Genetic Analysis Using a Multi-Parent Wheat Population Identifies Novel Sources of Septoria Tritici Blotch Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Trials
2.2.1. 2016 Field Trial
2.2.2. 2017 Field Trial
2.2.3. 2018 Field Trial
2.3. Phenotypic Data Analysis
2.4. QTL Mapping
2.5. Gene Annotation of Significant Markers
2.6. Alignment of QTL Identified in This Study with Previously Reported Stb Genes and QTL
3. Results
3.1. Field STB Response
3.2. QTL Mapping
3.3. In-Silico Gene Annotation Data and Alignment of QTL and Genes
4. Discussion
4.1. Field Trials and Natural Septoria Infection
4.2. QTL Analysis and Comparison with Previously Reported STB QTL and Resistance Genes
4.3. Breeding for Durable STB Resistance
4.4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cook, E. Agriculture, Forestry and Fishery Statistics, 2018 Edition; Publications Office of the European Union: Luxembourg, 2018. [Google Scholar]
- Trnka, M.; Rötter, R.P.; Ruiz-Ramos, M.; Kersebaum, K.C.; Olesen, J.E.; Žalud, Z.; Semenov, M.A. Adverse weather conditions for European wheat production will become more frequent with climate change. Nat. Clim. Chang. 2014, 4, 637–643. [Google Scholar] [CrossRef]
- Van der Bom, F.; Magid, J.; Jensen, L.S. Long-term P and K fertilisation strategies and balances affect soil availability indices, crop yield depression risk and N use. Eur. J. Agron. 2017, 86, 12–23. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.K.; Rutkoski, J.; Hodson, D.P.; He, X.; Jørgensen, L.N.; Hovmøller, M.S.; Huerta-Espino, J. Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 2016, 54, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, C.A.; Tewksbury, J.J.; Tigchelaar, M.; Battisti, D.S.; Merrill, S.C.; Huey, R.B.; Naylor, R.L. Increase in crop losses to insect pests in a warming climate. Science 2018, 361, 916–919. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Eyal, Z. The Septoria Diseases of Wheat: Concepts and Methods of Disease Management; CIMMYT: El Batán, Mexico, 1987. [Google Scholar]
- Fones, H.; Gurr, S. The impact of Septoria tritici Blotch disease on wheat: An EU perspective. Fungal Genet. Biol. 2015, 79, 3–7. [Google Scholar] [CrossRef]
- Eyal, Z.; Amiri, Z.; Wahl, I. Physiologic specialization of Septoria tritici. Phytopathology 1973, 63, 1087–1091. [Google Scholar] [CrossRef]
- Hardwick, N.; Jones, D.; Slough, J. Factors affecting diseases of winter wheat in England and Wales, 1989–98. Plant Pathol. 2001, 50, 453–462. [Google Scholar] [CrossRef]
- McKendry, A.; Henke, G.; Finney, P. Effects of Septoria leaf blotch on soft red winter wheat milling and baking quality. Cereal Chem. 1995, 72, 142–146. [Google Scholar]
- Quaedvlieg, W.; Kema, G.; Groenewald, J.; Verkley, G.; Seifbarghi, S.; Razavi, M.; Gohari, A.M.; Mehrabi, R.; Crous, P. Zymoseptoria gen. nov.: A new genus to accommodate Septoria-like species occurring on graminicolous hosts. Pers. Mol. Phylogeny Evol. Fungi 2011, 26, 57. [Google Scholar] [CrossRef]
- Kema, G.H.; Verstappen, E.C.; Todorova, M.; Waalwijk, C. Successful crosses and molecular tetrad and progeny analyses demonstrate heterothallism in Mycosphaerella graminicola. Curr. Genet. 1996, 30, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Suffert, F.; Sache, I.; Lannou, C. Early stages of septoria tritici blotch epidemics of winter wheat: Build-up, overseasoning, and release of primary inoculum. Plant Pathol. 2011, 60, 166–177. [Google Scholar] [CrossRef]
- Linde, C.C.; Zhan, J.; McDonald, B.A. Population structure of Mycosphaerella graminicola: From lesions to continents. Phytopathology 2002, 92, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, A.; Goodwin, S.B.; Kema, G.H. Septoria tritici blotch (STB) of wheat. Plant Health Instr. 2011. [Google Scholar] [CrossRef]
- Kema, G.; Annone, J.; Sayoud, R.; Van Silfhout, C. Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. I. Interactions between pathogen isolates and host cultivars. Phytopathology 1996. [Google Scholar] [CrossRef]
- Gough, F.; Lee, T. Moisture effects on the discharge and survival of conidia of Septoria tritici. Phytopathology 1985, 75, 180–182. [Google Scholar] [CrossRef]
- Steinberg, G. Cell biology of Zymoseptoria tritici: Pathogen cell organization and wheat infection. Fungal Genet. Biol. 2015, 79, 17–23. [Google Scholar] [CrossRef]
- Hehir, J.; Connolly, C.; O’driscoll, A.; Lynch, J.; Spink, J.; Brown, J.; Doohan, F.; Mullins, E. Temporal and spatial field evaluations highlight the importance of the presymptomatic phase in supporting strong partial resistance in Triticum aestivum against Zymoseptoria tritici. Plant Pathol. 2018, 67, 573–583. [Google Scholar] [CrossRef]
- Shaw, M. Effects of temperature, leaf wetness and cultivar on the latent period of Mycosphaerella graminicola on winter wheat. Plant Pathol. 1990, 39, 255–268. [Google Scholar] [CrossRef]
- Armour, T.; Viljanen-Rollinson, S.; Chng, S.; Butler, R.; Jamieson, P.; Zyskowski, R. Examining the latent period of Septoria tritici blotch in a field trial of winter wheat. N. Z. Plant Prot. 2004, 57, 116–120. [Google Scholar] [CrossRef]
- Lovell, D.; Hunter, T.; Powers, S.; Parker, S.; Van den Bosch, F. Effect of temperature on latent period of septoria leaf blotch on winter wheat under outdoor conditions. Plant Pathol. 2004, 53, 170–181. [Google Scholar] [CrossRef]
- Hess, D.E.; Shaner, G. Effect of moisture and temperature on development of Septoria tritici blotch in wheat. Phytopathology 1987, 77, 215–219. [Google Scholar] [CrossRef]
- Thomas, M.; Cook, R.; King, J. Factors affecting development of Septoria tritici in winter wheat and its effect on yield. Plant Pathol. 1989, 38, 246–257. [Google Scholar] [CrossRef]
- Viljanen-Rollinson, S.; Marroni, M.; Butler, R.; Deng, Y.; Armour, T. Latent periods of Septoria tritici blotch on ten cultivars of wheat. N. Z. Plant Prot. 2005, 58, 256–260. [Google Scholar] [CrossRef]
- Henze, M.; Beyer, M.; Klink, H.; Verreet, J.-A. Characterizing meteorological scenarios favorable for Septoria tritici infections in wheat and estimation of latent periods. Plant Dis. 2007, 91, 1445–1449. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rehfus, A.; Strobel, D.; Bryson, R.; Stammler, G. Mutations in sdh genes in field isolates of Zymoseptoria tritici and impact on the sensitivity to various succinate dehydrogenase inhibitors. Plant Pathol. 2018, 67, 175–180. [Google Scholar] [CrossRef]
- McDonald, M.C.; Renkin, M.; Spackman, M.; Orchard, B.; Croll, D.; Solomon, P.S.; Milgate, A. Rapid parallel evolution of azole fungicide resistance in Australian populations of the wheat pathogen Zymoseptoria tritici. Appl. Environ. Microbiol. 2019, 85, e01908–e01918. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, B.; Cools, H.; Fountaine, J.; Lovell, D.; Motteram, J.; West, J.; Lucas, J. Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology 2005, 95, 933–941. [Google Scholar] [CrossRef]
- Brown, J.K.; Chartrain, L.; Lasserre-Zuber, P.; Saintenac, C. Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol 2015, 79, 33–41. [Google Scholar] [CrossRef]
- Arraiano, L.; Brown, J. Identification of isolate-specific and partial resistance to septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 2006, 55, 726–738. [Google Scholar] [CrossRef]
- Saintenac, C.; Lee, W.-S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Marcel, T.C.; Hartmann, F.E.; Ma, X.; Plissonneau, C.; Zala, M.; Ducasse, A.; Confais, J.; Compain, J.; Lapalu, N. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 2017, 214, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Arraiano, L.; Balaam, N.; Fenwick, P.; Chapman, C.; Feuerhelm, D.; Howell, P.; Smith, S.; Widdowson, J.; Brown, J. Contributions of disease resistance and escape to the control of Septoria tritici blotch of wheat. Plant Pathol. 2009, 58, 910–922. [Google Scholar] [CrossRef]
- Ghaffary, S.M.T.; Faris, J.D.; Friesen, T.L.; Visser, R.G.; van der Lee, T.A.; Robert, O.; Kema, G.H. New broad-spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 2012, 124, 125–142. [Google Scholar] [CrossRef]
- Stukenbrock, E.H.; Jørgensen, F.G.; Zala, M.; Hansen, T.T.; McDonald, B.A.; Schierup, M.H. Whole-genome and chromosome evolution associated with host adaptation and speciation of the wheat pathogen Mycosphaerella graminicola. PLoS Genet. 2010, 6, e1001189. [Google Scholar] [CrossRef]
- Goodwin, S.B.; M’Barek, S.B.; Dhillon, B.; Wittenberg, A.H.; Crane, C.F.; Hane, J.K.; Foster, A.J.; Van der Lee, T.A.; Grimwood, J.; Aerts, A. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011, 7, e1002070. [Google Scholar] [CrossRef]
- Eriksen, L.; Borum, F.; Jahoor, A. Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor. Appl. Genet. 2003, 107, 515–527. [Google Scholar] [CrossRef]
- Zhan, J.; McDonald, B.A. The interaction among evolutionary forces in the pathogenic fungus Mycosphaerella graminicola. Fungal Genet. Biol. 2004, 41, 590–599. [Google Scholar] [CrossRef]
- Kildea, S.; Byrne, J.; Cucak, M.; Hutton, F. First report of virulence to the septoria tritici blotch resistance gene Stb16q in the Irish Zymoseptoria tritici population. New Dis. Rep. 2020, 41, 13. [Google Scholar] [CrossRef]
- Morais, D.; Duplaix, C.; Sache, I.; Laval, V.; Suffert, F.; Walker, A.-S. Overall stability in the genetic structure of a Zymoseptoria tritici population from epidemic to interepidemic stages at a small spatial scale. Eur. J. Plant Pathol. 2019, 154, 423–436. [Google Scholar] [CrossRef]
- Ladejobi, O.; Elderfield, J.; Gardner, K.A.; Gaynor, R.C.; Hickey, J.; Hibberd, J.M.; Mackay, I.J.; Bentley, A.R. Maximizing the potential of multi-parental crop populations. Appl. Transl. Genom. 2016, 11, 9–17. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.D.; Kresovich, S.; Villeda, H.S.; Bradbury, P.; Li, H.; Sun, Q.; Flint-Garcia, S.; Thornsberry, J.; Acharya, C.; Bottoms, C. Genetic properties of the maize nested association mapping population. Science 2009, 325, 737–740. [Google Scholar] [CrossRef]
- Huang, B.E.; George, A.W.; Forrest, K.L.; Kilian, A.; Hayden, M.J.; Morell, M.K.; Cavanagh, C.R. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 2012, 10, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, S.; Rakshit, A.; Patil, J. Multiparent intercross populations in analysis of quantitative traits. J. Genet. 2012, 91, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Kover, P.X.; Valdar, W.; Trakalo, J.; Scarcelli, N.; Ehrenreich, I.M.; Purugganan, M.D.; Durrant, C.; Mott, R. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000551. [Google Scholar] [CrossRef] [PubMed]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.L.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.-W.; McCouch, S.; Thomson, M.; Mauleon, R. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.J.; Bansept-Basler, P.; Barber, T.; Bentley, A.R.; Cockram, J.; Gosman, N.; Greenland, A.J.; Horsnell, R.; Howells, R.; O’Sullivan, D.M. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: Creation, properties, and validation. G3 Genes Genomes Genet. 2014, 4, 1603–1610. [Google Scholar] [CrossRef]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Léon, J. Multi-parent advanced generation inter-cross in barley: High-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Jiménez-Galindo, J.C.; Malvar, R.A.; Butrón, A.; Santiago, R.; Samayoa, L.F.; Caicedo, M.; Ordás, B. Mapping of resistance to corn borers in a MAGIC population of maize. BMC Plant Biol. 2019, 19, 431. [Google Scholar] [CrossRef]
- Pascual, L.; Desplat, N.; Huang, B.E.; Desgroux, A.; Bruguier, L.; Bouchet, J.P.; Le, Q.H.; Chauchard, B.; Verschave, P.; Causse, M. Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 2015, 13, 565–577. [Google Scholar] [CrossRef]
- Scott, M.F.; Ladejobi, O.; Amer, S.; Bentley, A.R.; Biernaskie, J.; Boden, S.A.; Clark, M.; Dell’Acqua, M.; Dixon, L.E.; Filippi, C.V. Multi-parent populations in crops: A toolbox integrating genomics and genetic mapping with breeding. Heredity 2020. [Google Scholar] [CrossRef] [PubMed]
- Cockram, J.; Mackay, I. Genetic mapping populations for conducting high-resolution trait mapping in plants. In Plant Genetics and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 109–138. [Google Scholar]
- Milner, S.G.; Maccaferri, M.; Huang, B.E.; Mantovani, P.; Massi, A.; Frascaroli, E.; Tuberosa, R.; Salvi, S. A multiparental cross population for mapping QTL for agronomic traits in durum wheat (Triticum turgidum ssp. durum). Plant Biotechnol. J. 2016, 14, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Sannemann, W.; Lisker, A.; Maurer, A.; Léon, J.; Kazman, E.; Cöster, H.; Holzapfel, J.; Kempf, H.; Korzun, V.; Ebmeyer, E. Adaptive selection of founder segments and epistatic control of plant height in the MAGIC winter wheat population WM-800. BMC Genom. 2018, 19, 559. [Google Scholar] [CrossRef] [PubMed]
- Stadlmeier, M.; Hartl, L.; Mohler, V. Usefulness of a Multiparent Advanced Generation Intercross Population With a Greatly Reduced Mating Design for Genetic Studies in Winter Wheat. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Cockram, J. MAGIC Map and Go: Deploying MAGIC Populations for Rapid Development and Dissemination of Genetic Markers for Yield Improvement in Elite UK Winter Wheat; AHDB: Kenilworth, UK, 2020; p. 2. [Google Scholar]
- Corsi, B.; Percival-Alwyn, L.; Downie, R.C.; Venturini, L.; Iagallo, E.M.; Mantello, C.C.; McCormick-Barnes, C.; See, P.T.; Oliver, R.P.; Moffat, C.S. Genetic analysis of wheat sensitivity to the ToxB fungal effector from Pyrenophora tritici-repentis, the causal agent of tan spot. Theor. Appl. Genet. 2020, 133, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Stadlmeier, M.; Jørgensen, L.N.; Corsi, B.; Cockram, J.; Hartl, L.; Mohler, V. Genetic dissection of resistance to the three fungal plant pathogens Blumeria graminis, Zymoseptoria tritici, and Pyrenophora tritici-repentis using a multiparental winter wheat population. G3 Genes Genomes Genet. 2019, 9, 1745–1757. [Google Scholar] [CrossRef]
- Lin, M.; Corsi, B.; Ficke, A.; Tan, K.-C.; Cockram, J.; Lillemo, M. Genetic mapping using a wheat multi-founder population reveals a locus on chromosome 2A controlling resistance to both leaf and glume blotch caused by the necrotrophic fungal pathogen Parastagonospora nodorum. Theor. Appl. Genet. 2020, 133, 785–808. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Richards, R.; Condon, A.; Rebetzke, G. Traits to Improve Yield in Dry Environments: Application of Physiology in Wheat Breeding; CIMMYT: El Batán, Mexico, 2001; pp. 88–100. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Found. Stat. Comput.: Vienna, Austria, 2017. [Google Scholar]
- Harrell, F.E., Jr.; Harrell, M.F.E., Jr.; Hmisc, D. Package ‘rms’; Vanderbilt University: Nashville, TN, USA, 2019; p. 229. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Statistical Software. 2015, 67, 48. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; Version 3.1. 2 [Computer Program]; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Gardner, K.A.; Wittern, L.M.; Mackay, I.J. A highly recombined, high-density, eight-founder wheat MAGIC map reveals extensive segregation distortion and genomic locations of introgression segments. Plant Biotechnol. J. 2016, 14, 1406–1417. [Google Scholar] [CrossRef]
- Broman, K.W.; Gatti, D.M.; Simecek, P.; Furlotte, N.A.; Prins, P.; Sen, Ś.; Yandell, B.S.; Churchill, G.A. R/qtl2: Software for mapping quantitative trait loci with high-dimensional data and multiparent populations. Genetics 2019, 211, 495–502. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, R.; Dubcovsky, J.; Rogers, W.; Morris, C.; Appels, R.; Xia, X.; Azul, B. Catalogue of Gene Symbols for Wheat: 2013–2014. In Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8–14 September 2013. [Google Scholar]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Blake, V.; Birkett, C.; Matthews, D.; Hane, D.; Bradbury, P. The Triticeae Toolbox: Combining Phenotype and Genotype Data to Advance Small-Grains Breeding. Plant Genome 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Yates, S.; Mikaberidze, A.; Krattinger, S.G.; Abrouk, M.; Hund, A.; Yu, K.; Studer, B.; Fouche, S.; Meile, L.; Pereira, D. Precision phenotyping reveals novel loci for quantitative resistance to septoria tritici blotch. Plant Phenomics 2019, 2019, 3285904. [Google Scholar] [CrossRef]
- Odilbekov, F.; Armoniene, R.; Koc, A.; Svensson, J.; Chawade, A. GWAS-Assisted Genomic Prediction to Predict Resistance to Septoria Tritici Blotch in Nordic Winter Wheat at Seedling Stage. Front. Genet. 2019, 10, 1224. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Zhao, Y.; Rodemann, B.; Plieske, J.; Ganal, M.W.; Röder, M.S. Genome-wide Association Mapping and Prediction of Adult Stage Septoria tritici Blotch Infection in European Winter Wheat via High-Density Marker Arrays. Plant Genome 2019, 12. [Google Scholar] [CrossRef]
- Ando, K.; Rynearson, S.; Muleta, K.T.; Gedamu, J.; Girma, B.; Bosque-Perez, N.A.; Chen, M.-S.; Pumphrey, M.O. Genome-wide associations for multiple pest resistances in a Northwestern United States elite spring wheat panel. PLoS ONE 2018, 13, e0191305. [Google Scholar] [CrossRef]
- Kidane, Y.G.; Hailemariam, B.N.; Mengistu, D.K.; Fadda, C.; Pè, M.E.; Dell‘Acqua, M. Genome-wide association study of Septoria tritici blotch resistance in Ethiopian durum wheat landraces. Front. Plant Sci. 2017, 8, 1586. [Google Scholar] [CrossRef]
- Naz, A.A.; Klaus, M.; Pillen, K.; Léon, J. Genetic analysis and detection of new QTL alleles for S eptoria tritici blotch resistance using two advanced backcross wheat populations. Plant Breed. 2015, 134, 514–519. [Google Scholar] [CrossRef]
- Gurung, S.; Mamidi, S.; Bonman, J.M.; Xiong, M.; Brown-Guedira, G.; Adhikari, T.B. Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS ONE 2014, 9, e108179. [Google Scholar] [CrossRef] [PubMed]
- Karlstedt, F.; Kopahnke, D.; Perovic, D.; Jacobi, A.; Pillen, K.; Ordon, F. Mapping of quantitative trait loci (QTL) for resistance against Zymoseptoria tritici in the winter spelt wheat accession HTRI1410 (Triticum aestivum subsp. spelta). Euphytica 2019, 215, 108. [Google Scholar] [CrossRef]
- Odilbekov, F.; He, X.; Armoniené, R.; Saripella, G.V.; Henriksson, T.; Singh, P.K.; Chawade, A. QTL Mapping and Transcriptome Analysis to Identify Differentially Expressed Genes Induced by Septoria Tritici Blotch Disease of Wheat. Agronomy 2019, 9, 510. [Google Scholar] [CrossRef]
- Vagndorf, N.; Nielsen, N.H.; Edriss, V.; Andersen, J.R.; Orabi, J.; Jørgensen, L.N.; Jahoor, A. Genomewide association study reveals novel quantitative trait loci associated with resistance towards Septoria tritici blotch in North European winter wheat. Plant Breed. 2017, 136, 474–482. [Google Scholar] [CrossRef]
- Scutari, M.; Howell, P.; Balding, D.J.; Mackay, I. Multiple quantitative trait analysis using Bayesian networks. Genetics 2014, 198, 129–137. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Anderson, J.M.; Goodwin, S.B. Identification and molecular mapping of a gene in wheat conferring resistance to Mycosphaerella graminicola. Phytopathology 2003, 93, 1158–1164. [Google Scholar] [CrossRef]
- Cowling, S.G. Identification and Mapping of Host Resistance Genes to Septoria Tritici Blotch of Wheat. Master’s Thesis, University of Manitoba, Winnipeg, Manitoba, 2006. [Google Scholar]
- Paveley, N.; Clark, B. The Wheat Disease Management Guide; Home Grown Cereals Authority: London, UK, 2000. [Google Scholar]
- Lynch, J.P.; Glynn, E.; Kildea, S.; Spink, J. Yield and optimum fungicide dose rates for winter wheat (Triticum aestivum L.) varieties with contrasting ratings for resistance to septoria tritici blotch. Field Crop. Res. 2017, 204, 89–100. [Google Scholar] [CrossRef]
- McDonald, B.A.; Martinez, J. DNA restriction fragment length polymorphisms among Mycosphaerella graminicola (anamorph Septoria tritici) isolates collected from a single wheat field. Phytopathology 1990, 80, 1368–1373. [Google Scholar] [CrossRef]
- Robert, C.; Garin, G.; Abichou, M.; Houlès, V.; Pradal, C.; Fournier, C. Plant architecture and foliar senescence impact the race between wheat growth and Zymoseptoria tritici epidemics. Ann. Bot. 2018, 121, 975–989. [Google Scholar] [CrossRef]
- Gerard, G.S.; Börner, A.; Lohwasser, U.; Simón, M.R. Genome-wide association mapping of genetic factors controlling Septoria tritici blotch resistance and their associations with plant height and heading date in wheat. Euphytica 2017, 213, 27. [Google Scholar] [CrossRef]
- Risser, P. Mapping of Quantitative-Trait Loci (QTL) for Adult-Plant Resistance to Septoria Tritici in Five Wheat Populations (Triticum aestivum L.). Master’s Thesis, Universität Hohenheim, Stuttgart, Germany, 2010. [Google Scholar]
- Juliana, P.; Singh, R.P.; Singh, P.K.; Crossa, J.; Rutkoski, J.E.; Poland, J.A.; Bergstrom, G.C.; Sorrells, M.E. Comparison of models and whole-genome profiling approaches for genomic-enabled prediction of Septoria tritici blotch, Stagonospora nodorum blotch, and tan spot resistance in wheat. Plant Genome 2017, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rosielle, A. Sources of resistance in wheat to speckled leaf blotch caused by Septoria tritici. Euphytica 1972, 21, 152–161. [Google Scholar] [CrossRef]
- Simón, M.; Worland, A.; Struik, P. Influence of plant height and heading date on the expression of the resistance to Septoria tritici blotch in near isogenic lines of wheat. Crop Sci. 2004, 44, 2078–2085. [Google Scholar] [CrossRef]
- Parlevliet, J. Plant pathosystems: An attempt to elucidate horizontal resistance. Euphytica 1977, 26, 553–556. [Google Scholar] [CrossRef]
- Jubene, M.; Gustafson, J.; Rajaram, S. A field disease evaluation method for selecting wheats resistant to Mycosphaerella graminicola. Plant Breed. 1992, 108, 26–32. [Google Scholar] [CrossRef]
- Parker, S.; Welham, S.; Paveley, N.; Foulkes, J.; Scott, R. Tolerance of septoria leaf blotch in winter wheat. Plant Pathol. 2004, 53, 1–10. [Google Scholar] [CrossRef]
- Arraiano, L.; Worland, A.; Ellerbrook, C.; Brown, J. Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in the hexaploid wheat’Synthetic 6x’. Theor. Appl. Genet. 2001, 103, 758–764. [Google Scholar] [CrossRef]
- Fradgley, N.; Gardner, K.A.; Cockram, J.; Elderfield, J.; Hickey, J.M.; Howell, P.; Jackson, R.; Mackay, I.J. A large-scale pedigree resource of wheat reveals evidence for adaptation and selection by breeders. PLoS Biol. 2019, 17, e3000071. [Google Scholar] [CrossRef]
- Ogbonnaya, F.C.; Abdalla, O.; Mujeeb-Kazi, A.; Kazi, A.G.; Xu, S.S.; Gosman, N.; Lagudah, E.S.; Bonnett, D.; Sorrells, M.E.; Tsujimoto, H. Synthetic hexaploids: Harnessing species of the primary gene pool for wheat improvement. Plant Breed. Rev. 2013, 37, 35–122. [Google Scholar]
- Sarinelli, J.M.; Murphy, J.P.; Tyagi, P.; Holland, J.B.; Johnson, J.W.; Mergoum, M.; Mason, R.E.; Babar, A.; Harrison, S.; Sutton, R. Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 2019, 132, 1247–1261. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]


| Test Environment | Experiment Type | RILs Tested (n) | Data Collected | Time Points | Leaves | Datasets | Fungicide Applied 1 |
|---|---|---|---|---|---|---|---|
| 2016 | Single row | 1076 | STB, plant height, days to flowering | T1 T2 | flag, flag-1, flag-2 | 2016_T1_flag, 2016_T1_flag-1, 2016_T1_flag-2, 2016_T2_flag | Jenton |
| 2017 | 6-row plot | 192 | STB, plant height, days to flowering | T1 T2 | flag, flag-1, flag-2 | 2017_T1_flag, 2017_T1_flag-1, 2017_T1_flag-2, 2017_T2_flag, 2017_T2_flag-1, 2017_T2_flag-2, | Jenton 2 |
| 2018 | Single row | 388 | STB | T1 | flag, flag-1, flag-2 | 2018_T1_flag, 2018_T1_flag-1, 2018_T1_flag-2 | Jenton/ Comet |
| Datasets | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | 2016 | 2017 | 2018 | ||||||||||
| Trait | T1_flag | T1_flag-1 | T1_flag-2 | T2_flag | T1_flag | T1_flag-1 | T1_flag-2 | T2_flag | T2_flag-1 | T2_flag-2 | flag | flag-1 | flag-2 |
| Flowering time | −0.17 | −0.25 | −0.29 | −0.24 | −0.19 | −0.24 | −0.29 | −0.30 | −0.17 | −0.09 | −0.14 | −0.23 | −0.23 |
| Plant height | −0.28 | −0.22 | −0.20 | −0.20 | −0.23 | −0.26 | −0.26 | −0.29 | −0.20 | −0.25 | −0.22 | −0.25 | −0.23 |
| Datasets 1,2,3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | ||||||||||
| T1_flag | T1_flag-1 | T1_flag-2 | T2_flag | T1_flag | T1_flag-1 | T1_flag-2 | T2_flag | T2_flag-1 | T2_flag-2 | flag | flag-1 | |
| 2016_T1_flag-1 | 0.54 | 1 | ||||||||||
| 2016_T1_flag-2 | 0.49 | 0.71 | 1 | |||||||||
| 2016_T2_flag | 0.34 | 0.52 | 0.51 | 1 | ||||||||
| 2017_T1_flag | 0.38 | 0.47 | 0.44 | 0.42 | 1 | |||||||
| 2017_T1_flag-1 | 0.43 | 0.66 | 0.5 | 0.48 | 0.65 | 1 | ||||||
| 2017_T1_flag-2 | 0.35 | 0.59 | 0.5 | 0.44 | 0.62 | 0.82 | 1 | |||||
| 2017_T2_flag | 0.4 | 0.56 | 0.52 | 0.55 | 0.55 | 0.7 | 0.61 | 1 | ||||
| 2017_T2_flag-1 | 0.42 | 0.61 | 0.6 | 0.56 | 0.58 | 0.77 | 0.69 | 0.84 | 1 | |||
| 2017_T2_flag-2 | 0.32 | 0.42 | 0.47 | 0.38 | 0.32 | 0.55 | 0.51 | 0.59 | 0.7 | 1 | ||
| 2018_flag | 0.32 | 0.35 | 0.41 | 0.4 | 0.39 | 0.4 | 0.35 | 0.51 | 0.51 | 0.41 | 1 | |
| 2018_flag-1 | 0.34 | 0.47 | 0.51 | 0.48 | 0.34 | 0.44 | 0.49 | 0.59 | 0.53 | 0.44 | 0.69 | 1 |
| 2018_flag-2 | 0.26 | 0.48 | 0.5 | 0.47 | 0.29 | 0.4 | 0.46 | 0.55 | 0.51 | 0.44 | 0.62 | 0.83 |
| Dataset 1 | MAGIC Population | Founders | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Lines | Range | Mean | Alchemy | Brompton | Claire | Hereward | Rialto | Robigus | Soissons | Xi19 | |
| (n) | (min–max) | (µ) | |||||||||
| 2016_T1_flag | 1072 | 0.11–17.09 | 1.75 | - 2 | 1.17 | - | 5.22 | - | 1.03 | 4.57 | 2.18 |
| 2016_T1_flag-1 | 1072 | 0.58–30.85 | 6.04 | - | 7.97 | 2.30 | 3.77 | 2.48 | 2.91 | 6.12 | 6.90 |
| 2016_T1_flag-2 | 1072 | 3.032–85.97 | 20.19 | - | 26.34 | 12.58 | 21.38 | 9.54 | 20.47 | 23.52 | 18.43 |
| 2016_T2_flag | 1072 | 0.68–86.34 | 17.52 | - | 15.85 | 16.31 | 19.01 | 34.24 | 22.56 | 38.52 | 12.19 |
| 2017_T1_flag | 191 | 0.35–8.98 | 2.42 | 0.75 | 2.50 | 1.75 | 2.12 | 3.99 | 0.75 | 2.53 | 1.77 |
| 2017_T1_flag-1 | 191 | 0.71–33.04 | 6.55 | 4.97 | 4.78 | 2.65 | 4.61 | 14.96 | 2.38 | 11.16 | 5.95 |
| 2017_T1_flag-2 | 191 | 2.63–65.69 | 17.21 | 9.86 | 9.95 | 11.62 | 9.70 | 61.04 | 8.55 | 35.01 | 18.11 |
| 2017_T2_flag | 191 | 3.42–79.96 | 16.51 | 6.71 | 23.35 | 7.55 | 11.12 | 15.34 | 7.37 | 43.35 | 17.23 |
| 2017_T2_flag-1 | 191 | 5.758–95.00 | 46.51 | 21.25 | 36.24 | 29.40 | 53.87 | 51.35 | 33.18 | 88.30 | 74.74 |
| 2017_T2_flag-2 | 191 | 13.51–92.90 | 68.91 | 63.98 | 67.01 | 79.29 | 85.90 | 55.70 | 37.99 | 92.79 | 84.23 |
| 2018_flag | 374 | 0.26–47.1 | 2.47 | - | 2.92 | 0.76 | 1.29 | 2.30 | 1.14 | - | - |
| 2018_flag-1 | 374 | 0.67–79.99 | 8.63 | - | 6.34 | 1.50 | 2.85 | 11.34 | 3.84 | - | - |
| 2018_flag-2 | 374 | 1.22–87.28 | 34.31 | - | 18.67 | 7.50 | 28.13 | 61.66 | 21.88 | - | - |
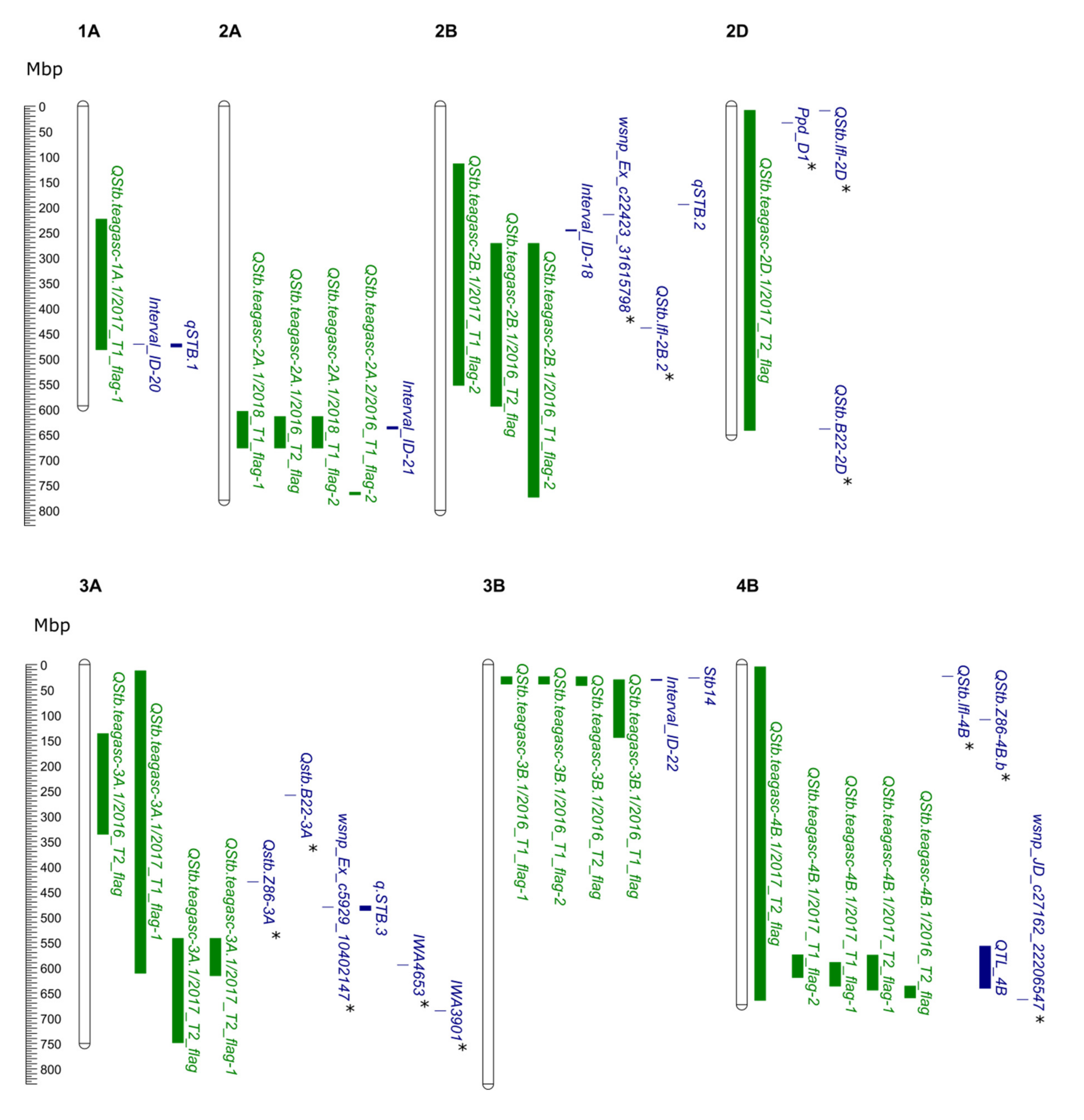
| QTL Name | Dataset | Chr. | Interval (cM) 1 | Interval (Mbp) 2 | Flanking Markers | Peak Marker | Pos (cM) | Phy.pos (Mbp) | LOD 4 | LOD thr 5 | R 2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| QStb.teagasc-1A.1 | 2017_T1_flag-1 | 1A | 66.54–106.72 | 224.83–482.97 | Ex_c21450_396- RFL_Contig3203_1971 | BobWhite_c38865_319 | 95.09 | 465.28 | 11.19 | 13.97 | 26.42 |
| QStb.teagasc-1B.1 | 2018_T1_flag-1 | 1B | 46.87–69.6 | 53.25–240.48 | Excalibur_c46902_92- RFL_Contig2852_1839 | Kukri_c18006_1568 | 56.55 | 563.11 | 10.74 | 12.46 | 13.8 |
| QStb.teagasc-1B.2 | 2017_T2_flag | 1B | 283.53–299.76 | 648.45–662.72 | BS00043666_51- BS00078414_51 | Kukri_rep_c102102_2733 | 291.21 | 563.11 | 10.19 | 15.4 | 24.36 |
| QStb.teagasc-1D.1 | 2017_T2_flag | 1D | 103.8–131.34 | 423.25–490.6 | Excalibur_c23473_451- Excalibur_c6476_811 | Excalibur_c6476_811 | 131.34 | 460.59 | 10.07 | 15.4 | 24.13 |
| QStb.teagasc-2A.1 | 2018_T1_flag-1 | 2A | 139.25–150.46 | 605.18–677.53 | BS00049644_51- IAAV4015 | RAC875_c38018_278 | 144.81 | 639.98 | 10.31 | 12.46 | 13.28 |
| 2016_T2_flag | 2A | 142.77–150.46 | 615.29–677.53 | BS00062679_51- IAAV4015 | BS00022241_51 | 146.83 | 663.32 | 12.38 | 14.03 | 9.18 | |
| 2018_T1_flag-2 | 2A | 142.77–150.46 | 615.29–677.53 | BS00062679_51- IAAV4015 | BS00022241_51 | 146.83 | 663.32 | 10.83 | 12.13 | 13.91 | |
| QStb.teagasc-2A.2 | 2016_T1_flag-2 | 2A | 249.27–259.39 | 765.26–770.02 | BS00107649_51- BS00064055_51 | BS00064055_51 | 259.39 | 765.26 | 10.9 | 14.28 | 8.13 |
| QStb.teagasc-2B.1 | 2017_T1_flag-2 | 2B | 216.09–259.25 | 115.11–553.62 | RFL_Contig4718_1323- IACX5941 | Kukri_c693_87 | 232.95 | 636.11 | 10.04 | 13.16 | 24.07 |
| 2016_T2_flag | 2B | 224.28–366.34 | 272.83–595.13 | BS00022800_51- tplb0042o21_419 | Excalibur_c105074_293 | 235.52 | 109.53 | 10.47 | 14.03 | 7.82 | |
| 2016_T1_flag-2 | 2B | 317.29–366.34 | 272.83–774.95 | RFL_Contig2751_1562- tplb0042o21_419 | BS00067878_51 | 336.02 | 784.62 | 11.93 | 14.28 | 8.86 | |
| QStb.teagasc-2D.1 | 2017_T2_flag | 2D | 0.5–21.94 | 9.343 –642.87 | tplb0060e06_1793- D_contig17313_245 | BS00063251_51 | 15.33 | - | 11.43 | 15.4 | 26.91 |
| QStb.teagasc-3A.1 | 2016_T2_flag | 3A | 1.51–107.22 | 137.583–336.78 | RAC875_c3141_214- BS00030876_51 | RFL_Contig4403_1034 | 98.98 | 176.55 | 10.01 | 14.03 | 7.49 |
| 2017_T1_flag-1 | 3A | 22.81–176.22 | 13.41–611.7 | BS00104401_51- wsnp_Ex_c26887_36107413 | Tdurum_contig10307_375 | 166.27 | 538.01 | 11.13 | 13.97 | 26.3 | |
| 2017_T2_flag | 3A | 139.3–308.8 | 543.18–749.33 | Tdurum_contig59531_914- Tdurum_contig27982_568 | wsnp_Ex_c1335_2556442 | 163.97 | 532.33 | 11.45 | 15.4 | 26.93 | |
| 2017_T2_flag-1 | 3A | 139.3–174.7 | 543.18–616.49 | Tdurum_contig59531_914- BS00056089_51 | wsnp_Ex_c1335_2556442 | 163.97 | 532.33 | 10.24 | 13.98 | 24.48 | |
| QStb.teagasc-3B.1 | 2016_T1_flag-1 | 3B | 49.33–60.59 | 24.94–39.46 | Tdurum_contig22897_107- Excalibur_c79902_439 | Excalibur_c24391_321 | 57.06 | 31.1 | 11.55 | 12.39 | 8.59 |
| 2016_T1_flag-2 | 3B | 49.33–60.59 | 24.94–39.46 | Tdurum_contig22897_107- Excalibur_c79902_439 | Excalibur_c24391_321 | 57.06 | 31.1 | 11.22 | 14.28 | 8.36 | |
| 2016_T2_flag | 3B | 49.33–63.63 | 24.94–42.34 | Tdurum_contig22897_107- BS00022242_51 | Excalibur_c24391_321 | 57.06 | 31.1 | 14.51 | 14.03 | 10.68 | |
| 2016_T1_flag | 3B | 57.06–95.19 | 31.11–145.31 | Excalibur_c24391_321- IACX971 | BS00073732_51 | 87.63 | 67.92 | 10.36 | 12.39 | 7.74 | |
| QStb.teagasc-3D.1 | 2016_T1_flag-2 | 3D | 100.88–119.23 | 274.49–524.87 | Ku_c2845_342- IAAV5136 | Ku_c6080_1667 | 107.27 | 488.85 | 11.07 | 14.28 | 8.25 |
| QStb.teagasc-4A.1 | 2017_T1_flag-1 | 4A | 33.88–74.98 | 16.19–68.56 | wsnp_Ex_c28429_37553452- wsnp_Ex_c10390_17007929 | BS00110459_51 | 66.77 | 41.94 | 11.2 | 13.97 | 26.44 |
| 2016_T1_flag-1 | 4A | 55.12–164.01 | 30.29–632.86 | Kukri_c16916_1073- BS00091752_51 | BS00108849_51 | 132.74 | 617.28 | 10.62 | 12.39 | 7.93 | |
| 2017_T2_flag | 4A | 91.12–103.23 | 583.95–597.69 | BS00010339_51- BS00077716_51 | BS00040648_51 | 96.69 | 594.66 | 12.09 | 15.4 | 28.21 | |
| QStb.teagasc-4B.1 | 2017_T2_flag | 4B | 0–173.22 | 5.47–665.6 | BS00039936_51- BS00009342_51 | wsnp_BF482960B_Ta_1_4 | 47.64 | 289.54 | 12.24 | 15.4 | 28.51 |
| 2017_T1_flag-2 | 4B | 124.46–158.5 | 575.6–620.06 | BobWhite_c11005_236- BobWhite_c27751_206 | Tdurum_contig93160_155 | 148.22 | 576.2 | 11.48 | 13.16 | 27.01 | |
| 2017_T1_flag-1 | 4B | 125.98–182.54 | 590.44–637.39 | Ku_c48056_436- wsnp_Ku_c12503_20174234 | RAC875_c1918_101 | 152.89 | 636.77 | 10.83 | 13.97 | 25.69 | |
| 2017_T2_flag-1 | 4B | 148.22–179.45 | 576.2–645.3 | Tdurum_contig93160_155- RAC875_c24515_602 | Ra_c10455_3226 | 166.59 | 590.44 | 12.19 | 13.98 | 28.4 | |
| 2016_T2_flag | 4B | 189.64–209.52 | 637.39–660.47 | BS00035426_51- IACX7540 | RAC875_c87897_333 | 196.07 | 650.94 | 10.2 | 14.03 | 7.62 | |
| QStb.teagasc-4D.1 | 2016_T1_flag-2 | 4D | 0–110.72 | 6.6–481.613 | BS00054978_51- IAAV5607 | BS00064176_51 | 104.49 | 483.29 | 10.85 | 14.28 | 8.09 |
| 2016_T2_flag | 4D | 0–9.87 | 1.24–6.6 | BS00054978_51- Excalibur_c26088_184 | BS00099053_51 | 4.1 | 3.61 | 10.32 | 14.03 | 7.72 | |
| 2017_T2_flag | 4D | 9.87–40.11 | 1.24–25.99 | Excalibur_c26088_184- RAC875_rep_c105718_304 | RAC875_c1673_663 | 32.24 | 16.59 | 11 | 15.4 | 26.03 | |
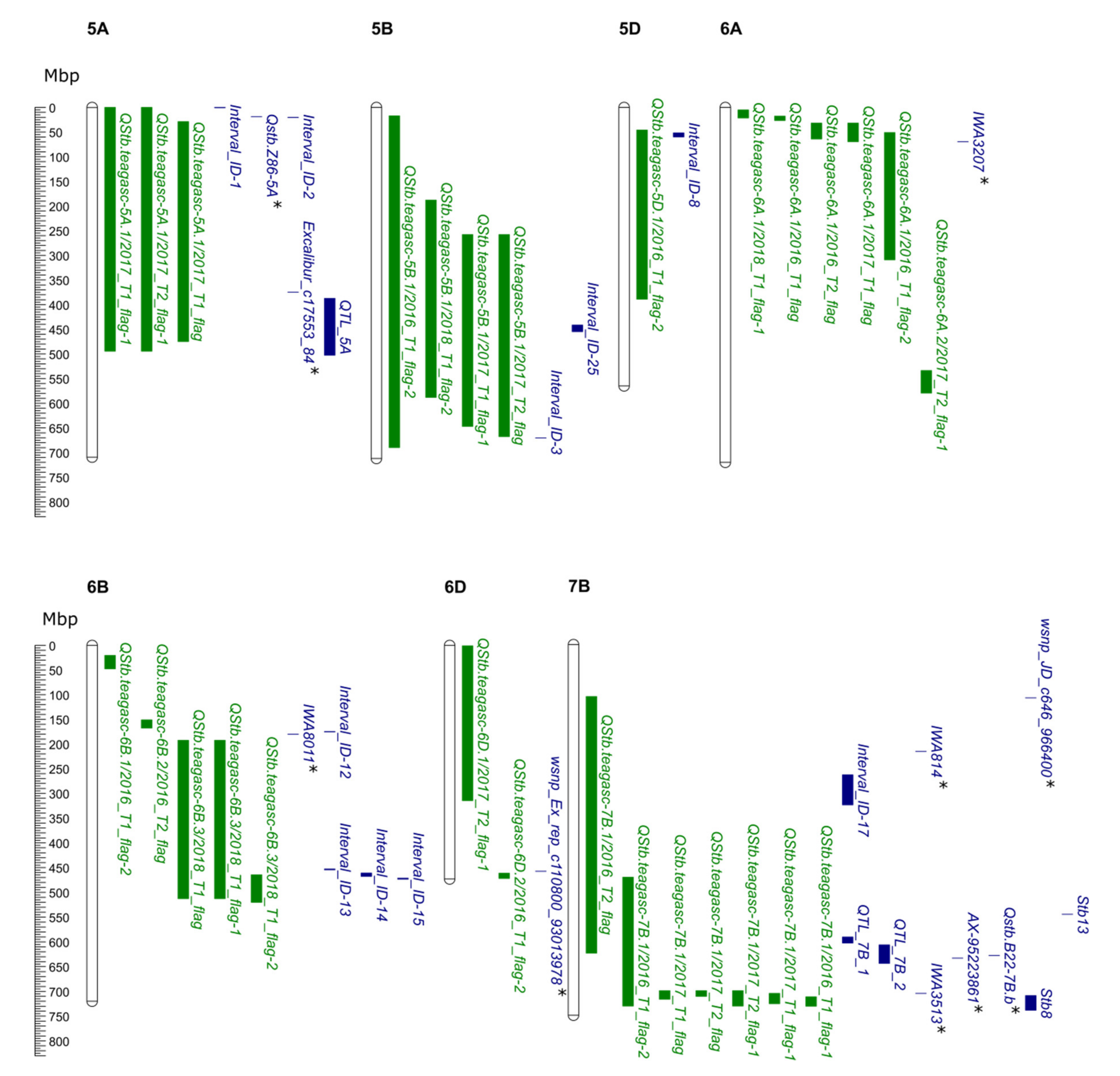
| QStb.teagasc-5A.1 | 2017_T1_flag-1 | 5A | 0–138.95 | 0.64–494.87 | BobWhite_c7114_237- wsnp_Ku_c35386_44598937 | Excalibur_rep_c104815_1181 | 6.08 | 3.4 | 10.43 | 13.97 | 24.86 |
| 2017_T2_flag-1 | 5A | 0–138.95 | 0.64–494.87 | BobWhite_c7114_237- wsnp_Ku_c35386_44598937 | BS00015653_51 | 98.27 | 474.5 | 10.16 | 13.98 | 24.32 | |
| 2017_T1_flag | 5A | 41.26–94.7 | 29.51–475.47 | Excalibur_rep_c90275_262- tplb0057m23_716 | BS00066499_51 | 82.55 | - | 10.12 | 12.47 | 24.24 | |
| QStb.teagasc-5B.1 | 2016_T1_flag-2 | 5B | 1.27–282.07 | 17.97–690.7 | BS00015136_51- BS00092233_51 | Kukri_c60322_490 | 6.88 | 19.44 | 10.62 | 14.28 | 7.93 |
| 2018_T1_flag-2 | 5B | 184–202.43 | 589.12 | RAC875_c30867_515-IAAV4388 | BS00061414_51 | 189.1 | 580.7 | 10.07 | 12.13 | 13 | |
| 2017_T1_flag-1 | 5B | 230.67–261.48 | 647.94 | BobWhite_c12883_272- Excalibur_c9563_1157 | BS00062972_51 | 258.92 | 670.54 | 10.66 | 13.97 | 25.34 | |
| 2017_T2_flag | 5B | 253.87–261.48 | 668.47 | RAC875_c45135_184- Excalibur_c9563_1157 | Excalibur_c17489_804 | 258.92 | 670.82 | 12.02 | 15.4 | 28.08 | |
| QStb.teagasc-5D.1 | 2016_T1_flag-2 | 5D | 12.57–49.43 | 47.02–389.93 | BS00003975_51- BobWhite_c7263_337 | IACX2960 | 28.11 | 347.48 | 11.92 | 14.28 | 8.85 |
| QStb.teagasc-6A.1 | 2018_T1_flag-1 | 6A | 21.25–65.6 | 6.2– 22.44 | Ra_c22493_190-BobWhite_c13839_135 | Excalibur_c7044_243 | 56.01 | 18.71 | 10.79 | 12.46 | 13.86 |
| 2016_T1_flag | 6A | 56.01–70.12 | 18.71–27.5 | Excalibur_c7044_243- RAC875_c18659_402 | BS00022951_51 | 61.07 | 18.71 | 12.77 | 12.39 | 9.45 | |
| 2016_T2_flag | 6A | 88.28–106.44 | 32.66–65.07 | wsnp_Ex_c31149_39976103- BS00021999_51 | Kukri_c27958_334 | 96.41 | 51.51 | 10.89 | 14.03 | 8.12 | |
| 2017_T1_flag | 6A | 88.28–106.95 | 32.66–70.4 | wsnp_Ex_c31149_39976103- wsnp_Ku_c18534_27848426 | Jagger_c2853_75 | 93.31 | 51.4 | 10.5 | 12.47 | 25.02 | |
| 2016_T1_flag-2 | 6A | 103.35–137.51 | 51.95–309.86 | Tdurum_contig50698_601- BS00065309_51 | BS00026558_51 | 127.92 | 94.2 | 11.31 | 14.28 | 8.42 | |
| QStb.teagasc-6A.2 | 2017_T2_flag-1 | 6A | 173.36–191.33 | 534.67–580.19 | BS00028263_51- wsnp_BE495143A_Ta_2_2 | BS00023893_51 | 176.94 | 557.93 | 11.31 | 13.98 | 26.66 |
| QStb.teagasc-6B.1 | 2016_T1_flag-2 | 6B | 35.81–57.49 | 21.41–48.35 | wsnp_Ex_c702_1382859- wsnp_Ex_c13352_21044607 | BS00090073_51 | 51.22 | 41.97 | 11.12 | 14.28 | 8.28 |
| QStb.teagasc-6B.2 | 2016_T2_flag | 6B | 92.34–109.98 | 152.09–168.473 | BS00063608_51- BobWhite_c686_387 | wsnp_Ex_c34011_42398362 | 104.92 | 462.13 | 10.3 | 14.03 | 7.7 |
| QStb.teagasc-6B.3 | 2018_T1_flag | 6B | 128.13–149.57 | 193.46 | Kukri_c23433_416- BobWhite_c14575_323 | BS00048295_51 | 133.69 | 513.69 | 14.3 | 13.55 | 17.94 |
| 2018_T1_flag-1 | 6B | 128.13–149.57 | 193.46 | Kukri_c23433_416- BobWhite_c14575_323 | BS00048293_51 | 134.69 | 513.69 | 12.38 | 12.46 | 15.74 | |
| 2018_T1_flag-2 | 6B | 129.66–144.9 | 465.68–521.03 | RFL_Contig3609_1678- wsnp_Ku_c24981_34948114 | BS00035381_51 | 141.88 | 542.22 | 11.4 | 12.13 | 14.59 | |
| QStb.teagasc-6D.1 | 2017_T2_flag-1 | 6D | 25.79–102.17 | 1.77–315.6 | Excalibur_c10358_1800- IACX10982 | BS00021970_51 | 53.79 | 24 | 12.2 | 13.98 | 28.43 |
| QStb.teagasc-6D.2 | 2016_T1_flag-2 | 6D | 196.75–215 | 462.2–472.79 | BobWhite_c34798_184- BS00070856_51 | wsnp_Ex_c13188_20825019 | 202.62 | 464.71 | 10.15 | 14.28 | 7.59 |
| QStb.teagasc-7B.1 | 2016_T2_flag | 7B | 73.93–242.34 | 106.34–625.52 | BobWhite_c14812_828- wsnp_Ex_c64848_63486322 | BobWhite_c24067_5193 | 213.89 | 713.36 | 11.9 | 14.03 | 8.84 |
| 2016_T1_flag-2 | 7B | 109.14–239.29 | 472.33–732.15 | Ra_c3470_1551- Kukri_c40149_436 | Tdurum_contig30163_1053 | 185.27 | 701.63 | 11.6 | 14.28 | 8.63 | |
| 2017_T1_flag | 7B | 190.34–225.24 | 701.34–718.36 | RAC875_rep_c78007_425- BS00022162_51 | Tdurum_contig67161_99 | 203.23 | 703.3 | 10.46 | 12.47 | 24.93 | |
| 2017_T2_flag | 7B | 193.88–216.43 | 701.34–712.73 | RAC875_c41938_471- BS00066124_51 | Tdurum_contig13268_1067 | 194.89 | 708.95 | 12.76 | 15.4 | 29.52 | |
| 2017_T2_flag-1 | 7B | 193.88–239.29 | 701.34–732.15 | RAC875_c41938_471- Kukri_c40149_436 | Tdurum_contig81587_90 | 195.39 | 706.81 | 11.55 | 13.98 | 27.14 | |
| 2017_T1_flag-1 | 7B | 208.8–240.81 | 706.81–727.63 | Tdurum_contig61856_900- wsnp_Ex_c4484_8065800 | BobWhite_c2892_167 | 233.61 | 721.21 | 10.06 | 13.97 | 24.1 | |
| 2016_T1_flag-1 | 7B | 212.37–250.49 | 713.66–732.65 | RAC875_c39269_312- wsnp_Ex_c32905_41484291 | BobWhite_c2892_167 | 233.61 | 721.21 | 13.8 | 12.39 | 10.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riaz, A.; KockAppelgren, P.; Hehir, J.G.; Kang, J.; Meade, F.; Cockram, J.; Milbourne, D.; Spink, J.; Mullins, E.; Byrne, S. Genetic Analysis Using a Multi-Parent Wheat Population Identifies Novel Sources of Septoria Tritici Blotch Resistance. Genes 2020, 11, 887. https://doi.org/10.3390/genes11080887
Riaz A, KockAppelgren P, Hehir JG, Kang J, Meade F, Cockram J, Milbourne D, Spink J, Mullins E, Byrne S. Genetic Analysis Using a Multi-Parent Wheat Population Identifies Novel Sources of Septoria Tritici Blotch Resistance. Genes. 2020; 11(8):887. https://doi.org/10.3390/genes11080887
Chicago/Turabian StyleRiaz, Adnan, Petra KockAppelgren, James Gerard Hehir, Jie Kang, Fergus Meade, James Cockram, Dan Milbourne, John Spink, Ewen Mullins, and Stephen Byrne. 2020. "Genetic Analysis Using a Multi-Parent Wheat Population Identifies Novel Sources of Septoria Tritici Blotch Resistance" Genes 11, no. 8: 887. https://doi.org/10.3390/genes11080887
APA StyleRiaz, A., KockAppelgren, P., Hehir, J. G., Kang, J., Meade, F., Cockram, J., Milbourne, D., Spink, J., Mullins, E., & Byrne, S. (2020). Genetic Analysis Using a Multi-Parent Wheat Population Identifies Novel Sources of Septoria Tritici Blotch Resistance. Genes, 11(8), 887. https://doi.org/10.3390/genes11080887






