Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Vector Construction
2.2. Plant Transformation
2.3. Genotyping
2.4. Estimation of Transgene Copy Numbers by Quantitative PCR
2.5. Seed Germination Assay for Transgenic Progeny
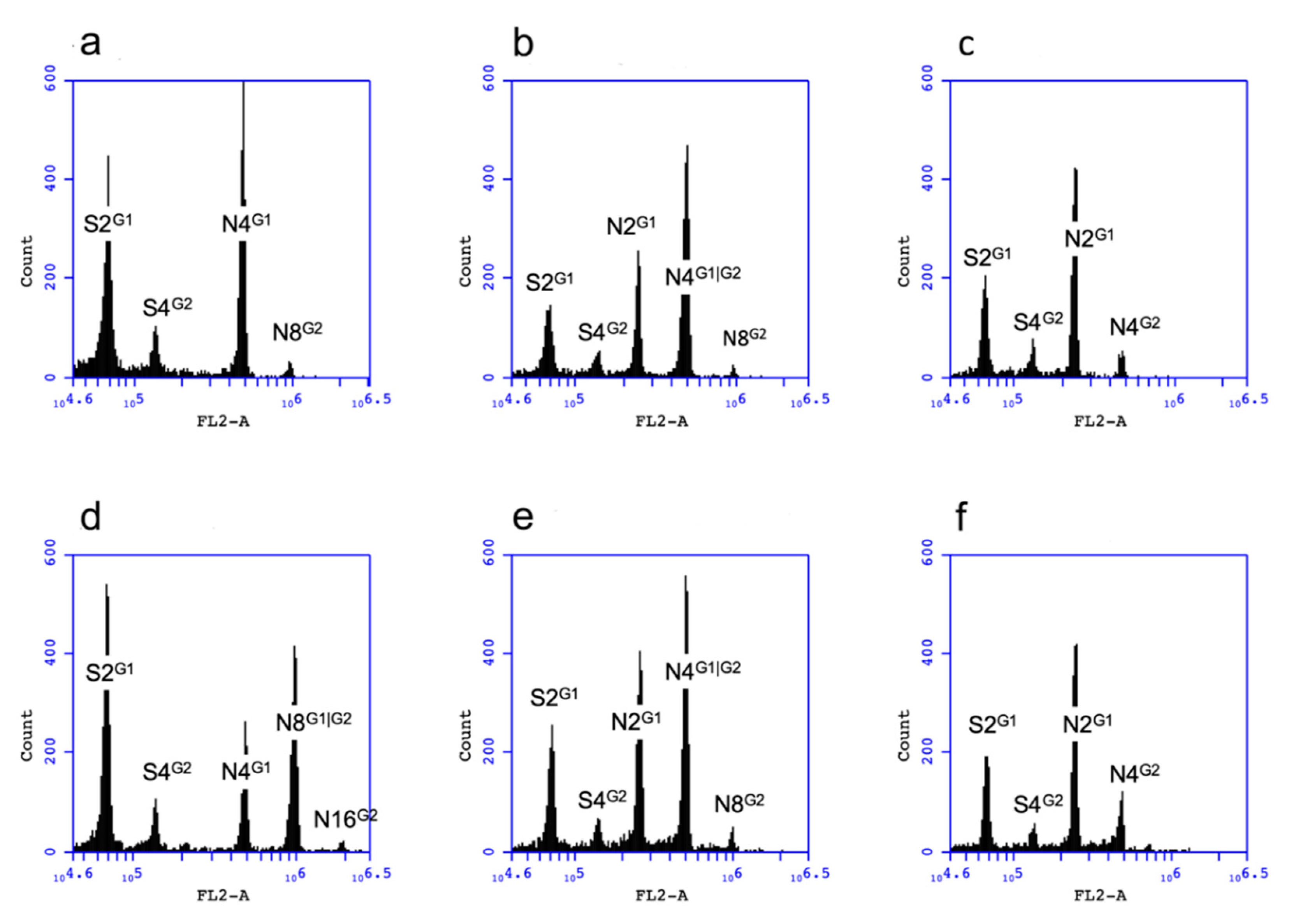
2.6. Flow Cytometry
2.7. Transcription Analysis of the PsASGR-BBML Transgene in Tobacco Ovules
2.8. Sequence Analysis of the PsASGR-BBML Transgene from Line RKD2gBB_6.1
2.9. Tobacco Crossing
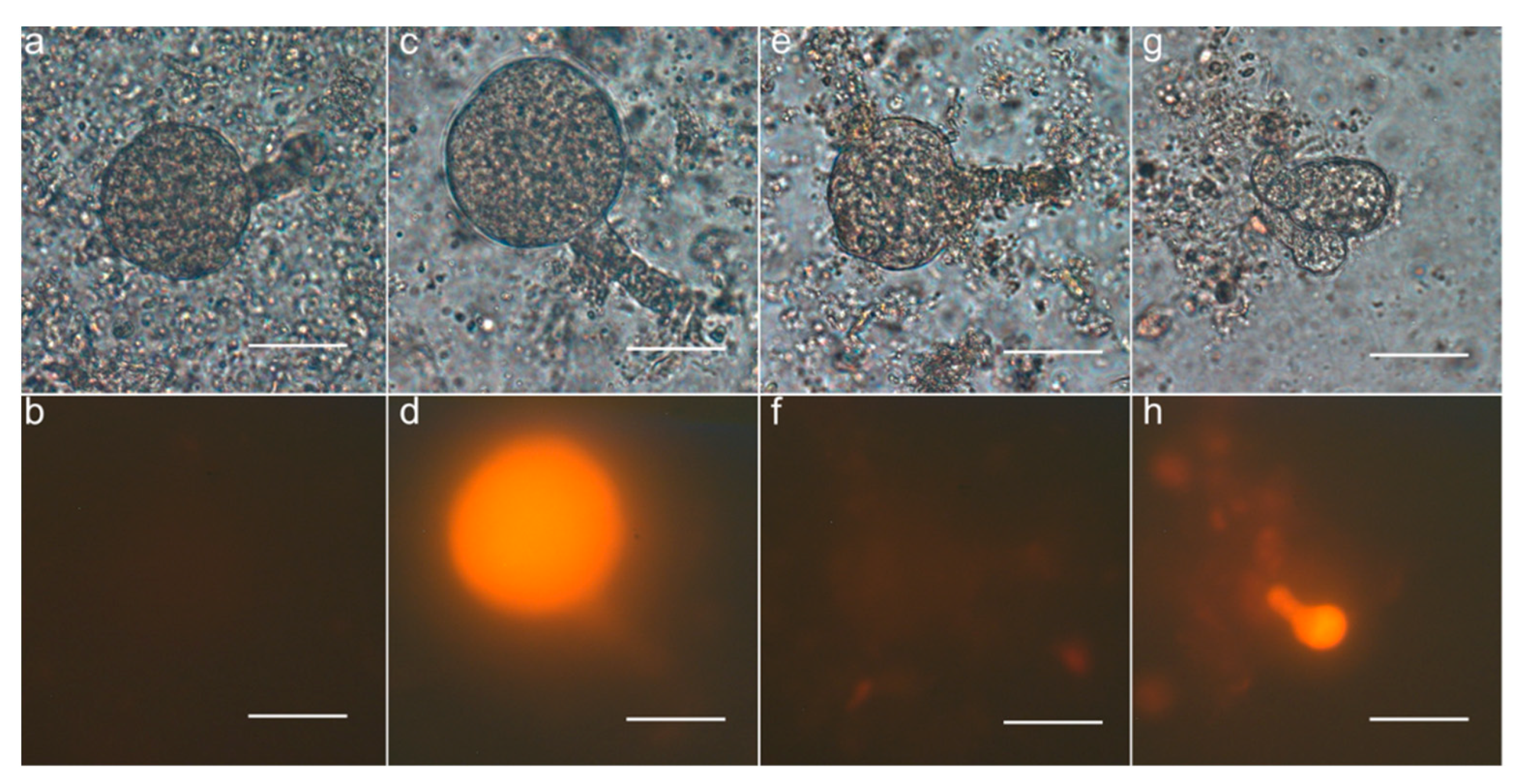
2.10. Embryo Isolation and Observation
3. Results
3.1. Recovery of Transgenic Tobacco
3.2. Haploid Production of T0 Transgenic Tobacco
3.3. Haploid Production of the Progeny Derived from Line RKD2gBB_6.1
3.4. Transcription of the PsASGR-BBML Transgene in the 4x T1 of Line RKD2gBB_6.1
3.5. Observation of Parthenogenetic Embryos
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johri, B.M. (Ed.) Embryology of Angiosperms, 1st ed.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA; Tokyo, Japan, 1984. [Google Scholar]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms, 1st ed.; Johri, B.M., Ed.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 475–518. [Google Scholar]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [PubMed]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef] [PubMed]
- Hand, M.L.; Koltunow, A.M. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Vijverberg, K.; Ozias-Akins, P.; Schranz, M.E. Identifying and engineering genes for parthenogenesis in plants. Front. Plant Sci. 2019, 10, 128–144. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Roche, D.; Hanna, W.W. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 1998, 95, 5127–5132. [Google Scholar] [CrossRef]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef]
- Steffen, J.G.; Kang, I.H.; Macfarlane, J.; Drews, G.N. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007, 51, 281–292. [Google Scholar] [CrossRef]
- Conner, J.A.; Podio, M.; Ozias-Akins, P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 2017, 30, 41–52. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef]
- de Nettancourt, D.D.; Stokes, G.W. Haploidy in tobacco. J. Hered. 1960, 51, 102–104. [Google Scholar] [CrossRef]
- Burk, L.G. Haploids in genetically marked progenies of tobacco. J. Hered. 1962, 53, 222–226. [Google Scholar] [CrossRef]
- Lewis, R.S.; Rose, C. Identification of tobacco haploids on the basis of transgenic overexpression of PAP1 from Arabidopsis thaliana. Crop. Sci. 2011, 51, 1491–1497. [Google Scholar] [CrossRef]
- Huo, H. Genetic Analysis of the Apospory-Specific Genomic Region. (ASGR) in Pennisetum squamulatum: From Mapping to Candidate Gene. Ph.D. Thesis, University of Georgia, Athens, GA, USA, August 2008; pp. 123–189. [Google Scholar]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Kőszegi, D.; Johnston, A.J.; Rutten, T.; Czihal, A.; Altschmied, L.; Kumlehn, J.; Wüst, S.E.; Kirioukhova, O.; Gheyselinck, J.; Grossniklaus, U.; et al. Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 2011, 67, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, G.; Conner, J.A.; Morishige, D.T.; Moore, L.D.; Mullet, J.E.; Ozias-Akins, P. A segment of the apospory-specific genomic region is highly microsyntenic not only between the apomicts Pennisetum squamulatum and Buffelgrass, but also with a rice chromosome 11 centromeric-proximal genomic region. Plant Physiol. 2006, 140, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Goel, S.; Gunawan, G.; Cordonnier-Pratt, M.M.; Johnson, V.E.; Liang, C.; Wang, H.; Pratt, L.H.; Mullet, J.E.; Debarry, J.; et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008, 147, 1396–1411. [Google Scholar] [CrossRef]
- Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). In Agrobacterium Protocols, 2nd ed.; Wang, K., Ed.; Humana Press: Totowa, NJ, USA, 2006; Volume 1, pp. 143–154. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- Głowacka, K.; Kromdijk, J.; Leonelli, L.; Niyogi, K.K.; Clemente, T.E.; Long, S.P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 2016, 39, 908–917. [Google Scholar] [CrossRef]
- Harrison, S.J.; Mott, E.K.; Parsley, K.; Aspinall, S.; Gray, J.C.; Cottage, A. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2006, 2, 1–7. [Google Scholar] [CrossRef]
- Odell, J.T.; Hoopes, J.L.; Vermerris, W. Seed-Specific gene activation mediated by the Cre/lox site-specific recombination system. Plant Physiol. 1994, 106, 447–458. [Google Scholar] [CrossRef]
- Matzk, F.; Meister, A.; Schubert, I. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 2000, 21, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Koltunow, A.M.; Truettner, J.; Cox, K.H.; Wallroth, M.; Goldberg, R.B. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 1990, 2, 1201–1224. [Google Scholar] [CrossRef]
- Fu, C.; Sun, M.; Zhou, C.; Yang, H. Isolation of fertilized embryo sacs and zygotes and triggering of zygote division in Nicotiana tabacum. Acta Bot. Sin. 1996, 38, 262–267. [Google Scholar]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Begcy, K.; Dresselhaus, T.; Sun, M.X. Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiol. 2017, 173, 130–142. [Google Scholar] [CrossRef]
- Lawit, S.J.; Chamberlin, M.A.; Agee, A.; Caswell, E.S.; Albertsen, M.C. Transgenic manipulation of plant embryo sacs tracked through cell-type-specific fluorescent markers: Cell labeling, cell ablation, and adventitious embryos. Plant. Reprod. 2013, 26, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Rövekamp, M.; Bowman, J.L.; Grossniklaus, U. Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 2016, 26, 1–8. [Google Scholar] [CrossRef]
- Kirioukhova, O.; Shah, J.N.; Larsen, D.S.; Tayyab, M.; Mueller, N.E.; Govind, G.; Baroux, C.; Federer, M.; Gheyselinck, J.; Barrell, P.J.; et al. Aberrant imprinting may underlie evolution of parthenogenesis. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Costa, L.M.; Marshall, E.; Tesfaye, M.; Silverstein, K.A.; Mori, M.; Umetsu, Y.; Otterbach, S.L.; Papareddy, R.; Dickinson, H.G.; Boutiller, K.; et al. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 2014, 344, 168–172. [Google Scholar] [CrossRef]
- Bayer, M.; Nawy, T.; Giglione, C.; Galli, M.; Meinnel, T.; Lukowitz, W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science 2009, 323, 1485–1488. [Google Scholar] [CrossRef]
- Ueda, M.; Aichinger, E.; Gong, W.; Groot, E.; Verstraeten, I.; Dai Vu, L.; De Smet, I.; Higashiyama, T.; Umeda, M.; Laux, T. Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 2017, 31, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.Q.; Russell, S.D. Calcium distribution in fertilized and unfertilized ovules and embryo sacs of Nicotiana tabacum L. Planta 1997, 202, 93–105. [Google Scholar] [CrossRef]
| T0 line ID | Ploidy | # Total Seedlings | # Seedlings Screened 1 | # Haploid T1 (%) |
|---|---|---|---|---|
| ddBR1_13.1 | 4x | 1065 | 186 | 0 (0) |
| ddBR1_204.1 | 4x | 785 | 101 | 0 (0) |
| ddBR1_204.2 | 4x | 585 | 153 | 4 (0.7%) |
| ddBR1_205.1 | 4x | 582 | 68 | 1 (0.2%) |
| ddBR1_240.2 | 4x | 575 | 123 | 5 (0.9%) |
| RKD2gBB_3.2 | 4x | 127 | 80 | 0 (0) |
| RKD2gBB_6.1 | 8x | 129 | 117 | 12 (9.3%) |
| RKD2gBB_7.1 | 8x | 223 | 65 | 0 (0) |
| T1 Plant ID | Ploidy | # Total Seedlings | # Green Resistant Seedlings | # Seedlings Screened | # Haploid T2 (%) |
|---|---|---|---|---|---|
| K6.1_11 | 4x | 103 | 57 | 56 | 13 (12.6%) |
| K6.1_14 1 | 4x | - | - | - | - |
| K6.1_20 | 4x | 387 | 111 | 101 | 14 (3.6%) |
| K6.1_23 | 4x | 211 | 80 | 52 | 12 (5.7%) |
| K6.1_1 | 8x | 51 | 40 | 40 | 8 (15.7%) |
| K6.1_7 1 | 8x | - | - | - | - |
| K6.1_12 | 8x | 224 | 173 | 150 | 50 (22.3%) |
| K6.1_17 | 8x | 22 | 21 | 20 | 6 (27.3%) |
| K6.1_22 | 8x | 75 | 73 | 60 | 2 (2.7%) |
| ♀ | ♂ | # Total Seedlings | # Green Resistant Seedlings | # Seedlings Screened | # Haploid Progeny (%) |
|---|---|---|---|---|---|
| K6.1_20 | wildtype | 285 | 71 | 56 | 16 (5.6%) |
| Maternal Parent | # Crosses Investigated | # Total Embryos Observed | # DsRed + Embryos | # DsRed − Embryos | % Parthenogenetic (DsRed − Embryos) |
|---|---|---|---|---|---|
| wildtype | 9 | 1497 | 1497 | 0 | 0 |
| K6.1_1_35 | 4 | 446 | 428 | 18 | 4.0% |
| K6.1_12_26 | 8 | 537 | 504 | 33 | 6.1% |
| K6.1_12_75 | 1 | 250 | 242 | 8 | 3.2% |
| K6.1_12_104 | 1 | 155 | 150 | 5 | 3.2% |
| K6.1_17_7 | 6 | 550 | 506 | 44 | 8.0% |
| Maternal Parent | # Total Seedlings | # DsRed + Seedlings | # DsRed − Seedlings | # DsRed − Seedlings with Reduced Ploidy (Parthenogenesis %) |
|---|---|---|---|---|
| wildtype | 220 | 220 | 0 | 0 |
| K6.1_1_35 | 216 | 212 | 4 | 4 (1.8%) |
| K6.1_12_35 | 23 | 22 | 1 | 1 (4.3%) |
| K6.1_12_26 | 152 | 148 | 4 | 4 (2.6%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Conner, J.; Guo, Y.; Ozias-Akins, P. Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis. Genes 2020, 11, 1072. https://doi.org/10.3390/genes11091072
Zhang Z, Conner J, Guo Y, Ozias-Akins P. Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis. Genes. 2020; 11(9):1072. https://doi.org/10.3390/genes11091072
Chicago/Turabian StyleZhang, Zhifen, Joann Conner, Yinping Guo, and Peggy Ozias-Akins. 2020. "Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis" Genes 11, no. 9: 1072. https://doi.org/10.3390/genes11091072
APA StyleZhang, Z., Conner, J., Guo, Y., & Ozias-Akins, P. (2020). Haploidy in Tobacco Induced by PsASGR-BBML Transgenes via Parthenogenesis. Genes, 11(9), 1072. https://doi.org/10.3390/genes11091072





