al. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett. 2017, 20. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.; Pocheville, A.; Rey, O.; Pujol, B.; Blanchet, S. Epigenetically facilitated mutational assimilation: Epigenetics as a hub within the inclusive evolutionary synthesis: Epigenetics as a hub for genetic assimilation. Biol. Rev. 2019, 94, 259–282. [Google Scholar] [CrossRef]
- Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y.; Brunet, A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011, 479, 365–371. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Segal, D.L.; Hoyle, G.L.; Schrey, A.W.; Verhoeven, K.J.F.; Richards, C.L. Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecol. Evol. 2015, 5, 634–647. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; vonHoldt, B.M.; Sork, V.L. Epigenetics in ecology and evolution: What we know and what we need to know. Mol. Ecol. 2016, 25, 1631–1638. [Google Scholar] [CrossRef]
- Hu, J.; Barrett, R.D.H. Epigenetics in natural animal populations. J. Evol. Biol. 2017, 30, 1612–1632. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Larsen, G.; Manikkam, M.; Guerrero-Bosagna, C.; Savenkova, M.I.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Ovarian Disease. PLoS ONE 2012, 7, e36129. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, S.; Fernald, R.D.; Storey, K.B.; Szyf, M. The Dynamic Nature of DNA Methylation: A Role in Response to Social and Seasonal Variation. Integr. Comp. Biol. 2014, 54, 68–76. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lillycrop, K.A.; Burdge, G.C. Maternal diet as a modifier of offspring epigenetics. J. Dev. Orig. Health Dis. 2015, 6, 88–95. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef]
- Jablonka, E.; Raz, G. Transgenerational Epigenetic Inheritance: Prevalence, Mechanisms, and Implications for the Study of Heredity and Evolution. Q. Rev. Biol. 2009, 84, 131–176. [Google Scholar] [CrossRef]
- Bollati, V.; Baccarelli, A. Environmental epigenetics. Heredity 2010, 105, 105–112. [Google Scholar] [CrossRef]
- Danchin, É.; Charmantier, A.; Champagne, F.A.; Mesoudi, A.; Pujol, B.; Blanchet, S. Beyond DNA: Integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 2011, 12, 475–486. [Google Scholar] [CrossRef]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.-S.; Osborne, E.J.; Paolo Casale, F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjálmsson, B.; et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 2015, 4, e05255. [Google Scholar] [CrossRef] [PubMed]
- Taudt, A.; Colomé-Tatché, M.; Johannes, F. Genetic sources of population epigenomic variation. Nat. Rev. Genet. 2016, 17, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Lee, Y.-S.; Sung, S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013, 21, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Harvey, Z.H.; Chen, Y.; Jarosz, D.F. Protein-Based Inheritance: Epigenetics beyond the Chromosome. Mol. Cell 2018, 69, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Baruah, K.; Vanrompay, D.; Bossier, P. Can epigenetics translate environmental cues into phenotypes? Sci. Total Environ. 2019, 647, 1281–1293. [Google Scholar] [CrossRef]
- Herrera, C.M.; Medrano, M.; Bazaga, P. Comparative spatial genetics and epigenetics of plant populations: Heuristic value and a proof of concept. Mol. Ecol. 2016, 25, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Trucchi, E.; Mazzarella, A.B.; Gilfillan, G.D.; Lorenzo, M.T.; Schönswetter, P.; Paun, O. BsRADseq: Screening DNA methylation in natural populations of non-model species. Mol. Ecol. 2016, 25, 1697–1713. [Google Scholar] [CrossRef]
- Richards, C.L.; Bossdorf, O.; Verhoeven, K.J.F. Understanding natural epigenetic variation: Commentary. New Phytol. 2010, 187, 562–564. [Google Scholar] [CrossRef]
- Cubas, P.; Vincent, C.; Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999, 401, 157–161. [Google Scholar] [CrossRef]
- Issa, J.-P. Aging and epigenetic drift: A vicious cycle. J. Clin. Investig. 2014, 124, 24–29. [Google Scholar] [CrossRef]
- Shan, X.H.; Li, Y.D.; Liu, X.M.; Wu, Y.; Zhang, M.Z.; Guo, W.L.; Liu, B.; Yuan, Y.P. Comparative analyses of genetic/epigenetic diversities and structures in a wild barley species (Hordeum brevisubulatum) using MSAP, SSAP and AFLP. Genet. Mol. Res. 2012, 11, 2749–2759. [Google Scholar] [CrossRef] [PubMed]
- Lele, L.; Ning, D.; Cuiping, P.; Xiao, G.; Weihua, G. Genetic and epigenetic variations associated with adaptation to heterogeneous habitat conditions in a deciduous shrub. Ecol. Evol. 2018, 8, 2594–2606. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, K.; Jiang, T.; Ho, J.P.; Liu, B.; Feng, J. Natural epigenetic variation in the female great roundleaf bat (Hipposideros armiger) populations. Mol. Genet. Genom. 2012, 287, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, E.L.; Schrey, A.; Andrew, S.C.; Ragsdale, A.; Griffith, S.C. Epigenetic and genetic variation among three separate introductions of the house sparrow (Passer domesticus) into Australia. R. Soc. Open Sci. 2018, 5, 172185. [Google Scholar] [CrossRef] [PubMed]
- Foust, C.M.; Preite, V.; Schrey, A.W.; Alvarez, M.; Robertson, M.H.; Verhoeven, K.J.F.; Richards, C.L. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol. Ecol. 2016, 25, 1639–1652. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Li, S.; Lin, Y.; Xiong, W.; Huang, X.; Zhan, A. Methylation divergence of invasive Ciona ascidians: Significant population structure and local environmental influence. Ecol. Evol. 2018, 8, 10272–10287. [Google Scholar] [CrossRef]
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2007, 11, 106–115. [Google Scholar] [CrossRef]
- Fourtune, L.; Paz-Vinas, I.; Loot, G.; Prunier, J.G.; Blanchet, S. Lessons from the fish: A multi-species analysis reveals common processes underlying similar species-genetic diversity correlations. Freshw. Biol. 2016, 61, 1830–1845. [Google Scholar] [CrossRef]
- Fourtune, L.; Prunier, J.G.; Mathieu-Begne, E.; Canto, N.; Veyssiere, C.; Loot, G.; Blanchet, S. Intraspecific genetic and phenotypic diversity: Parallel processes and correlated patterns? BioRxiv 2018, arXiv:288357. [Google Scholar] [CrossRef]
- Buisson, L.; Blanc, L.; Grenouillet, G. Modelling stream fish species distribution in a river network: The relative effects of temperature versus physical factors. Ecol. Freshw. Fish 2008, 17, 244–257. [Google Scholar] [CrossRef]
- Crispo, E.; Chapman, L.J. Population genetic structure across dissolved oxygen regimes in an African cichlid fish: POPULATION STRUCTURE ACROSS OXYGEN REGIMES. Mol. Ecol. 2008, 17, 2134–2148. [Google Scholar] [CrossRef] [PubMed]
- Gama-Sosa, M.A.; Midgett, R.M.; Slagel, V.A.; Githens, S.; Kuo, K.C.; Gehrke, C.W.; Ehrlich, M. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta Gene Struct. Expr. 1983, 740, 212–219. [Google Scholar] [CrossRef]
- Sun, Y.; Hou, R.; Fu, X.; Sun, C.; Wang, S.; Wang, C.; Li, N.; Zhang, L.; Bao, Z. Genome-Wide Analysis of DNA Methylation in Five Tissues of Zhikong Scallop, Chlamys farreri. PLoS ONE 2014, 9, e86232. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, Y.; Liu, W.; Lu, X.; Li, C. Genome-wide analysis of DNA methylation in five tissues of sika deer (Cervus nippon). Gene 2018, 645, 48–54. [Google Scholar] [CrossRef]
- Blanchet, S.; Páez, D.J.; Bernatchez, L.; Dodson, J.J. An integrated comparison of captive-bred and wild Atlantic salmon (Salmo salar): Implications for supportive breeding programs. Biol. Conserv. 2008, 141, 1989–1999. [Google Scholar] [CrossRef]
- Paez, D.J.; Hedger, R.; Bernatchez, L.; Dodson, J.J. The morphological plastic response to water current velocity varies with age and sexual state in juvenile Atlantic salmon, Salmo salar. Freshw. Biol. 2008, 53, 1544–1554. [Google Scholar] [CrossRef]
- Pella, H.; Lejot, J.; Lamouroux, N.; Snelder, T. Le réseau hydrographique théorique (RHT) français et ses attributs environnementaux. Géomorphologie Relief Process. Environ. 2012, 18, 317–336. [Google Scholar] [CrossRef]
- Prunier, J.G.; Dubut, V.; Loot, G.; Tudesque, L.; Blanchet, S. The relative contribution of river network structure and anthropogenic stressors to spatial patterns of genetic diversity in two freshwater fishes: A multiple-stressors approach. Freshw. Biol. 2018, 63, 6–21. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Soft. 2007, 22. [Google Scholar] [CrossRef]
- Aljanabi, S. Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Schlötterer, C.; Tobler, R.; Kofler, R.; Nolte, V. Sequencing pools of individuals—mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 2014, 15, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Prunier, J.G.; Chevalier, M.; Raffard, A.; Loot, G.; Poulet, N.; Blanchet, S. Contemporary loss of genetic diversity in wild fish populations reduces biomass stability over time. BioRxiv 2019, arXiv:884734. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Res. 2013, 13, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.Z.; Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. MGG 1999, 261, 439–446. [Google Scholar] [CrossRef]
- Smith, T.A.; Martin, M.D.; Nguyen, M.; Mendelson, T.C. Epigenetic divergence as a potential first step in darter speciation. Mol. Ecol. 2016, 25, 1883–1894. [Google Scholar] [CrossRef]
- Caballero, A.; Quesada, H.; Rolán-Alvarez, E. Impact of Amplified Fragment Length Polymorphism Size Homoplasy on the Estimation of Population Genetic Diversity and the Detection of Selective Loci. Genetics 2008, 179, 539–554. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures: Invited technical review. Mol. Ecol. Res. 2011, 11, 5–18. [Google Scholar] [CrossRef]
- Prunier, J.G.; Poesy, C.; Dubut, V.; Veyssière, C.; Loot, G.; Poulet, N.; Blanchet, S. Quantifying the individual impact of artificial barriers in freshwaters: A standardized and absolute genetic index of fragmentation. Evol. Appl. 2020, 13, 2566–2581. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Gutiérrez-Velázquez, M.V.; Almaraz-Abarca, N.; Herrera-Arrieta, Y.; Ávila-Reyes, J.A.; González-Valdez, L.S.; Torres-Ricario, R.; Uribe-Soto, J.N.; Monreal-García, H.M. Comparison of the phenolic contents and epigenetic and genetic variability of wild and cultivated watercress (Rorippa nasturtium var. aquaticum L.). Electron. J. Biotechnol. 2018, 34, 9–16. [Google Scholar] [CrossRef]
- Avramidou, E.V.; Ganopoulos, I.V.; Doulis, A.G.; Tsaftaris, A.S.; Aravanopoulos, F.A. Beyond population genetics: Natural epigenetic variation in wild cherry (Prunus avium). Tree Genet. Genomes 2015, 11, 95. [Google Scholar] [CrossRef]
- Herrera, C.M.; Medrano, M.; Bazaga, P. Comparative epigenetic and genetic spatial structure of the perennial herb Helleborus foetidus: Isolation by environment, isolation by distance, and functional trait divergence. Am. J. Bot. 2017, 104, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Choi, J.-Y.; Lim, M.-J.; Lee, S.-I.; Choi, H.-J.; Kim, N.-S. Genetic and epigenetic diversity among dent, waxy, and sweet corns. Genes Genom. 2015, 37, 865–874. [Google Scholar] [CrossRef]
- Guo, W.; Hussain, N.; Wu, R.; Liu, B. High hypomethylation and epigenetic variation in fragmented populations of wild barley (Hordeum brevisubulatum). Pak. J. Bot. 2018, 50, 1379–1386. [Google Scholar]
- Duchemin, W.; Anselmetti, Y.; Patterson, M.; Ponty, Y.; Bérard, S.; Chauve, C.; Scornavacca, C.; Daubin, V.; Tannier, E. DeCoSTAR: Reconstructing the Ancestral Organization of Genes or Genomes Using Reconciled Phylogenies. Genome Biol. Evol. 2017, 9, 1312–1319. [Google Scholar] [CrossRef]
- DeFaveri, J.; Viitaniemi, H.; Leder, E.; Merilä, J. Characterizing genic and nongenic molecular markers: Comparison of microsatellites and SNPs. Mol. Ecol. Resour. 2013, 13, 377–392. [Google Scholar] [CrossRef]
- Nosil, P.; Funk, D.J.; Ortiz-Barrientos, D. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 2009, 18, 375–402. [Google Scholar] [CrossRef]
- Fourtune, L.; Prunier, J.G.; Paz-Vinas, I.; Loot, G.; Veyssière, C.; Blanchet, S. Inferring Causalities in Landscape Genetics: An Extension of Wright’s Causal Modeling to Distance Matrices. Am. Nat. 2018, 191, 491–508. [Google Scholar] [CrossRef]
- Smith, G.; Ritchie, M.G. How might epigenetics contribute to ecological speciation? Curr. Zool. 2013, 59, 686–696. [Google Scholar] [CrossRef]
- McMahon, B.J.; Teeling, E.C.; Ho, J. How and why should we implement genomics into conservation? Evol. Appl. 2014, 7, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Shafer, A.B.A.; Wolf, J.B.W.; Alves, P.C.; Bergström, L.; Bruford, M.W.; Brännström, I.; Colling, G.; Dalén, L.; De Meester, L.; Ekblom, R.; et al. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 2015, 30, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Eizaguirre, C.; Baltazar-Soares, M. Evolutionary conservation-evaluating the adaptive potential of species. Evol. Appl. 2014, 7, 963–967. [Google Scholar] [CrossRef]
- Massicotte, R.; Angers, B. General-Purpose Genotype or How Epigenetics Extend the Flexibility of a Genotype. Genet. Res. Int. 2012, 2012, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, Q.; Yu, H.; Kong, L.-F. Genetic and epigenetic variation in mass selection populations of Pacific oyster Crassostrea gigas. Genes Genom. 2013, 35, 641–647. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Kong, L.; Yu, H. Epigenetic variation of wild populations of the Pacific oyster Crassostrea gigas determined by methylation-sensitive amplified polymorphism analysis. Fish. Sci. 2018, 84, 61–70. [Google Scholar] [CrossRef]
- Liebl, A.L.; Schrey, A.W.; Richards, C.L.; Martin, L.B. Patterns of DNA Methylation Throughout a Range Expansion of an Introduced Songbird. Integr. Comp. Biol. 2013, 53, 351–358. [Google Scholar] [CrossRef]
- Schrey, A.W.; Alvarez, M.; Foust, C.M.; Kilvitis, H.J.; Lee, J.D.; Liebl, A.L.; Martin, L.B.; Richards, C.L.; Robertson, M. Ecological Epigenetics: Beyond MS-AFLP. Integr. Comp. Biol. 2013, 53, 340–350. [Google Scholar] [CrossRef]
- Wenzel, M.A.; Piertney, S.B. Fine-scale population epigenetic structure in relation to gastrointestinal parasite load in red grouse (Lagopus lagopus scotica). Mol. Ecol. 2014, 23, 4256–4273. [Google Scholar] [CrossRef]
- Wogan, G.O.U.; Yuan, M.L.; Mahler, D.L.; Wang, I.J. Genome-wide epigenetic isolation by environment in a widespread Anolis lizard. Mol. Ecol. 2020, 29, 40–55. [Google Scholar] [CrossRef]
- Johnson, K.M.; Kelly, M.W. Population epigenetic divergence exceeds genetic divergence in the Eastern oyster Crassostrea virginica in the Northern Gulf of Mexico. Evol. Appl. 2020, 13, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Baldanzi, S.; Watson, R.; McQuaid, C.D.; Gouws, G.; Porri, F. Epigenetic variation among natural populations of the South African sandhopper Talorchestia capensis. Evol. Ecol. 2017, 31, 77–91. [Google Scholar] [CrossRef]
- Watson, R.G.A.; Baldanzi, S.; Pérez-Figueroa, A.; Gouws, G.; Porri, F. Morphological and epigenetic variation in mussels from contrasting environments. Mar. Biol. 2018, 165, 50. [Google Scholar] [CrossRef]
- Hawes, N.A.; Amadoru, A.; Tremblay, L.A.; Pochon, X.; Dunphy, B.; Fidler, A.E.; Smith, K.F. Epigenetic patterns associated with an ascidian invasion: A comparison of closely related clades in their native and introduced ranges. Sci. Rep. 2019, 9, 14275. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Guo, W.; Huang, Y. Genetic and epigenetic regulation of phenotypic variation in invasive plants—linking research trends towards a unified framework. NB 2019, 49, 77–103. [Google Scholar] [CrossRef]
- Robertson, M.; Richards, C. Opportunities and challenges of next-generation sequencing applications in ecological epigenetics. Mol. Ecol. 2015, 24, 3799–3801. [Google Scholar] [CrossRef] [PubMed]
- Schield, D.R.; Walsh, M.R.; Card, D.C.; Andrew, A.L.; Adams, R.H.; Castoe, T.A. EpiRADseq: Scalable analysis of genomewide patterns of methylation using next-generation sequencing. Methods Ecol. Evol. 2016, 7, 60–69. [Google Scholar] [CrossRef]
- van Gurp, T.P.; Wagemaker, N.C.A.M.; Wouters, B.; Vergeer, P.; Ouborg, J.N.J.; Verhoeven, K.J.F. epiGBS: Reference-free reduced representation bisulfite sequencing. Nat. Methods 2016, 13, 322–324. [Google Scholar] [CrossRef]
- Paun, O.; Verhoeven, K.J.F.; Richards, C.L. Opportunities and Limitations of Reduced Representation Bisulfite Sequencing in Plant Ecological Epigenomics. New Phytol. 2019, 221, 738–742. [Google Scholar] [CrossRef]
- Hirsch, S.; Baumberger, R.; Grossniklaus, U. Epigenetic Variation, Inheritance, and Selection in Plant Populations. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 97–104. [Google Scholar] [CrossRef]


| – | Component 1 | Component 2 | Component 3 |
|---|---|---|---|
| Part of total variance (%) | 37.03 | 20.87 | 14.06 |
| River flow | −0.273 | 0.888 | −0.056 |
| River width | −0.295 | 0.864 | 0.037 |
| Slope | −0.434 | −0.407 | 0.614 |
| Altitude | −0.538 | −0.442 | 0.596 |
| Conductivity | 0.878 | 0.258 | 0.212 |
| BOD | 0.555 | −0.020 | 0.045 |
| MS | 0.262 | 0.060 | 0.529 |
| Nitrate | 0.748 | 0.166 | 0.344 |
| Nitrite | 0.855 | 0.199 | 0.266 |
| Orthophosphate | 0.354 | 0.303 | 0.757 |
| Oxygen | −0.903 | 0.267 | 0.191 |
| pH | −0.341 | 0.719 | 0.075 |
| Oxygen saturation | −0.848 | 0.304 | 0.238 |
| Temperature | 0.577 | 0.339 | −0.208 |
| Global characteristic | Oligotrophic water–Eutrophic water | Small river–Large river | Low altitude and nutrient–High altitude and nutrient |
| Degrees of Freedom | Sum of Squares | Variance Components | Variation (%) | Phi-st | p-value | |
|---|---|---|---|---|---|---|
| G. occitaniae | ||||||
| Genetic markers | 12 | 217.182 | 0.481 | 10.34 | 0.103 | <0.001 |
| Epigenetic markers | 12 | 5726.943 | 17.210 | 20.15 | 0.202 | <0.001 |
| P. phoxinus | ||||||
| Genetic markers | 12 | 484.297 | 1.160 | 16.75 | 0.168 | <0.001 |
| Epigenetic markers | 12 | 6369.97 | 19.478 | 19.59 | 0.196 | <0.001 |
| Microsatellites | SNP | MS-AFLP | |
|---|---|---|---|
| G. occitaniae | |||
| Microsatellites | – | 0.616 | 0.363 |
| SNP | 0.009 | – | 0.531 |
| MS-AFLP | 0.011 | <0.001 | – |
| P. phoxinus | |||
| Microsatellites | – | 0.894 | 0.287 |
| SNP | <0.001 | – | 0.294 |
| MS-AFLP | 0.089 | 0.121 | – |
| Component 1 | Component 2 | Component 3 | Riparian Distance | |||||
|---|---|---|---|---|---|---|---|---|
| r | p-value | r | p-value | r | p-value | r | p-value | |
| G. occitaniae | ||||||||
| MS-AFLP | 0.395 | <0.01 | 0.033 | 0.397 | −0.027 | 0.570 | −0.060 | 0.670 |
| SNP | 0.562 | <0.05 | −0.142 | 0.670 | −0.104 | 0.661 | −0.117 | 0.666 |
| Microsatellites | 0.161 | 0.195 | −0.066 | 0.538 | −0.162 | 0.765 | 0.095 | 0.333 |
| P. phoxinus | ||||||||
| MS-AFLP | −0.154 | 0.844 | −0.114 | 0.669 | −0.143 | 0.797 | −0.080 | 0.639 |
| SNP | −0.217 | 0.872 | −0.072 | 0.539 | −0.035 | 0.479 | 0.107 | 0.328 |
| Microsatellites | −0.11 | 0.663 | −0.180 | 0.747 | −0.001 | 0.436 | 0.186 | 0.228 |
| G. occitaniae | P. phoxinus | |||
|---|---|---|---|---|
| Coefficients | p-value | Coefficients | p-value | |
| Intercept | 0.197 | 0.945 | 0.312 | 0.362 |
| Component 1 | 0.007 | 0.317 | −0.008 | 0.370 |
| Component 2 | 0.006 | 0.473 | −0.006 | 0.672 |
| Component 3 | <0.001 | 0.925 | −0.009 | 0.559 |
| Riparian Dist. | <0.001 | 0.817 | −0.001 | 0.534 |
| Genetics (SNP) | 0.290 | 0.003 | 0.307 | 0.220 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fargeot, L.; Loot, G.; Prunier, J.G.; Rey, O.; Veyssière, C.; Blanchet, S. Patterns of Epigenetic Diversity in Two Sympatric Fish Species: Genetic vs. Environmental Determinants. Genes 2021, 12, 107. https://doi.org/10.3390/genes12010107
Fargeot L, Loot G, Prunier JG, Rey O, Veyssière C, Blanchet S. Patterns of Epigenetic Diversity in Two Sympatric Fish Species: Genetic vs. Environmental Determinants. Genes. 2021; 12(1):107. https://doi.org/10.3390/genes12010107
Chicago/Turabian StyleFargeot, Laura, Géraldine Loot, Jérôme G. Prunier, Olivier Rey, Charlotte Veyssière, and Simon Blanchet. 2021. "Patterns of Epigenetic Diversity in Two Sympatric Fish Species: Genetic vs. Environmental Determinants" Genes 12, no. 1: 107. https://doi.org/10.3390/genes12010107
APA StyleFargeot, L., Loot, G., Prunier, J. G., Rey, O., Veyssière, C., & Blanchet, S. (2021). Patterns of Epigenetic Diversity in Two Sympatric Fish Species: Genetic vs. Environmental Determinants. Genes, 12(1), 107. https://doi.org/10.3390/genes12010107








