Genome-Wide Identification and Comparative Analysis of Myosin Gene Family in Four Major Cotton Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Myosins in Gossypium hirsutum, Gossypium barbadense, Gossypium raimondii and Gossypium arboretum
2.2. The Analysis of Myosin Proteins Characteristic
2.3. Phylogenetic Tree Construction and Evolution Analysis
2.4. Plant Material, Treatment and qRT-PCR Analysis
3. Results
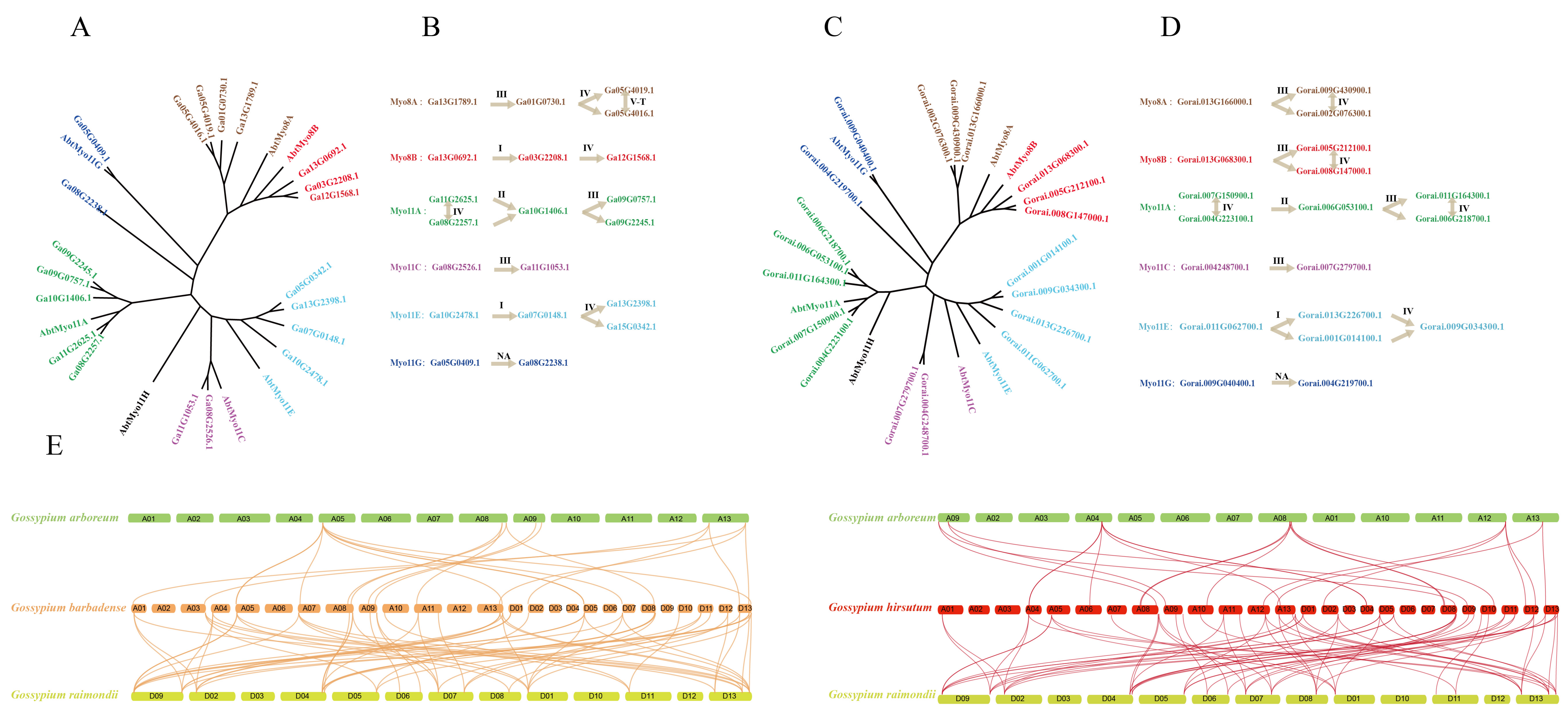
3.1. Identification of Myosin Genes in Four Cotton Species
3.2. Phylogenetic, Conserved Domain/Motifs, and Gene Structures Analysis in Four Cotton Species
3.3. Gene Duplication, Collinearity, and Evolutionary Analysis
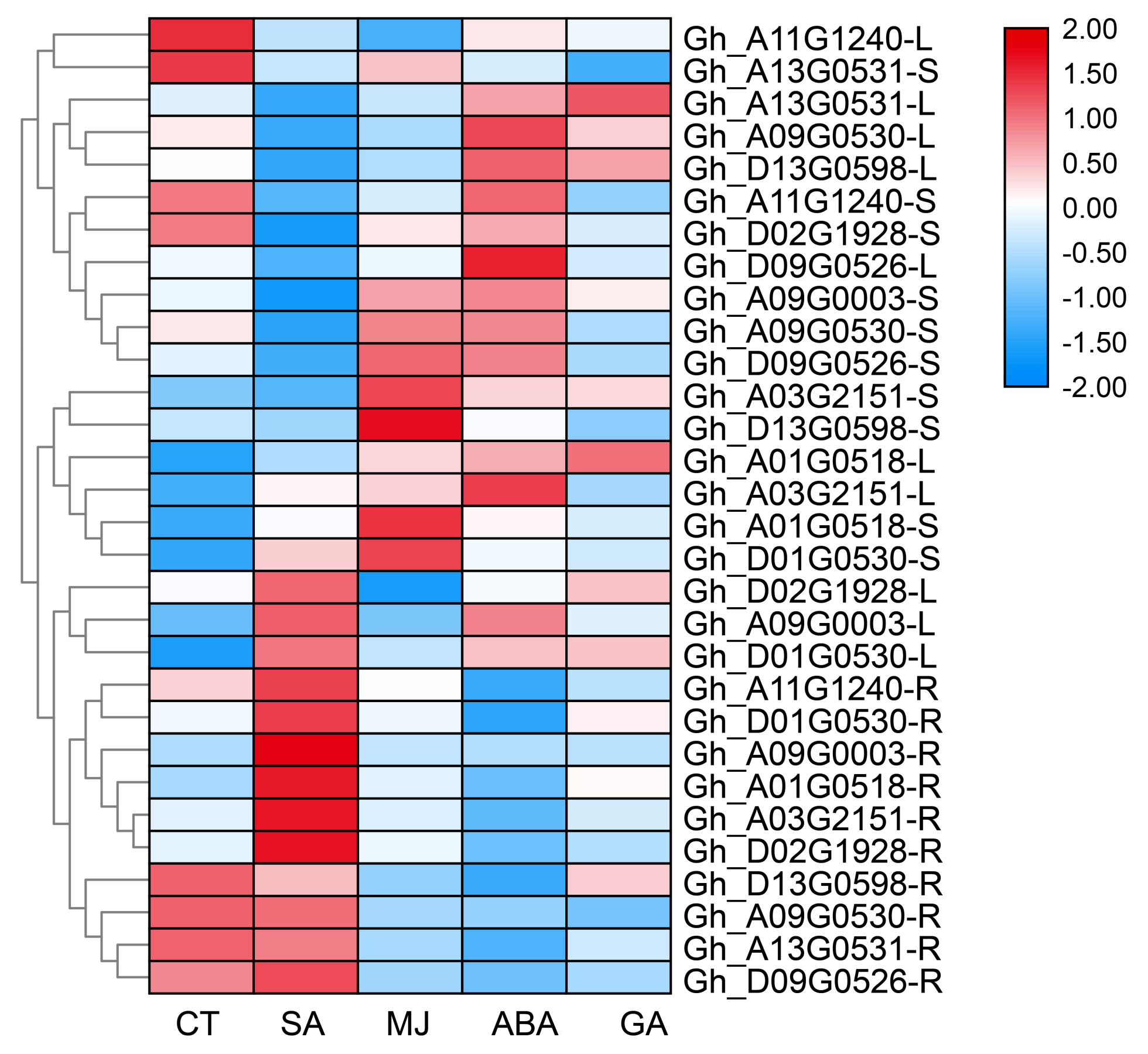
3.4. Cis-Element Analysis, Expression Pattern, and Hormone Treatment Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tominaga, M.; Nakano, A. Plant-specific myosin XI, a molecular perspective. Front. Plant Sci. 2012, 3, 211. [Google Scholar] [CrossRef] [Green Version]
- Kollmar, M.; Muhlhausen, S. Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 2017, 17, 211. [Google Scholar] [CrossRef] [Green Version]
- Krendel, M.; Mooseker, M.S. Myosins: Tails (and heads) of functional diversity. Physiology (Bethesda) 2005, 20, 239–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.Y.; Cai, M.; Ramachandran, S. ORYZA SATIVA MYOSIN XI B controls pollen development by photoperiod-sensitive protein localizations. Dev. Biol. 2007, 304, 579–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.F.; Zhong, M.Y.; Wang, J.J.; Zhang, J.S.; Tang, Y.P.; Wang, G.; Song, R.T. Genome-wide identification, splicing, and expression analysis of the myosin gene family in maize (Zea mays). J. Exp. Bot. 2014, 65, 923–938. [Google Scholar] [CrossRef] [Green Version]
- Duan, Z.R.; Tominaga, M. Actin-myosin XI: An intracellular control network in plants. Biochem. Biophys. Res. Commun. 2018, 506, 403–408. [Google Scholar] [CrossRef]
- Shimmen, T. The sliding theory of cytoplasmic streaming: Fifty years of progress. J. Plant Res. 2007, 120, 31–43. [Google Scholar] [CrossRef]
- Tominaga, M.; Kimura, A.; Yokota, E.; Haraguchi, T.; Shimmen, T.; Yamamoto, K.; Nakano, A.; Ito, K. Cytoplasmic Streaming Velocity as a Plant Size Determinant. Dev. Cell 2013, 27, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Tominaga, M.; Yokota, E.; Sonobe, S.; Shimmen, T. Mechanism of inhibition of cytoplasmic streaming by a myosin inhibitor, 2,3-butanedione monoxime. Protoplasma 2000, 213, 46–54. [Google Scholar] [CrossRef]
- Cai, C.; Henty-Ridilla, J.L.; Szymanski, D.B.; Staiger, C.J. Arabidopsis Myosin XI: A Motor Rules the Tracks. Plant Physiol. 2014, 166, 1359–1370. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Prokhnevsky, A.I.; Dolja, V.V. Class XI Myosins Are Required for Development, Cell Expansion, and F-Actin Organization in Arabidopsis. Plant Cell 2010, 22, 1883–1897. [Google Scholar] [CrossRef] [Green Version]
- Prokhnevsky, A.I.; Peremyslov, V.V.; Dolja, V.V. Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. USA 2008, 105, 19744–19749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.N.; Day, I.S. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001, 2. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, T.; Tominaga, M.; Matsumoto, R.; Sato, K.; Nakano, A.; Yamamoto, K.; Ito, K. Molecular Characterization and Subcellular Localization of Arabidopsis Class VIII Myosin, ATM1. J. Biol. Chem. 2014, 289, 12343–12355. [Google Scholar] [CrossRef] [Green Version]
- Golomb, L.; Abu-Abied, M.; Belausov, E.; Sadot, E. Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 2008, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mühlhausen, S.; Kollmar, M. Whole genome duplication events in plant evolution reconstructed and predicted using myosin motor proteins. BMC Evol. Biol. 2013, 13, 202. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, S.; Nowak, J.; Wang, G.; Han, L.; Feng, Z.; Mendrinna, A.; Ma, Y.; Wang, H.; Zhang, X.; et al. Live-cell imaging of the cytoskeleton in elongating cotton fibres. Nat. Plants 2019, 5, 498–504. [Google Scholar] [CrossRef]
- Thyssen, G.N.; Fang, D.D.; Turley, R.B.; Florane, C.B.; Li, P.; Mattison, C.P.; Naoumkina, M. A Gly65Val substitution in an actin, GhACT_LI1, disrupts cell polarity and F-actin organization resulting in dwarf, lintless cotton plants. Plant J. 2017, 90, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Odronitz, F.; Kollmar, M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007, 8, R196. [Google Scholar] [CrossRef] [Green Version]
- Henn, A.; Sadot, E. The unique enzymatic and mechanistic properties of plant myosins. Curr. Opin. Plant Biol. 2014, 22, 65–70. [Google Scholar] [CrossRef]
- Sattarzadeh, A.; Franzen, R.; Schmelzer, E. The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil. Cytoskelet. 2008, 65, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Peremyslov, V.; Mockler, T.C.; Filichkin, S.A.; Fox, S.E.; Jaiswal, P.; Makarova, K.S.; Koonin, E.V.; Dolja, V.V. Expression, Splicing, and Evolution of the Myosin Gene Family in Plants. Plant Physiol. 2011, 155, 1191–1204. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, J.D.; Fang, L.; Zhang, Z.Y.; Ma, W.; Niu, Y.C.; Ju, L.Z.; Deng, J.Q.; Zhao, T.; Lian, J.M.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.Z.; Hu, Y.; Jiang, W.K.; Fang, L.; Guan, X.Y.; Chen, J.D.; Zhang, J.B.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.Y.; Ramachandran, S. Identification and molecular characterization of myosin gene family in Oryza sativa genome. Plant Cell Physiol. 2004, 45, 590–599. [Google Scholar] [CrossRef] [Green Version]
- Haraguchi, T.; Ito, K.; Duan, Z.; Rula, S.; Takahashi, K.; Shibuya, Y.; Hagino, N.; Miyatake, Y.; Nakano, A.; Tominaga, M. Functional Diversity of Class XI Myosins in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2268–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.G.; Fan, G.Y.; Lu, C.R.; Xiao, G.H.; Zou, C.S.; Kohel, R.J.; Ma, Z.Y.; Shang, H.H.; Ma, X.F.; Wu, J.Y.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tingler, M.; Kurz, S.; Maerker, M.; Ott, T.; Fuhl, F.; Schweickert, A.; LeBlanc-Straceski, J.M.; Noselli, S.; Blum, M. A Conserved Role of the Unconventional Myosin 1d in Laterality Determination. Curr. Biol. 2018, 28, 810–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peremyslov, V.V.; Cole, R.A.; Fowler, J.E.; Dolja, V.V. Myosin-Powered Membrane Compartment Drives Cytoplasmic Streaming, Cell Expansion and Plant Development. PLoS ONE 2015, 10, e0139331. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Morgun, E.A.; Kurth, E.G.; Makarova, K.S.; Koonin, E.V.; Dolja, V.V. Identification of Myosin XI Receptors in Arabidopsis Defines a Distinct Class of Transport Vesicles. Plant Cell 2013, 25, 3022–3038. [Google Scholar] [CrossRef] [Green Version]
- Sparkes, I.A. Motoring around the plant cell: Insights from plant myosins. Biochem. Soc. Trans. 2010, 38, 833–838. [Google Scholar] [CrossRef] [Green Version]
- Lv, F.N.; Han, M.Y.; Ge, D.D.; Dong, H.; Zhang, X.T.; Li, L.F.; Zhang, P.P.; Zhang, Z.Q.; Sun, J.; Liu, K.; et al. GhVLN4 is involved in cell elongation via regulation of actin organization. Planta 2017, 246, 687–700. [Google Scholar] [CrossRef]
- Wang, J.A.; Wang, H.Y.; Zhao, P.M.; Han, L.B.; Jiao, G.L.; Zheng, Y.Y.; Huang, S.J.; Xia, G.X. Overexpression of a Profilin (GhPFN2) Promotes the Progression of Developmental Phases in Cotton Fibers. Plant Cell Physiol. 2010, 51, 1276–1290. [Google Scholar] [CrossRef] [Green Version]
- Li, X.B.; Fan, X.P.; Wang, X.L.; Cai, L.; Yang, W.C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 2005, 17, 859–875. [Google Scholar] [CrossRef] [Green Version]
- Murthy, K.; Wadsworth, P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 2005, 15, 724–731. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.Z.; Ritchie, J.A.; Pan, A.H.; Quatrano, R.S.; Bezanilla, M. Myosin VIII Regulates Protonemal Patterning and Developmental Timing in the Moss Physcomitrella patens. Mol. Plant 2011, 4, 909–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Zhao, Z.; Wang, N.; Azhar, M.T.; Du, X. Genome-Wide Identification and Comparative Analysis of Myosin Gene Family in Four Major Cotton Species. Genes 2020, 11, 731. https://doi.org/10.3390/genes11070731
Ma C, Zhao Z, Wang N, Azhar MT, Du X. Genome-Wide Identification and Comparative Analysis of Myosin Gene Family in Four Major Cotton Species. Genes. 2020; 11(7):731. https://doi.org/10.3390/genes11070731
Chicago/Turabian StyleMa, Chenhui, Zibo Zhao, Na Wang, Muhammad Tehseen Azhar, and Xiongming Du. 2020. "Genome-Wide Identification and Comparative Analysis of Myosin Gene Family in Four Major Cotton Species" Genes 11, no. 7: 731. https://doi.org/10.3390/genes11070731
APA StyleMa, C., Zhao, Z., Wang, N., Azhar, M. T., & Du, X. (2020). Genome-Wide Identification and Comparative Analysis of Myosin Gene Family in Four Major Cotton Species. Genes, 11(7), 731. https://doi.org/10.3390/genes11070731




