Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution
Abstract
1. Introduction
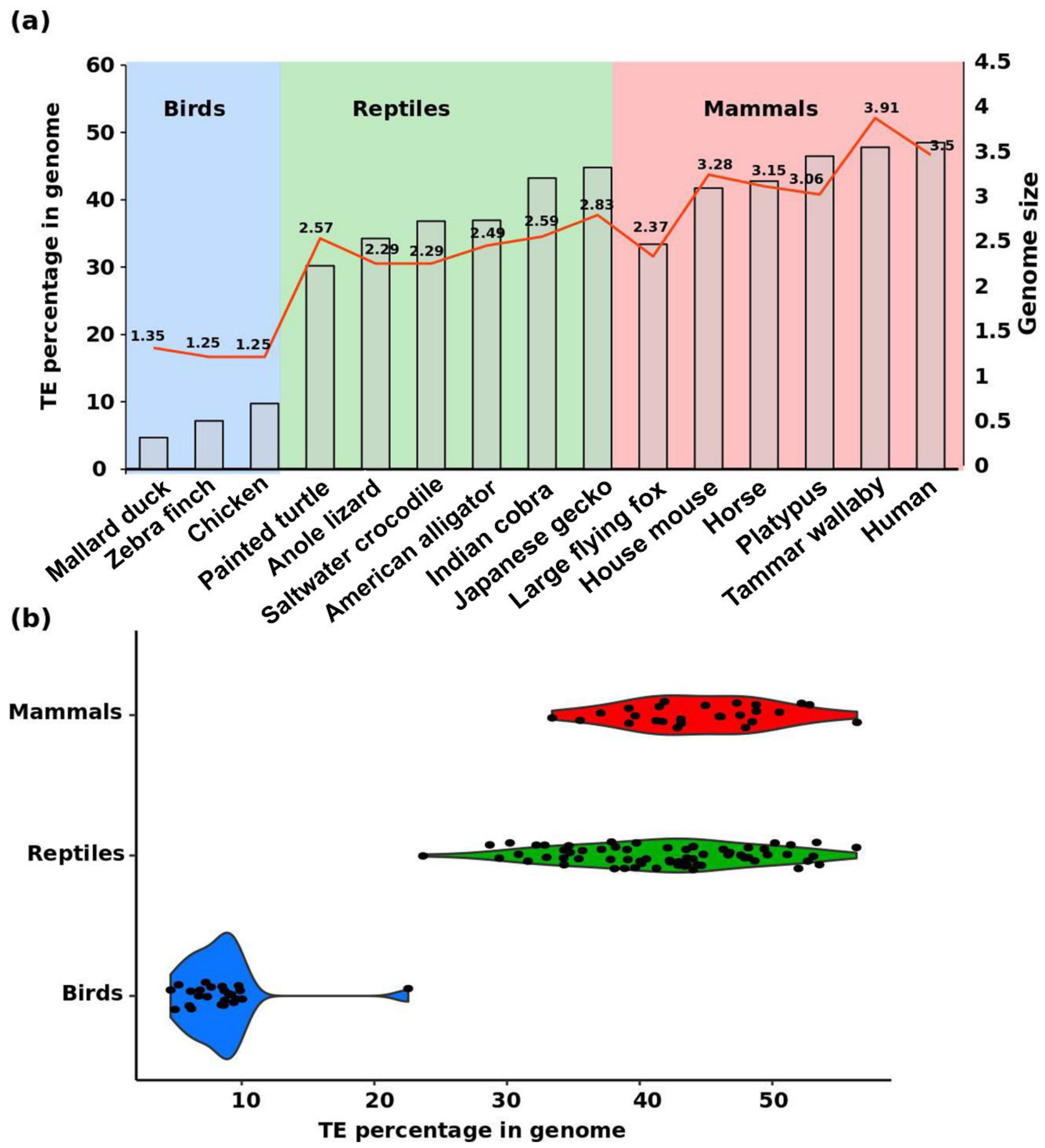
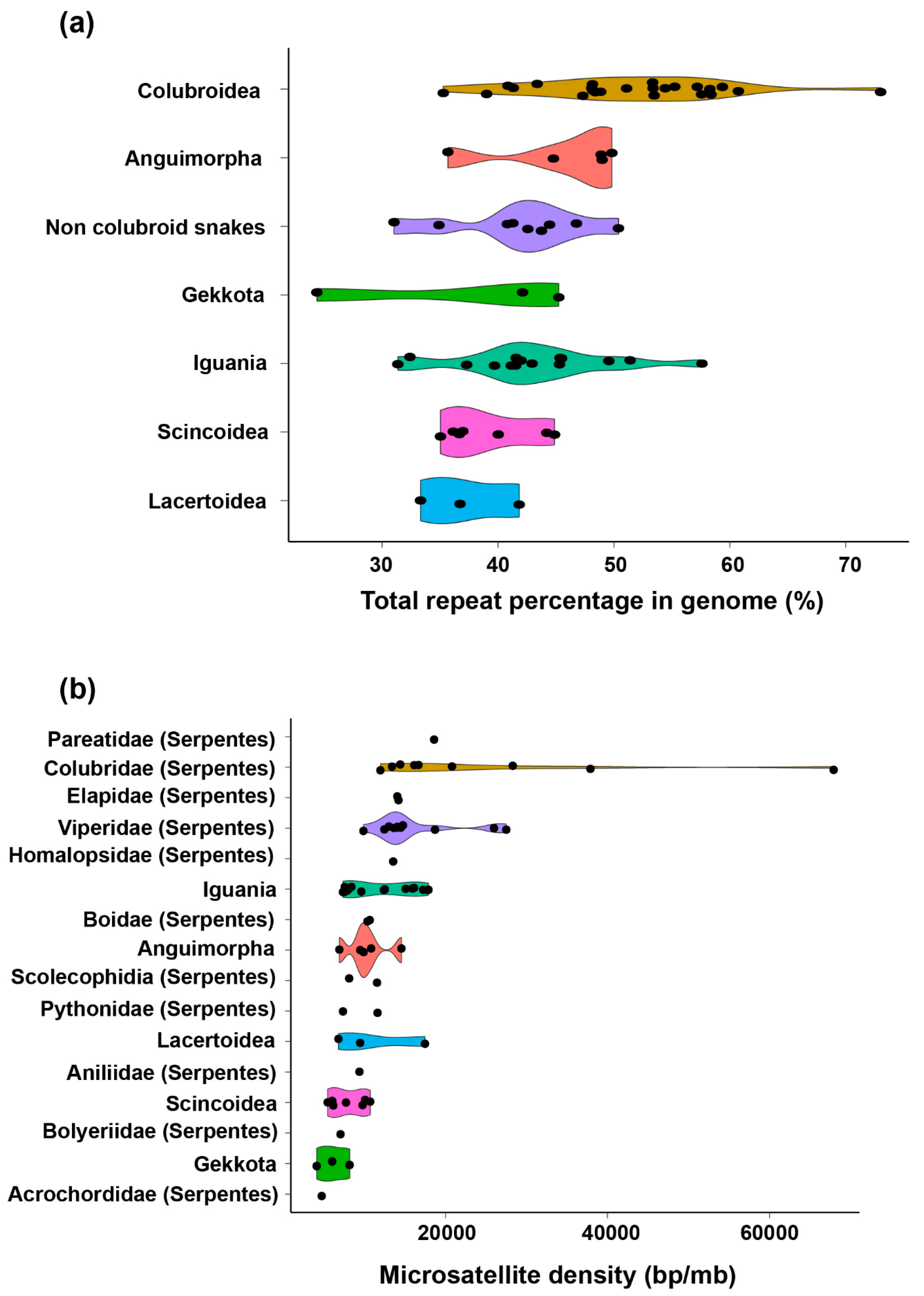
2. Diversity of Repeats in Reptiles Versus Other Amniotes
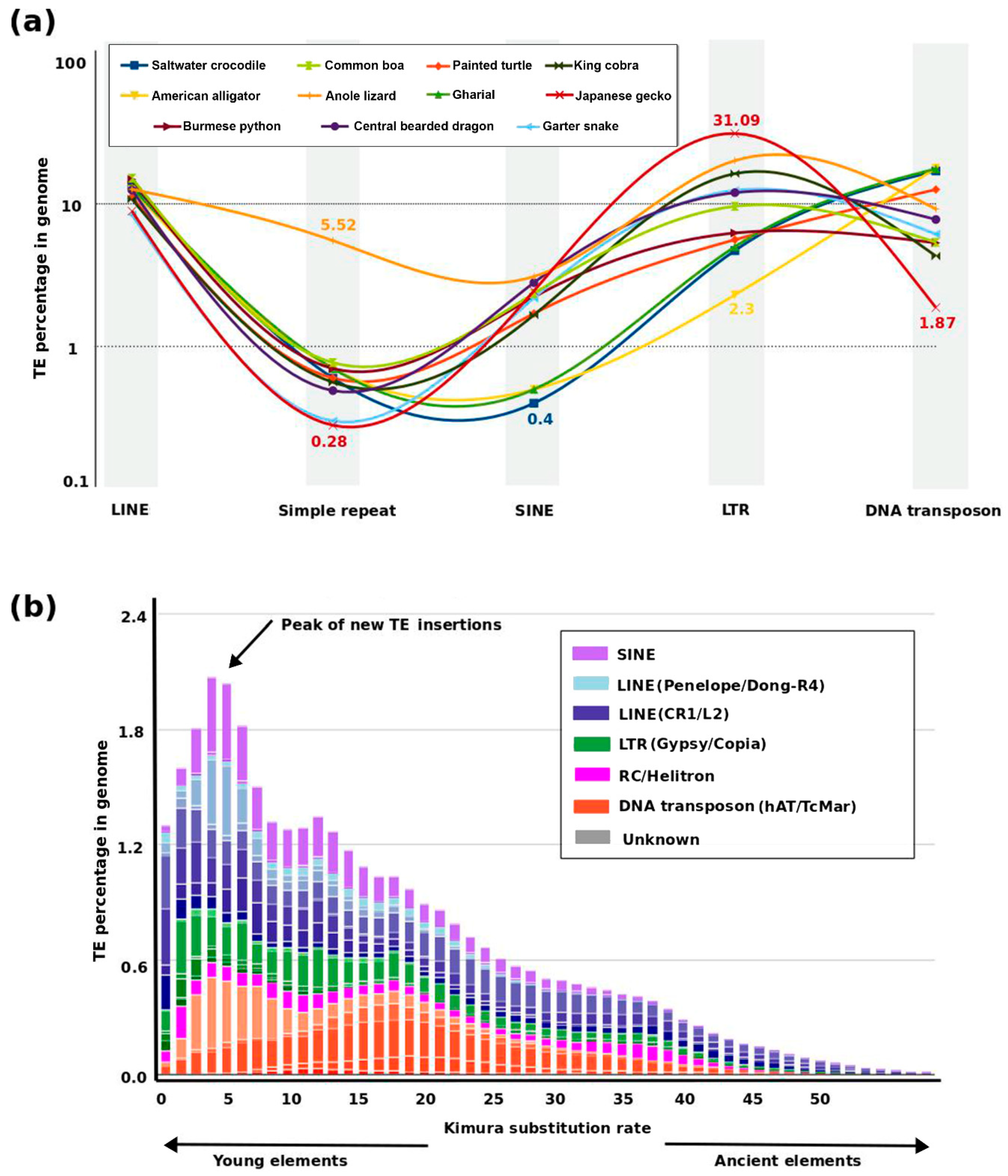
3. Dynamics of TE and Satellite Landscapes in Different Reptilian Lineages
4. Evolutionary Impact of Repeats in Reptiles: Mediators of Chromosomal Rearrangements to Drive Genome Reorganization
5. Repeatome and Genome Complexity with Evolutionary Breakpoint Regions
6. Repeats with Sex Chromosomes in Relation to an Ancestral Amniote Super-Sex Chromosome Evolution Hypothesis
7. Evolutionary Products of Micro- and Macrochromosomal Rearrangements in Reptiles
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Darwin, C.; Wallace, A. On the Tendency of Species to form Varieties; and on the Perpetuation of Varieties and Species by Natural Means of Selection. J. Proc. Linn. Soc. Lond. Zool. 1858, 3, 45–62. [Google Scholar] [CrossRef]
- Darwin, C. The Origin of Species, by Means of Natural Selection, or the Preservation of Favored Races in the Struggle for Life, 2nd ed.; John Murray: London, UK, 1860; pp. 149–150. [Google Scholar] [CrossRef]
- Dobzhansky, T. Genetics of Natural Populations. XXVI. Chromosomal Variability in Island and Continental Populations of Drosophila willistoni from Central America and the West Indies. Evolution 1957, 11, 280. [Google Scholar] [CrossRef]
- Crombach, A.; Hogeweg, P. Chromosome rearrangements and the evolution of genome structuring and adaptability. Mol. Biol. Evol. 2007, 24, 1130–1139. [Google Scholar] [CrossRef]
- Stebbins, G.L. The Inviability, Weakness, and Sterility of Interspecific Hybrids. Adv. Genet. 1958, 9, 147–215. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.G.; Singhal, S.; Rabosky, D.L. Beyond Reproductive Isolation: Demographic Controls on the Speciation Process. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 75–95. [Google Scholar] [CrossRef]
- Lynch, M. The Genetic Interpretation of Inbreeding Depression and Outbreeding Depression. Evolution 1991, 45, 622. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Thapana, W.; Muangmai, N. Role of Chromosome Changes in Crocodylus Evolution and Diversity. Genom. Inform. 2015, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Ponjarat, J.; Singchat, W.; Monkheang, P.; Suntronpong, A.; Tawichasri, P.; Sillapaprayoon, S.; Ogawa, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; et al. Evidence of dramatic sterility in F 1 male hybrid catfish [male Clarias gariepinus (Burchell, 1822) × female C. macrocephalus (Günther, 1864)] resulting from the failure of homologous chromosome pairing in meiosis I. Aquaculture 2019, 505, 84–91. [Google Scholar] [CrossRef]
- Donoghue, P.C.J.; Benton, M.J. Rocks and clocks: Calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 2007, 22, 424–431. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Nishida, C.; Matsuda, Y.; Apisitwanich, S. Genetic relationship of three butterfly lizard species (Leiolepis reevesii rubritaeniata, Leiolepis belliana belliana, Leiolepis boehmei, Agamidae, Squamata) inferred from nuclear gene sequence analyses. Kasetsart J. Nat. Sci. 2010, 44, 424–435. [Google Scholar]
- Brocklehurst, N.; Kammerer, C.; Frobisch, J. The early evolution of synapsids, and the influence of sampling on their fossil record. Paleobiology 2013, 39, 470–490. [Google Scholar] [CrossRef]
- Chromorep: A Reptile Chromosomes Database. Available online: http://chromorep.univpm.it/ (accessed on 13 May 2020).
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y.; Nishida, C. Karyological characterization of the butterfly lizard (leiolepis reevesii rubritaeniata, agamidae, squamata) by molecular cytogenetic approach. Cytogenet. Genome Res. 2009, 125, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Nishida, C.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y. Karyotypic evolution in squamate reptiles: Comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res. 2009, 17, 975. [Google Scholar] [CrossRef]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Matsuda, Y. Karyotype evolution in monitor lizards: Cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 2013, 21, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Nishida, C.; Olsson, M.; Matsuda, Y. Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 2014, 123, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Ota, H.; Matsuda, Y. Karyotype reorganization in the Hokou Gecko (Gekko hokouensis, Gekkonidae): The process of microchromosome disappearance in Gekkota. PLoS ONE 2015, 10, e0134829. [Google Scholar] [CrossRef] [PubMed]
- Singchat, W.; O’Connor, R.E.; Tawichasri, P.; Suntronpong, A.; Sillapaprayoon, S.; Suntrarachun, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genom. 2018, 19, 939. [Google Scholar] [CrossRef]
- Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Indananda, C.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”? Chromosome Res. 2020, 28, 209–228. [Google Scholar] [CrossRef]
- O’Connor, R.E.; Kiazim, L.; Skinner, B.; Fonseka, G.; Joseph, S.; Jennings, R.; Larkin, D.M.; Griffin, D.K. Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 2019, 128, 21–29. [Google Scholar] [CrossRef]
- Belterman, R.H.R.; De Boer, L.E.M. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 1984, 65, 39–82. [Google Scholar] [CrossRef]
- Matsuda, Y.; Nishida-Umehara, C.; Tarui, H.; Kuroiwa, A.; Yamada, K.; Isobe, T.; Ando, J.; Fujiwara, A.; Hirao, Y.; Nishimura, O.; et al. Highly conserved linkage homology between birds and turtles: Bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005, 13, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Kawagoshi, T.; Uno, Y.; Matsubara, K.; Matsuda, Y.; Nishida, C. The ZW micro-sex chromosomes of the chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet. Genome Res. 2009, 125, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Kawagoshi, T.; Nishida, C.; Matsuda, Y. The origin and differentiation process of X and y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res. 2012, 20, 95–110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cohen, M.M.; Clark, H.F. The somatic chromosomes of five crocodilian species. Cytogenet. Genome Res. 1967, 6, 193–203. [Google Scholar] [CrossRef]
- Kawai, A.; Nishida-Umehara, C.; Ishijima, J.; Tsuda, Y.; Ota, H.; Matsuda, Y. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet. Genome Res. 2007, 117, 92–102. [Google Scholar] [CrossRef]
- Kawagoshi, T.; Nishida, C.; Ota, H.; Kumazawa, Y.; Endo, H.; Matsuda, Y. Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae, Crocodylia). Chromosome Res. 2008, 16, 1119–1132. [Google Scholar] [CrossRef]
- Kasai, F.; O’Brien, P.C.M.; Ferguson-Smith, M.A. Reassessment of genome size in turtle and crocodile based on chromosome measurement by flow karyotyping: Close similarity to chicken. Biol. Lett. 2012, 8, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Valleley, E.M.A.; Harrison, C.J.; Cook, Y.; Ferguson, M.W.J.; Sharpe, P.T. The karyotype of Alligator mississippiensis, and chromosomal mapping of the ZFY/X homologue, Zfc. Chromosoma 1994, 103, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Syvänen, A.C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001, 2, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Völker, M.; Backström, N.; Skinner, B.M.; Langley, E.J.; Bunzey, S.K.; Ellegren, H.; Griffin, D.K. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010, 20, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Bradley, F.H.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K.; et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013, 14, 1–23. [Google Scholar] [CrossRef]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schröder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Thongchum, R.; Singchat, W.; Laopichienpong, N.; Tawichasri, P.; Kraichak, E.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Suntrarachun, S.; et al. Diversity of PBI-DdeI satellite DNA in snakes correlates with rapid independent evolution and different functional roles. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Suh, A.; Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA 2017, 114, E1460–E1469. [Google Scholar] [CrossRef]
- Matsubara, K.; Tarui, H.; Toriba, M.; Yamada, K.; Nishida-Umehara, C.; Agata, K.; Matsuda, Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 18190–18195. [Google Scholar] [CrossRef]
- Matsubara, K.; O’Meally, D.; Azad, B.; Georges, A.; Sarre, S.D.; Graves, J.A.M.; Matsuda, Y.; Ezaz, T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 2016, 125, 111–123. [Google Scholar] [CrossRef]
- Chaiprasertsri, N.; Uno, Y.; Peyachoknagul, S.; Prakhongcheep, O.; Baicharoen, S.; Charernsuk, S.; Nishida, C.; Matsuda, Y.; Koga, A.; Srikulnath, K. Highly species-specific centromeric repetitive DNA sequences in lizards: Molecular cytogenetic characterization of a novel family of satellite DNA sequences isolated from the water monitor lizard (Varanus salvator macromaculatus, Platynota). J. Hered. 2013, 104, 798–806. [Google Scholar] [CrossRef]
- Prakhongcheep, O.; Hirai, Y.; Hara, T.; Srikulnath, K.; Hirai, H.; Koga, A. Two types of Alpha satellite DNA in distinct chromosomal locations in Azara’s owl monkey. DNA Res. 2013, 20, 235–240. [Google Scholar] [CrossRef]
- Prakhongcheep, O.; Chaiprasertsri, N.; Terada, S.; Hirai, Y.; Srikulnath, K.; Hirai, H.; Koga, A. Heterochromatin blocks constituting the entire short arms of acrocentric chromosomes of Azara’s owl monkey: Formation processes inferred from chromosomal locations. DNA Res. 2013, 20, 461–470. [Google Scholar] [CrossRef]
- Thapana, W.; Sujiwattanarat, P.; Srikulnath, K.; Hirai, H.; Koga, A. Reduction in the structural instability of cloned eukaryotic tandem-repeat DNA by low-temperature culturing of host bacteria. Genet. Res. 2014, 96. [Google Scholar] [CrossRef]
- Sujiwattanarat, P.; Thapana, W.; Srikulnath, K.; Hirai, Y.; Hirai, H.; Koga, A. Higher-order repeat structure in Alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci. Rep. 2015, 5, 10315. [Google Scholar] [CrossRef]
- Srikulnath, K.; Azad, B.; Singchat, W.; Ezaz, T. Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS ONE 2019, 14, e0212683. [Google Scholar] [CrossRef] [PubMed]
- Suntronpong, A.; Singchat, W.; Kruasuwan, W.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Somyong, S.; Indananda, C.; Kraichak, E.; Peyachoknagul, S.; et al. Characterization of centromeric satellite DNAs (MALREP) in the Asian swamp eel (Monopterus albus) suggests the possible origin of repeats from transposable elements. Genomics 2020, 112, 3097–3107. [Google Scholar] [CrossRef]
- Enukashvily, N.I.; Ponomartsev, N.V. Mammalian satellite DNA: A speaking dumb. Adv. Protein Chem. Struct. Biol. 2013, 9, 31–65. [Google Scholar]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012, 13, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Boissinot, S.; Bourgeois, Y.; Manthey, J.D.; Ruggiero, R.P. The mobilome of reptiles: Evolution, structure, and function. Cytogenet. Genome Res. 2019, 157, 21–33. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; Fitzhugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef]
- Tollis, M.; Hutchins, E.D.; Kusumi, K. Reptile genomes open the frontier for comparative analysis of amniote development and regeneration. Int. J. Dev. Biol. 2014, 58, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Tollis, M.; Boissinot, S. The evolutionary dynamics of transposable elements in eukaryote genomes. Genome Dyn. 2012, 7, 68–91. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef]
- Siefert, J.L. Defining the mobilome. Methods Mol. Biol. 2009, 532, 13–27. [Google Scholar] [PubMed]
- Warren, I.A.; Naville, M.; Chalopin, D.; Levin, P.; Berger, C.S.; Galiana, D.; Volff, J.N. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 2015, 23, 505–531. [Google Scholar] [CrossRef]
- Trizzino, M.; Park, Y.S.; Holsbach-Beltrame, M.; Aracena, K.; Mika, K.; Caliskan, M.; Perry, G.H.; Lynch, V.J.; Brown, C.D. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017, 27, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Pasyukova, E.G.; Nuzhdin, S.V.; Morozova, T.V.; Mackay, T.F.C. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J. Hered. 2004, 95, 284–290. [Google Scholar] [CrossRef]
- Boissinot, S.; Davis, J.; Entezam, A.; Petrov, D.; Furano, A.V. Fitness cost of LINE-1 (L1) activity in humans. Proc. Natl. Acad. Sci. USA 2006, 103, 9590–9594. [Google Scholar] [CrossRef]
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Suh, A.; Churakov, G.; Ramakodi, M.P.; Platt, R.N.; Jurka, J.; Kojima, K.K.; Caballero, J.; Smit, A.F.; Vliet, K.A.; Hoffmann, F.G.; et al. Multiple lineages of ancient CR1 retroposons shaped the early genome evolution of amniotes. Genome Biol. Evol. 2014, 7, 205–217. [Google Scholar] [CrossRef]
- Volff, J.N.; Körting, C.; Schartl, M. Ty3/Gypsy retrotransposon fossils in mammalian genomes: Did they evolve into new cellular functions? Mol. Biol. Evol. 2001, 18, 266–270. [Google Scholar] [CrossRef][Green Version]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Mazzuchelli, J.; Ferreira, I.A.; Poletto, A.B.; Fantinatti, B.E.A.; Martins, C. Cytogenetic mapping of the retroelements Rex1, Rex3 and Rex6 among cichlid Fish: New insights on the chromosomal distribution of transposable elements. Cytogenet. Genome Res. 2011, 133, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Splendore de Borba, R.; Lourenço da Silva, E.; Parise-Maltempi, P.P. Chromosome mapping of retrotransposable elements Rex1 and Rex3 in Leporinus Spix, 1829 species (Characiformes: Anostomidae) and its relationships among heterochromatic segments and W sex chromosome. Mob. Genet. Elem. 2013, 3, e27460. [Google Scholar] [CrossRef]
- Wichman, H.A.; Van Den Bussche, R.A.; Hamilton, M.J.; Baker, R.J. Transposable elements and the evolution of genome organization in mammals. Genetica 1992, 86, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Seki, R.; Nishida, C.; Matsuda, Y. Molecular cloning and characterization of satellite DNA sequences from constitutive heterochromatin of the habu snake (Protobothrops flavoviridis, Viperidae) and the Burmese python (Python bivittatus, Pythonidae). Chromosoma 2015, 124, 529–539. [Google Scholar] [CrossRef]
- Prakhongcheep, O.; Thapana, W.; Suntronpong, A.; Singchat, W.; Pattanatanang, K.; Phatcharakullawarawat, R.; Muangmai, N.; Peyachoknagul, S.; Matsubara, K.; Ezaz, T.; et al. Lack of satellite DNA species-specific homogenization and relationship to chromosomal rearrangements in monitor lizards (Varanidae, Squamata). BMC Evol. Biol. 2017, 17, 193. [Google Scholar] [CrossRef]
- Yamada, K.; Nishida-Umehara, C.; Matsuda, Y. Molecular and cytogenetic characterization of site-specific repetitive DNA sequences in the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae). Chromosome Res. 2005, 13, 33–46. [Google Scholar] [CrossRef]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef]
- Dover, G.A. Molecular drive in multigene families: How biological novelties arise, spread and are assimilated. Trends Genet. 1986, 2, 159–165. [Google Scholar] [CrossRef]
- Adams, R.H.; Blackmon, H.; Reyes-Velasco, J.; Schield, D.R.; Card, D.C.; Andrew, A.L.; Waynewood, N.; Castoe, T.A. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Neff, B.D.; Gross, M.R. Microsatellite evolution in vertebrates: Inference from AC dinucleotide repeats. Evolution 2001, 55, 1717–1733. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Knopp, T.; Sarre, S.D.; Georges, A.; Ezaz, T. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink-tailed worm-lizard (Aprasia parapulchella, Pygopodidae, Squamata). Mol. Cytogenet. 2013, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Pokorná, M.J. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef] [PubMed]
- Augstenová, B.; Mazzoleni, S.; Kratochvíl, L.; Rovatsos, M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes 2018, 9, 5. [Google Scholar] [CrossRef]
- Singh, L.; Purdom, I.F.; Jones, K.W. Sex chromosome associated satellite DNA: Evolution and conservation. Chromosoma 1980, 79, 137–157. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Johnson Pokorná, M.; Augstenová, B.; Kratochvíl, L. Cytogenetics of the Javan file snake (Acrochordus javanicus) and the evolution of snake sex chromosomes. J. Zool. Syst. Evol. Res. 2018, 56, 117–125. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Castoe, T.A.; Hall, K.T.; Guibotsy Mboulas, M.L.; Gu, W.; Jason De Koning, A.P.; Fox, S.E.; Poole, A.W.; Vemulapalli, V.; Daza, J.M.; Mockler, T.; et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol. Evol. 2011, 3, 641–653. [Google Scholar] [CrossRef]
- Castoe, T.A.; De Koning, A.P.J.; Hall, K.T.; Card, D.C.; Schield, D.R.; Fujita, M.K.; Ruggiero, R.P.; Degner, J.F.; Daza, J.M.; Gu, W.; et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl. Acad. Sci. USA 2013, 110, 20645–20650. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, B.; Li, C.; Gilbert, M.T.P.; Jarvis, E.D.; Wang, J. Comparative genomic data of the avian phylogenomics project. Gigascience 2014, 3. [Google Scholar] [CrossRef]
- Tollis, M.; Boissinot, S. The transposable element profile of the Anolis genome. Mob. Genet. Elem. 2011, 1, 107–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kordis, D. Transposable elements in reptilian and avian (Sauropsida) genomes. Cytogenet. Genome Res. 2010, 127, 94–111. [Google Scholar] [CrossRef] [PubMed]
- Novick, P.A.; Smith, J.D.; Floumanhaft, M.; Ray, D.A.; Stéphane, B. The evolution and diversity of DNA transposons in the genome of the lizard Anolis carolinensis. Genome Biol. Evol. 2011, 3, 1–14. [Google Scholar] [CrossRef]
- Piskurek, O.; Nishihara, H.; Okada, N. The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene 2009, 441, 111–118. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Z.J.; Li, Q.Y.; Lian, J.M.; Zhou, Y.; Lu, B.Z.; Jin, L.J.; Qiu, P.X.; Zhang, P.; Zhu, W.B.; et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Green, R.E.; Braun, E.L.; Armstrong, J.; Earl, D.; Nguyen, N.; Hickey, G.; Vandewege, M.W.; St John, J.A.; Capella-Gutiérrez, S.; Castoe, T.A.; et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 2014, 346. [Google Scholar] [CrossRef]
- Rice, E.S.; Kohno, S.; St John, J.; Pham, S.; Howard, J.; Lareau, L.F.; O’Connell, B.L.; Hickey, G.; Armstrong, J.; Deran, A.; et al. Improved genome assembly of American alligator genome reveals conserved architecture of estrogen signaling. Genome Res. 2017, 27, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Organ, C.L.; Shedlock, A.M.; Meade, A.; Pagel, M.; Edwards, S.V. Origin of avian genome size and structure in non-avian dinosaurs. Nature 2007, 446, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.H.; Pan, S.K.; Hu, L.; Zhu, Y.; Xu, P.W.; Xia, J.Q.; Chen, H.; He, G.Y.; He, J.; Ni, X.W.; et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013, 23, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.Y.; Kojima, K.K.; Jurka, J.; Ray, D.A.; Smit, A.F.A.; Isberg, S.R.; Gongora, J. Evolution and gene capture in ancient endogenous retroviruses—Insights from the crocodilian genomes. Retrovirology 2014, 11, 71. [Google Scholar] [CrossRef]
- Kojima, K.K. A new class of SINEs with snRNA gene-derived heads. Genome Biol. Evol. 2015, 7, 1702–1712. [Google Scholar] [CrossRef]
- Endoh, H.; Okada, N. Total DNA transcription in vitro: A procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc. Natl. Acad. Sci. USA 1986, 83, 251–255. [Google Scholar] [CrossRef]
- Kajikawa, M.; Ohshima, K.; Okada, N. Determination of the entire sequence of turtle CR1: The first open reading frame of the turtle CR1 element encodes a protein with a novel zinc finger motif. Mol. Biol. Evol. 1997, 14, 1206–1217. [Google Scholar] [CrossRef]
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef]
- Tollis, M.; DeNardo, D.F.; Cornelius, J.A.; Dolby, G.A.; Edwards, T.; Henen, B.T.; Karl, A.E.; Murphy, R.W.; Kusumi, K. The Agassiz’s desert tortoise genome provides a resource for the conservation of a threatened species. PLoS ONE 2017, 12, e0177708. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Caputo, V.; Olmo, E. Characterisation of a GC-rich telomeric satellite DNA in Eumeces schneideri Daudin (Reptilia, Scincidae). Cytogenet. Genome Res. 2009, 125, 272–278. [Google Scholar] [CrossRef]
- Capriglione, T.; Cardone, A.; Odierna, G.; Olmo, E. Evolution of a centromeric satellite DNA and phylogeny of lacertid lizards. Comp. Biochem. Physiol. Part B Biochem. 1991, 100, 641–645. [Google Scholar] [CrossRef]
- Capriglione, T.; Cardone, A.; Odierna, G.; Olmo, E. Further data on the occurrence and evolution of satellite DNA families in the lacertid genome. Chromosome Res. 1994, 2, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Capriglione, T.; De Santo, M.G.; Odierna, G.; Olmo, E. An alphoid-like satellite DNA sequence is present in the genome of a lacertid lizard. J. Mol. Evol. 1998, 46, 240–244. [Google Scholar] [CrossRef]
- Ciobanu, D.; Grechko, V.V.; Darevsky, I.S.; Kramerov, D.A. New satellite DNA in Lacerta s. str. lizards (Sauria: Lacertidae): Evolutionary pathways and phylogenetic impact. J. Exp. Zool. Part B Mol. Dev. Evol. 2004, 302, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Grechko, V.V.; Ciobanu, D.G.; Darevsky, I.S.; Kramerov, D.A. Satellite DNA of lizards of the genus Lacerta s. str. (the group L. agilis), the family Lacertidae. Dokl. Biochem. Biophys. 2005, 400, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Giovannotti, M.; Cerioni, P.N.; Splendiani, A.; Ruggeri, P.; Olmo, E.; Barucchi, V.C. Slow evolving satellite DNAs: The case of a centromeric satellite in Chalcides ocellatus (Forskål, 1775) (Reptilia, Scincidae). Amphib. Reptil. 2014, 34, 401–411. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Rojo, V.; Olmo, E.; Slimani, T.; Splendiani, A.; Caputo Barucchi, V. Characterization of a satellite DNA in the genera Lacerta and Timon (Reptilia, Lacertidae) and its role in the differentiation of the W chromosome. J. Exp. Zool. Part B Mol. Dev. Evol. 2018, 330, 83–95. [Google Scholar] [CrossRef]
- Giovannotti, M.; S’Khifa, A.; Nisi Cerioni, P.; Splendiani, A.; Slimani, T.; Fioravanti, T.; Olmo, E.; Caputo Barucchi, V. Isolation and characterization of two satellite DNAs in Atlantolacerta andreanskyi (Werner, 1929) (Reptilia, Lacertidae). J. Exp. Zool. Part B Mol. Dev. Evol. 2020, 334, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Rojo, V.; Martínez-Lage, A.; Giovannotti, M.; González-Tizón, A.M.; Cerioni, P.N.; Barucchi, V.C.; Galán, P.; Olmo, E.; Naveira, H. Evolutionary dynamics of two satellite DNA families in rock lizards of the genus Iberolacerta (Squamata, Lacertidae): Different histories but common traits. Chromosome Res. 2015, 23, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Ciobanu, D.G.; Grechko, V.V.; Kramerov, D.A.; Darevsky, I.S. A new subfamily of the satellite DNA, CLsatIV, of the lizard Darevskia lindholmi (Sauria, Laceridae): Structure and evolution. Dokl. Biochem. Biophys. 2003, 392, 263–267. [Google Scholar] [CrossRef]
- Ciobanu, D.G.; Grechko, V.V.; Darevsky, I.S. Molecular evolution of satellite DNA CLsat in lizards from the genus Darevskia (Sauria: Lacertidae): Correlation with species diversity. Russ. J. Genet. 2003, 39, 1292–1305. [Google Scholar] [CrossRef]
- Grechko, V.V.; Ciobanu, D.G.; Darevsky, I.S.; Kosushkin, S.A.; Kramerov, D.A. Molecular evolution of satellite DNA repeats and speciation of lizards of the genus Darevskia (Sauria: Lacertidae). Genome 2006, 49, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Capriglione, T.; Olmo, E.; Odierna, G.; Smith, D.I.; Miller, O.J. Genome composition and tandemly repetitive sequence at some centromeres in the lizard Podarcis s. sicula Raf. Genetica 1989, 79, 85–91. [Google Scholar] [CrossRef]
- Gatesy, J.; Amato, G.; Norell, M.; DeSalle, R.; Hayashi, C. Combined support for wholesale taxic atavism in gavialine crocodylians. Syst. Biol. 2003, 52, 403–422. [Google Scholar] [CrossRef]
- Harshman, J.; Huddleston, C.J.; Bollback, J.P.; Parsons, T.J.; Braun, M.J. True and false gharials: A nuclear gene phylogeny of Crocodylia. Syst. Biol. 2003, 52, 386–402. [Google Scholar] [CrossRef]
- Kawagoshi, T.; Uno, Y.; Nishida, C.; Matsuda, Y. The Staurotypus turtles and aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS ONE 2014, 9, e105315. [Google Scholar] [CrossRef][Green Version]
- Badenhorst, D.; Hillier, L.D.W.; Literman, R.; Montiel, E.E.; Radhakrishnan, S.; Shen, Y.; Minx, P.; Janes, D.E.; Warren, W.C.; Edwards, S.V.; et al. Physical mapping and refinement of the painted turtle genome (chrysemys picta) inform amniote genome evolution and challenge turtle-bird chromosomal conservation. Genome Biol. Evol. 2015, 7, 2038–2050. [Google Scholar] [CrossRef][Green Version]
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 2018, 26, 5–23. [Google Scholar] [CrossRef]
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef]
- Pritham, E.J.; Feschotte, C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci. USA 2007, 104, 1895–1900. [Google Scholar] [CrossRef]
- Böhne, A.; Brunet, F.; Galiana-Arnoux, D.; Schultheis, C.; Volff, J.N. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008, 16, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. Chromosomal variation and evolution. Science 1966, 152, 1463–14693. [Google Scholar] [CrossRef] [PubMed]
- Ålund, M. Gametes and Speciation: From Prezygotic to Postzygotic Isolation; Department of Animal Ecology, Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2012; Volume 100. [Google Scholar]
- Islam, F.B.; Ishishita, S.; Uno, Y.; Mollah, M.B.R.; Srikulnath, K.; Matsuda, Y. Male hybrid sterility in the mule duck is associated with meiotic arrest in primary spermatocytes. Poult. Sci. J. 2013, 50, 311–320. [Google Scholar] [CrossRef]
- Hurst, G.D.D.; Schilthuizen, M. Selfish genetic elements and speciation. Heredity (Edinburgh) 1998, 80, 2–8. [Google Scholar] [CrossRef]
- Jurka, J.; Bao, W.; Kojima, K.K. Families of transposable elements, population structure and the origin of species. Biol. Direct 2011, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Belyayev, A. Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Matute, D.R. The role of transposable elements in speciation. Genes (Basel) 2018, 9, 254. [Google Scholar] [CrossRef]
- Verneau, O.; Catzeflis, F.; Furano, A.V. Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc. Natl. Acad. Sci. USA 1998, 95, 11284–11289. [Google Scholar] [CrossRef] [PubMed]
- Dobigny, G.; Ozouf-Costaz, C.; Waters, P.D.; Bonillo, C.; Coutanceau, J.P.; Volobouev, V. LINE-1 amplification accompanies explosive genome repatterning in rodents. Chromosome Res. 2004, 12, 787–793. [Google Scholar] [CrossRef]
- Ray, D.A.; Feschotte, C.; Pagan, H.J.T.; Smith, J.D.; Pritham, E.J.; Arensburger, P.; Atkinson, P.W.; Craig, N.L. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008, 18, 717–728. [Google Scholar] [CrossRef] [PubMed]
- De Boer, J.G.; Yazawa, R.; Davidson, W.S.; Koop, B.F. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genom. 2007, 8, 422. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Hernandez, S.S.; Flores-Benabib, J.; Smith, E.N.; Feschotte, C. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol. Biol. Evol. 2012, 29, 503–515. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kraaijeveld, K. Genome Size and Species Diversification. Evol. Biol. 2010, 37, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, L.; Raab, M.; Sperlich, D. Satellite DNA and speciation: A species specific satellite DNA of Drosophila guanche. J. Zool. Syst. Evol. Res. 1989, 27, 84–93. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef]
- Adega, F.; Guedes-Pinto, H.; Chaves, R. Satellite DNA in the karyotype evolution of domestic animals—Clinical considerations. Cytogenet. Genome Res. 2009, 126, 12–20. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Castresana, J.; Robinson, T.J. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006, 7. [Google Scholar] [CrossRef]
- Farré, M.; Bosch, M.; López-Giráldez, F.; Ponsà, M.; Ruiz-Herrera, A. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Vieira-Da-Silva, A.; Louzada, S.; Adega, F.; Chaves, R. A high-resolution comparative chromosome map of Cricetus cricetus and Peromyscus eremicus reveals the involvement of constitutive heterochromatin in breakpoint regions. Cytogenet. Genome Res. 2015, 145, 59–67. [Google Scholar] [CrossRef]
- De La Fuente, R.; Baumann, C.; Viveiros, M.M. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development 2015, 142, 1806–1817. [Google Scholar] [CrossRef]
- Giunta, S.; Funabiki, H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl. Acad. Sci. USA 2017, 114, 1928–1933. [Google Scholar] [CrossRef]
- Srikulnath, K.; Uno, Y.; Matsubara, K.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Nishida, C.; Matsuda, Y. Chromosomal localization of the 18S–28S and 5s rRNA genes and (TTAGGG)n sequences of butterfly lizards (Leiolepis belliana belliana and Leiolepis boehmei, Agamidae, Squamata). Genet. Mol. Biol. 2011, 34, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes (Basel) 2020, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Feliciello, I.; Akrap, I.; Brajkovi, J.; Zlatar, I.; Ugarkovic, D. Satellite DNA as a driver of population divergence in the red flour beetle tribolium castaneum. Genome Biol. Evol. 2014, 7, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Farré, M.; Robinson, T.J.; Ruiz-Herrera, A. An integrative breakage model of genome architecture, reshuffling and evolution: The integrative breakage model of genome evolution, a novel multidisciplinary hypothesis for the study of genome plasticity. BioEssays 2015, 37, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Chaves, R.; Louzada, S.; Meles, S.; Wienberg, J.; Adega, F. Praomys tullbergi (Muridae, Rodentia) genome architecture decoded by comparative chromosome painting with Mus and Rattus. Chromosome Res. 2012, 20, 673–683. [Google Scholar] [CrossRef]
- Farré, M.; Narayan, J.; Slavov, G.T.; Damas, J.; Auvil, L.; Li, C.; Jarvis, E.D.; Burt, D.W.; Griffin, D.K.; Larkin, D.M. Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol. Evol. 2016, 8, 2442–2451. [Google Scholar] [CrossRef]
- Badenhorst, D.; Stanyon, R.; Engstrom, T.; Valenzuela, N. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 2013, 21, 137–147. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Tarui, H.; Ishishita, S.; Takagi, C.; Nishimura, O.; Ishijima, J.; Ota, H.; Kosaka, A.; Matsubara, K.; et al. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 2012, 7, e0053027. [Google Scholar] [CrossRef]
- Giovannotti, M.; Caputo, V.; O’Brien, P.C.M.; Lovell, F.L.; Trifonov, V.; Nisi Cerioni, P.; Olmo, E.; Ferguson-Smith, M.A.; Rens, W. Skinks (reptilia: Scincidae) have highly conserved karyotypes as revealed by chromosome painting. Cytogenet. Genome Res. 2010, 127, 224–231. [Google Scholar] [CrossRef]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. genome evolution in reptilia, the sister group of mammals. Annu. Rev. Genom. Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef]
- Trifonov, V.A.; Paoletti, A.; Caputo Barucchi, V.; Kalinina, T.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Giovannotti, M. Comparative chromosome painting and NOR distribution suggest a complex hybrid origin of triploid Lepidodactylus lugubris (Gekkonidae). PLoS ONE 2015, 10, e0132380. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; O’Connor, R.; Farré, M.; Lenis, V.P.E.; Martell, H.J.; Mandawala, A.; Fowler, K.; Joseph, S.; Swain, M.T.; Griffin, D.K.; et al. Upgrading short-read animal genome assemblies to chromosome level using comparative genomics and a universal probe set. Genome Res. 2017. [Google Scholar] [CrossRef]
- O’Connor, R.E.; Romanov, M.N.; Kiazim, L.G.; Barrett, P.M.; Farré, M.; Damas, J.; Ferguson-Smith, M.; Valenzuela, N.; Larkin, D.M.; Griffin, D.K. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat. Commun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Matsubara, K.; Sakaizumi, M. Molecular cytogenetic identification and characterization of Robertsonian chromosomes in the large Japanese field mouse (Apodemus speciosus) using FISH. Zoolog. Sci. 2012, 29, 709–713. [Google Scholar] [CrossRef]
- Lönnig, W.-E.; Saedler, H. Chromosome Rearrangements and Transposable Elements. Annu. Rev. Genet. 2002, 36, 389–410. [Google Scholar] [CrossRef]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity (Edinburgh) 2012, 108, 59–67. [Google Scholar] [CrossRef]
- Sankoff, D. The where and wherefore of evolutionary breakpoints. J. Biol. 2009, 8, 66. [Google Scholar] [CrossRef]
- Longo, M.S.; Carone, D.M.; Green, E.D.; O’Neill, M.J.; O’Neill, R.J. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genom. 2009, 10. [Google Scholar] [CrossRef]
- Coghlan, A.; Eichler, E.E.; Oliver, S.G.; Paterson, A.H.; Stein, L. Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 2005, 21, 673–682. [Google Scholar] [CrossRef]
- Olmo, E. Rate of chromosome changes and speciation in reptiles. Genetica 2005, 125, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Hall, W.P. Chromosome variation, genomics, speciation and evolution in sceloporus lizards. Cytogenet. Genome Res. 2010, 127, 143–165. [Google Scholar] [CrossRef]
- Lu, W.; Zhang, Y.; Liu, D.; Songyang, Z.; Wan, M. Telomeres-structure, function, and regulation. Exp. Cell Res. 2013, 319, 133–141. [Google Scholar] [CrossRef]
- Singchat, W.; Kraichak, E.; Tawichasri, P.; Tawan, T.; Suntronpong, A.; Sillapaprayoon, S.; Phatcharakullawarawat, R.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; et al. Dynamics of telomere length in captive Siamese cobra (Naja kaouthia) related to age and sex. Ecol. Evol. 2019, 9, 6366–6377. [Google Scholar] [CrossRef]
- Bolzán, A.D.; Bianchi, M.S. Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat. Res. Rev. Mutat. Res. 2006, 612, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, A.; Nergadze, S.G.; Santagostino, M.; Giulotto, E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2009, 122, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Slijepcevic, P.; Xiao, Y.; Dominguez, I.; Natarajan, A.T. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma 1996, 104, 596–604. [Google Scholar] [CrossRef]
- Slijepcevic, P. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 1998, 107, 136–140. [Google Scholar] [CrossRef]
- Lee, B.; Sasi, R.; Lin, C.C. Interstitial localization of telomeric dima sequences in the indian muntjac chromosomes: Further evidence for tandem chromosome fusions in the karyotypic evolution of the asian muntjacs. Cytogenet. Genome Res. 1993, 63, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, K.C.M.; Bertolotto, C.E.V.; Yonenaga-Yassuda, Y.; Rodrigues, M.T. Banding patterns, heteromorphic sex chromosomes and Agstained NORs after pachytene stage in the meiosis of the Brazilian lizard Urostrophus vautieri (Squamata, Polychrotidae). Caryologia 1999, 52, 21–26. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710. [Google Scholar] [CrossRef]
- Ezaz, T.; Deakin, J.E. Repetitive sequence and sex chromosome evolution in vertebrates. Adv. Evol. Biol. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Deakin, J.E.; Ezaz, T. Tracing the evolution of amniote chromosomes. Chromosoma 2014, 123, 201–216. [Google Scholar] [CrossRef]
- Steinemann, S.; Steinemann, M. Retroelements: Tools for sex chromosome evolution. Cytogenet. Genome Res. 2005, 110, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Stiglec, R.; Veyrunes, F.; Marshall Graves, J.A. Relationships between Vertebrate ZW and XY Sex Chromosome Systems. Curr. Biol. 2006, 16, R736–R743. [Google Scholar] [CrossRef] [PubMed]
- Janzen, F.J.; Phillips, P.C. Exploring the evolution of environmental sex determination, especially in reptiles. J. Evol. Biol. 2006, 19, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Quinn, A.E.; Sarre, S.D.; O’Meally, D.; Georges, A.; Marshall Graves, J.A. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosom. Res. 2009, 17, 91–98. [Google Scholar] [CrossRef]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Matsuda, Y.; Miller, E.; Olsson, M. No interstitial telomeres on autosomes but remarkable amplification of telomeric repeats on the W sex chromosome in the sand lizard (Lacerta agilis). J. Hered. 2015, 106, 753–757. [Google Scholar] [CrossRef]
- O’Neill, M.J.; O’Neill, R.J. Sex chromosome repeats tip the balance towards speciation. Mol. Ecol. 2018, 27, 3783–3798. [Google Scholar] [CrossRef] [PubMed]
- O’Meally, D.; Patel, H.R.; Stiglec, R.; Sarre, S.D.; Georges, A.; Marshall Graves, J.A.; Ezaz, T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosom. Res. 2010, 18, 787–800. [Google Scholar] [CrossRef]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of Bkm sequences (GATA repeats): Predominant association with sex chromosomes and potential role in higher order chromatin organization and function. Bioinformatics 2003, 19, 681–685. [Google Scholar] [CrossRef]
- Demas, S.; Duronslet, M.; Wachtel, S.; Caillouet, C.; Nakamura, D. Sex-specific DNA in reptiles with temperature sex determination. J. Exp. Zool. 1990, 253, 319–324. [Google Scholar] [CrossRef]
- Ezaz, T.; Srikulnath, K.; Graves, J.A.M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements? J. Hered. 2017, 108, 94–105. [Google Scholar] [CrossRef]
- Matsubara, K.; O’Meally, D.; Sarre, S.D.; Georges, A.; Srikulnath, K.; Ezaz, T. ZW sex chromosomes in Australian dragon lizards (Agamidae) originated from a combination of duplication and translocation in the nucleolar organising region. Genes (Basel) 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.A.; Haiduk, M.W.; de Queiroz, K. Evolution and phylogenetic significance of ribosomal gene location in chromosomes of squamate reptiles. Copeia 1994, 1994, 302. [Google Scholar] [CrossRef]
- Ezaz, T.; Azad, B.; O’Meally, D.; Young, M.J.; Matsubara, K.; Edwards, M.J.; Zhang, X.; Holleley, C.E.; Deakin, J.E.; Marshall Graves, J.A.; et al. Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; O’Meally, D.; Sarre, S.D.; Georges, A.; Ezaz, T. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 2013, 21, 361–374. [Google Scholar] [CrossRef]
- Vicoso, B.; Emerson, J.J.; Zektser, Y.; Mahajan, S.; Bachtrog, D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013, 11, e1001643. [Google Scholar] [CrossRef]
- Matsubara, K.; Sarre, S.D.; Georges, A.; Matsuda, Y.; Marshall Graves, J.A.; Ezaz, T. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE 2014, 9, e95226. [Google Scholar] [CrossRef] [PubMed]
- Grützner, F.; Rens, W.; Tsend-Ayush, E.; El-Mogharbel, N.; O’Brien, P.C.; Jones, R.C.; Ferguson-Smith, M.A.; Graves, J.A.M. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 2004, 432, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Veyrunes, F.; Waters, P.D.; Miethke, P.; Rens, W.; McMillan, D.; Alsop, A.E.; Grützner, F.; Deakin, J.E.; Whittington, C.M.; Schatzkamer, K.; et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008, 18, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Kawai, A.; Ishijima, J.; Nishida, C.; Kosaka, A.; Ota, H.; Kohno, S.; Matsuda, Y. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 2009, 118, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Lai, Y.; Mostovoy, Y.; Holloway, A.K.; Iannucci, A.; Mak, A.; Fondi, M.; Orlandini, V.; Eckalbar, W.L.; Milan, M.; et al. Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 2019, 3, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Montiel, E.E.; Badenhorst, D.; Tamplin, J.; Burke, R.L.; Valenzuela, N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 2016, 126, 105–113. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef]
- Simakov, O.; Marlétaz, F.; Yue, J.X.; O’Connell, B.; Jenkins, J.; Brandt, A.; Calef, R.; Tung, C.H.; Huang, T.K.; Schmutz, J.; et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020, 4, 1–11. [Google Scholar] [CrossRef]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, E.; Webster, M.T.; Smith, N.G.C.; Burt, D.W.; Ellegren, H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005, 15, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Painter, T.S. The Y-chromosome in mammals. Science 1921, 53, 503–504. [Google Scholar] [CrossRef]
- Matthey, R.; Bovey, R. The chromosomal formula in five species of Chiroptera. Experientia 1948, 4, 26–27. [Google Scholar]
- Peccinini, D.; Frota-Pessoa, O.; Ferrari, I. Sex determination of the “pseudo-xo/xx” type in the brazilian lizard polychrus sp. (sauria, iguanidae). Caryologia 1971, 24, 129–139. [Google Scholar] [CrossRef]
- Fillon, V.; Morisson, M.; Zoorob, R.; Auffray, C.; Douaire, M.; Gellin, J.; Vignal, A. Identification of 16 chicken microchromosomes by molecular markers using two-colour fluorescence in situ hybridization (FISH). Chromosome Res. 1998, 6, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Solinhac, R.; Leroux, S.; Galkina, S.; Chazara, O.; Feve, K.; Vignoles, F.; Morisson, M.; Derjusheva, S.; Bed’hom, B.; Vignal, A.; et al. Integrative mapping analysis of chicken microchromosome 16 organization. BMC Genom. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Kuraku, S.; Tarui, H.; Nishimura, O.; Nishida, C.; Agata, K.; Kumazawa, Y.; Matsuda, Y. Intra-genomic GC heterogeneity in sauropsids: Evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genom. 2012, 13, 1–14. [Google Scholar] [CrossRef]
- Pokorná, M.; Kratochvíl, L.; Kejnovský, E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Shedlock, A.M.; Edwards, S. V Amniotes (Amniota). In The Timetree of Life; Oxford University Press: New York, NY, USA, 2009. [Google Scholar]
- Pokorná, M.; Altmanová, M.; Kratochvíl, L. Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrates. Chromosome Res. 2014, 22, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Singh, L. Evolution of karyotypes in snakes. Chromosoma 1972, 38, 185–236. [Google Scholar] [CrossRef] [PubMed]
- Altmanová, M.; Rovatsos, M.; Kratochvíl, L.; Johnson Pokorná, M. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae). Biol. J. Linn. Soc. 2016, 118, 618–633. [Google Scholar] [CrossRef]
- Tegelström, H.; Ryttman, H. Chromosomes in birds (Aves): Evolutionary implications of macro-and microchromosome numbers and lengths. Hereditas 1981, 94, 225–233. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution. Genes 2020, 11, 827. https://doi.org/10.3390/genes11070827
Ahmad SF, Singchat W, Jehangir M, Panthum T, Srikulnath K. Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution. Genes. 2020; 11(7):827. https://doi.org/10.3390/genes11070827
Chicago/Turabian StyleAhmad, Syed Farhan, Worapong Singchat, Maryam Jehangir, Thitipong Panthum, and Kornsorn Srikulnath. 2020. "Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution" Genes 11, no. 7: 827. https://doi.org/10.3390/genes11070827
APA StyleAhmad, S. F., Singchat, W., Jehangir, M., Panthum, T., & Srikulnath, K. (2020). Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution. Genes, 11(7), 827. https://doi.org/10.3390/genes11070827







