Transcriptomic Analysis of Gill and Kidney from Asian Seabass (Lates calcarifer) Acclimated to Different Salinities Reveals Pathways Involved with Euryhalinity
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
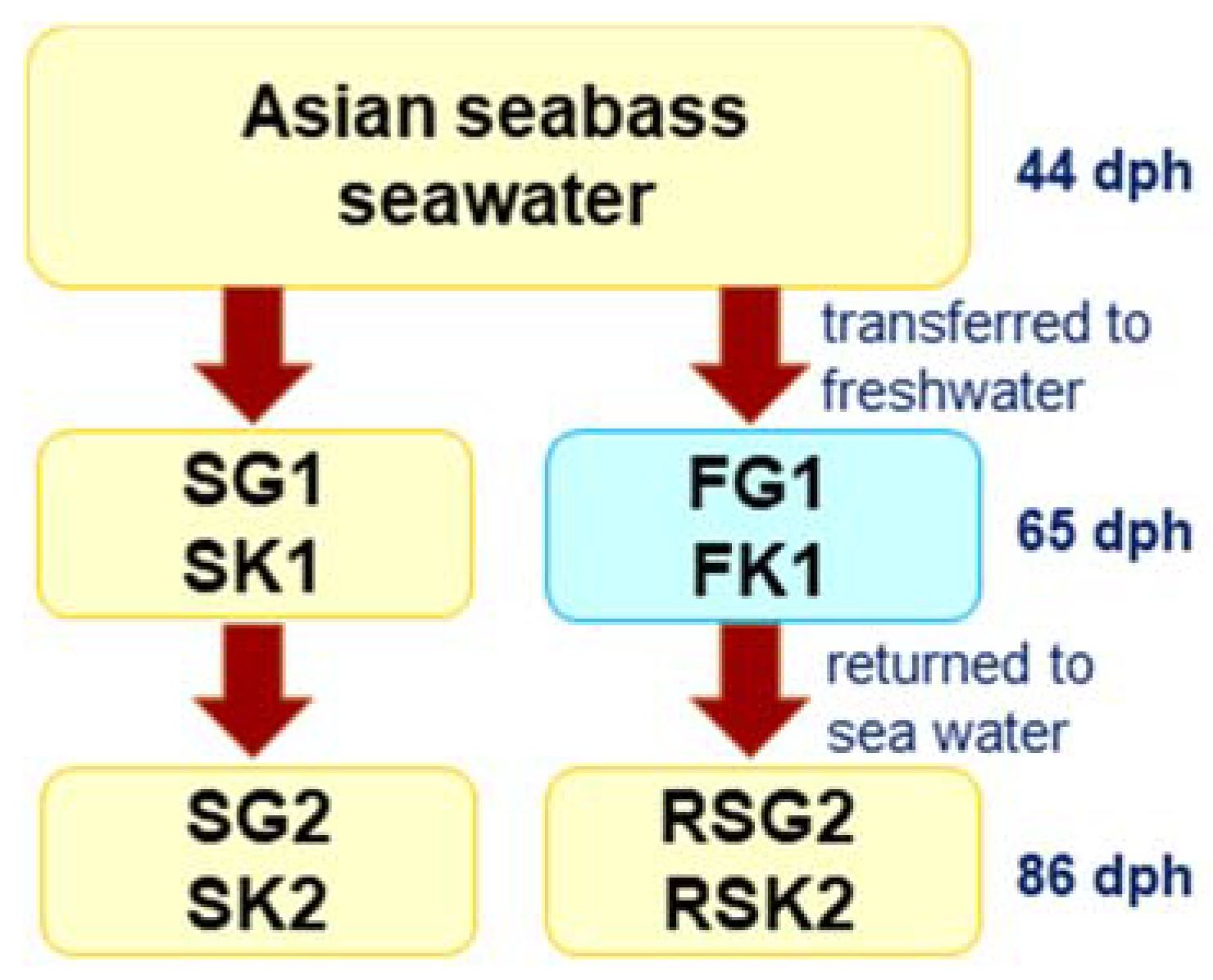
2.2. Experimental Setup and Sampling
2.3. RNA Extraction, Library Construction, and Sequencing
2.4. Mapping of Reads and Differential Expression Analyses
2.5. Gene Ontology and Pathway Enrichment Analyses
2.6. Histology
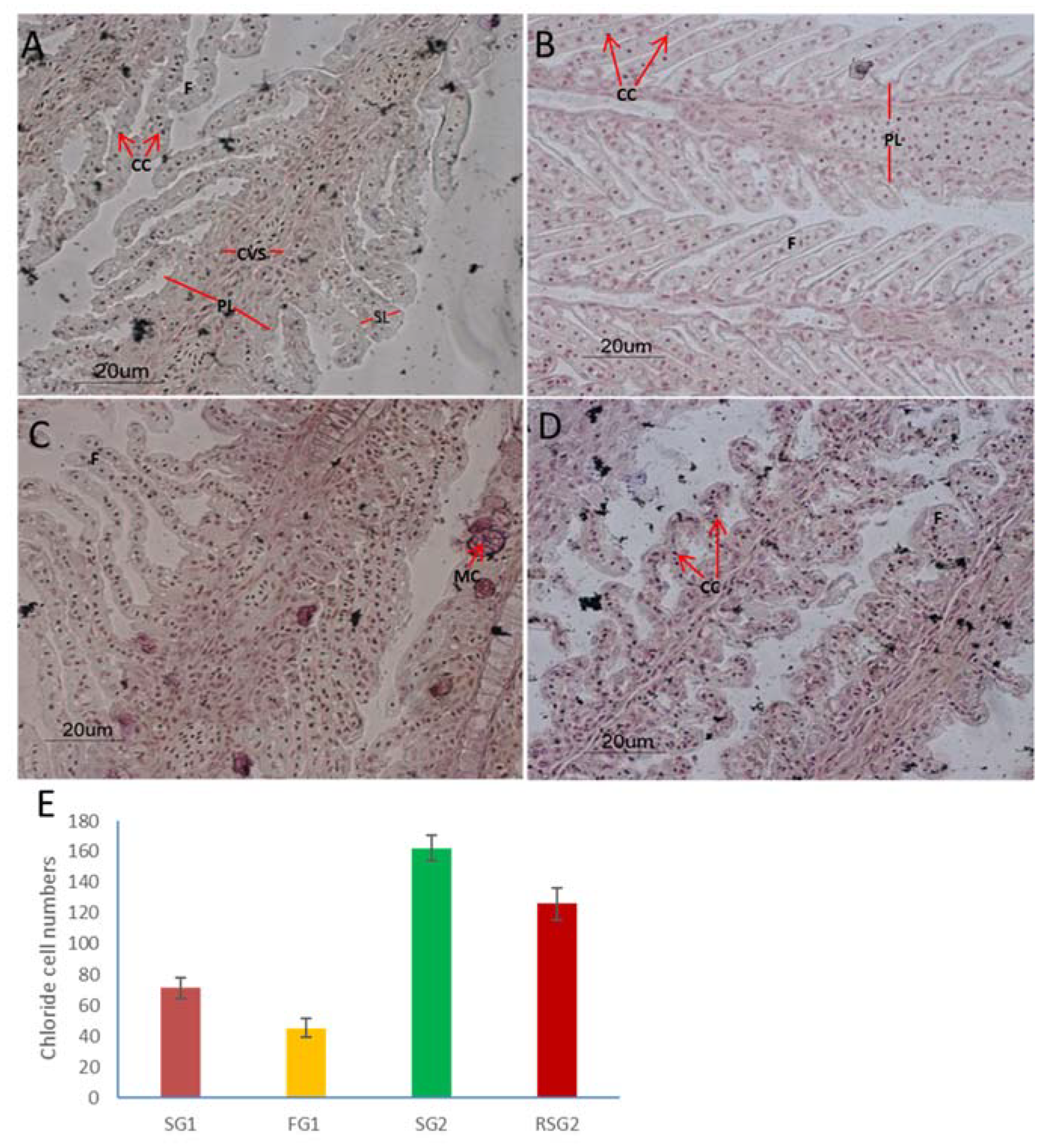
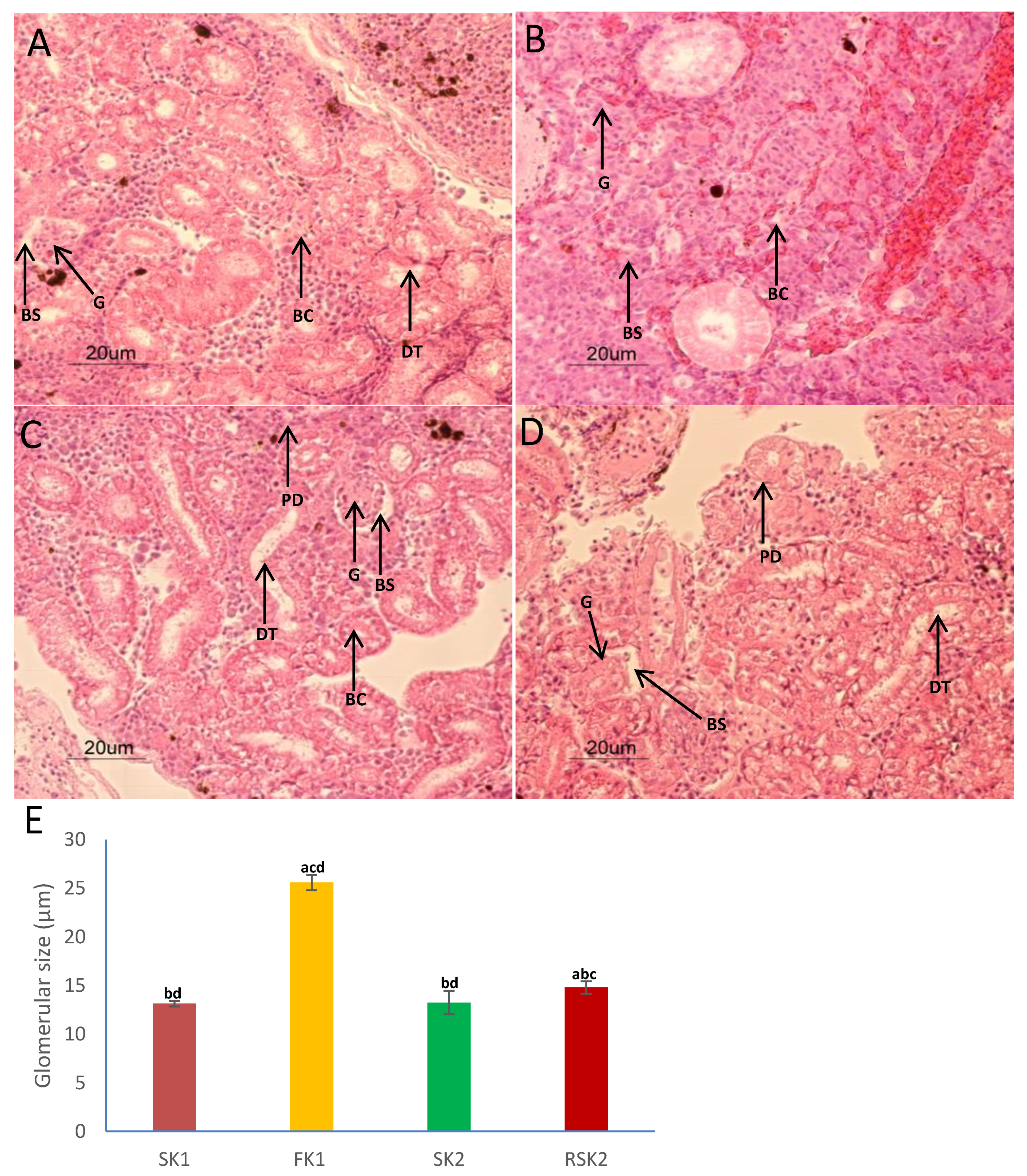
2.7. Counting Gill Chloride Cell Numbers and Kidney Glomerular Size
3. Results and Discussion
3.1. Changes in Gills and Kidney Histology Accompanied the Acclimation Response to Different Salinities
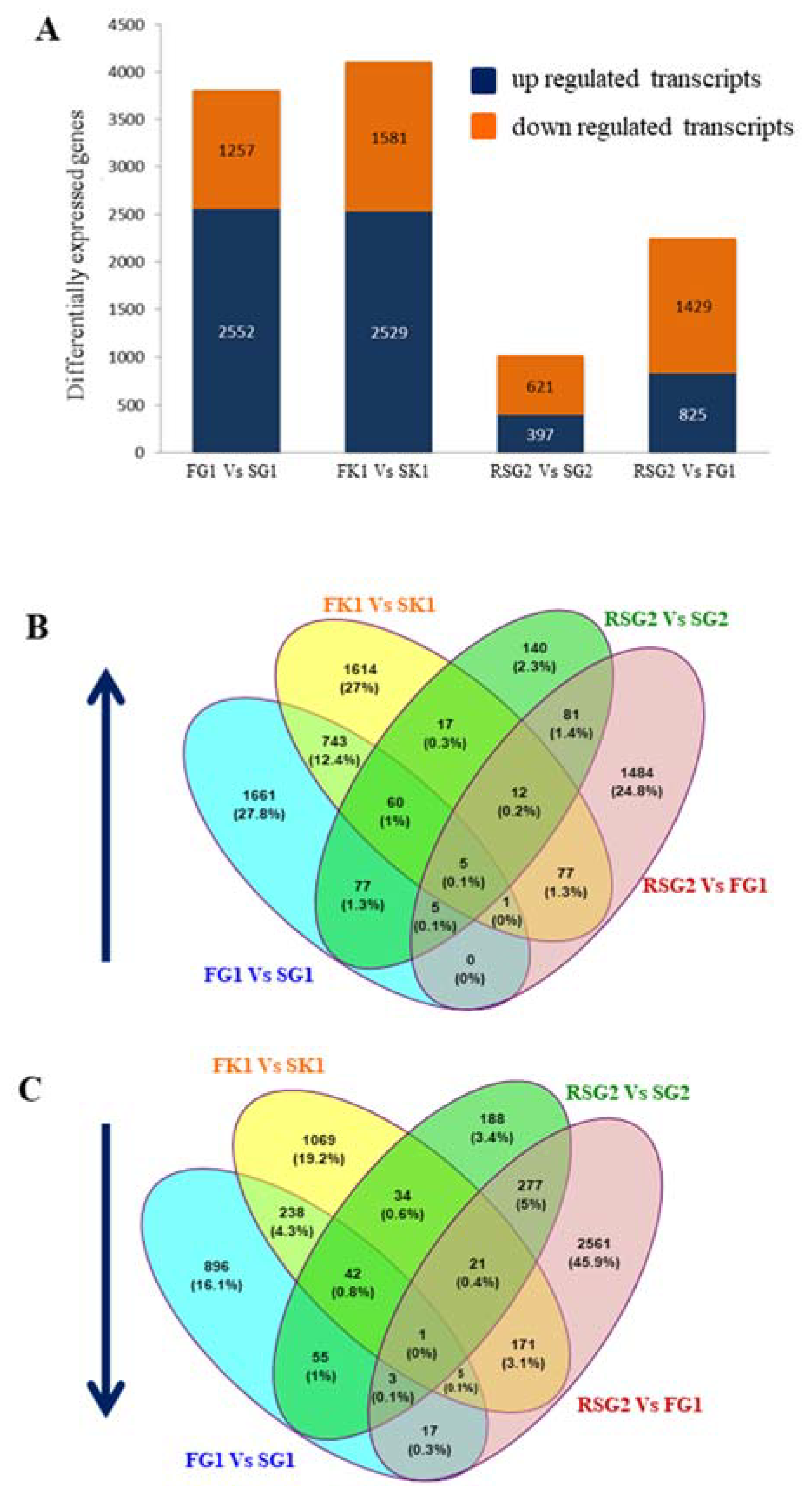
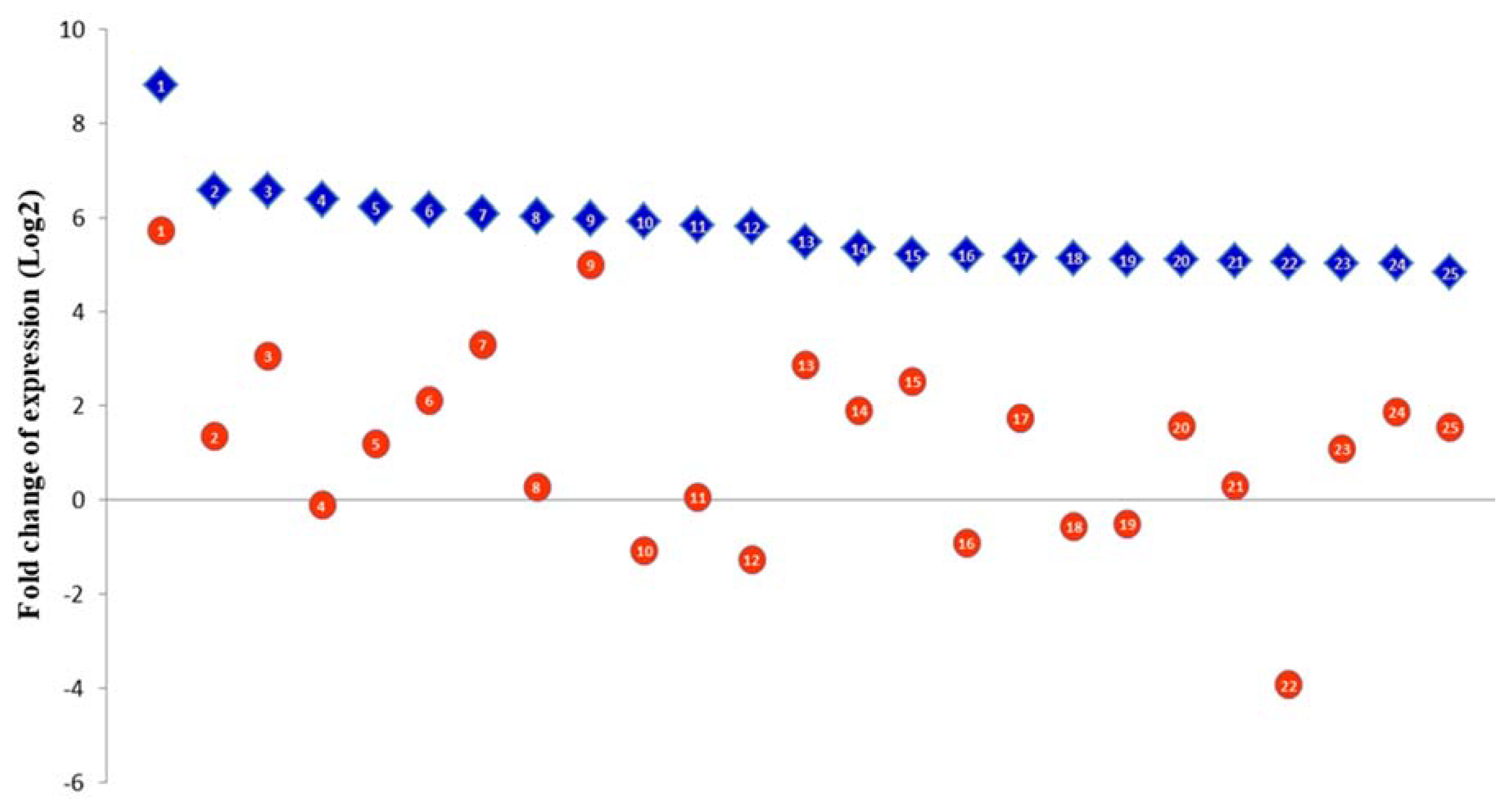
3.2. RNA-Seq Analyses
3.3. The Euryhaline Trait is Pliable
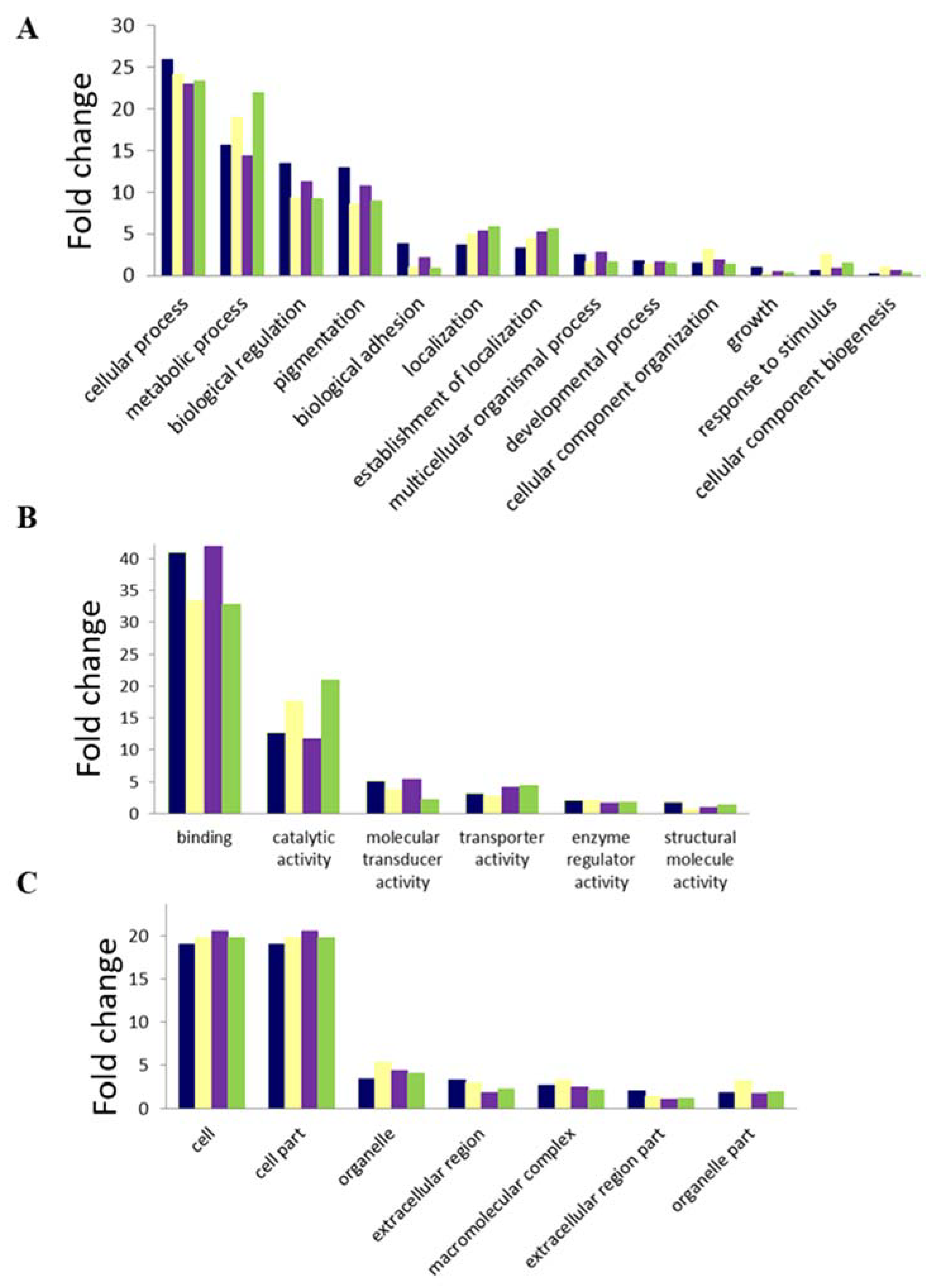
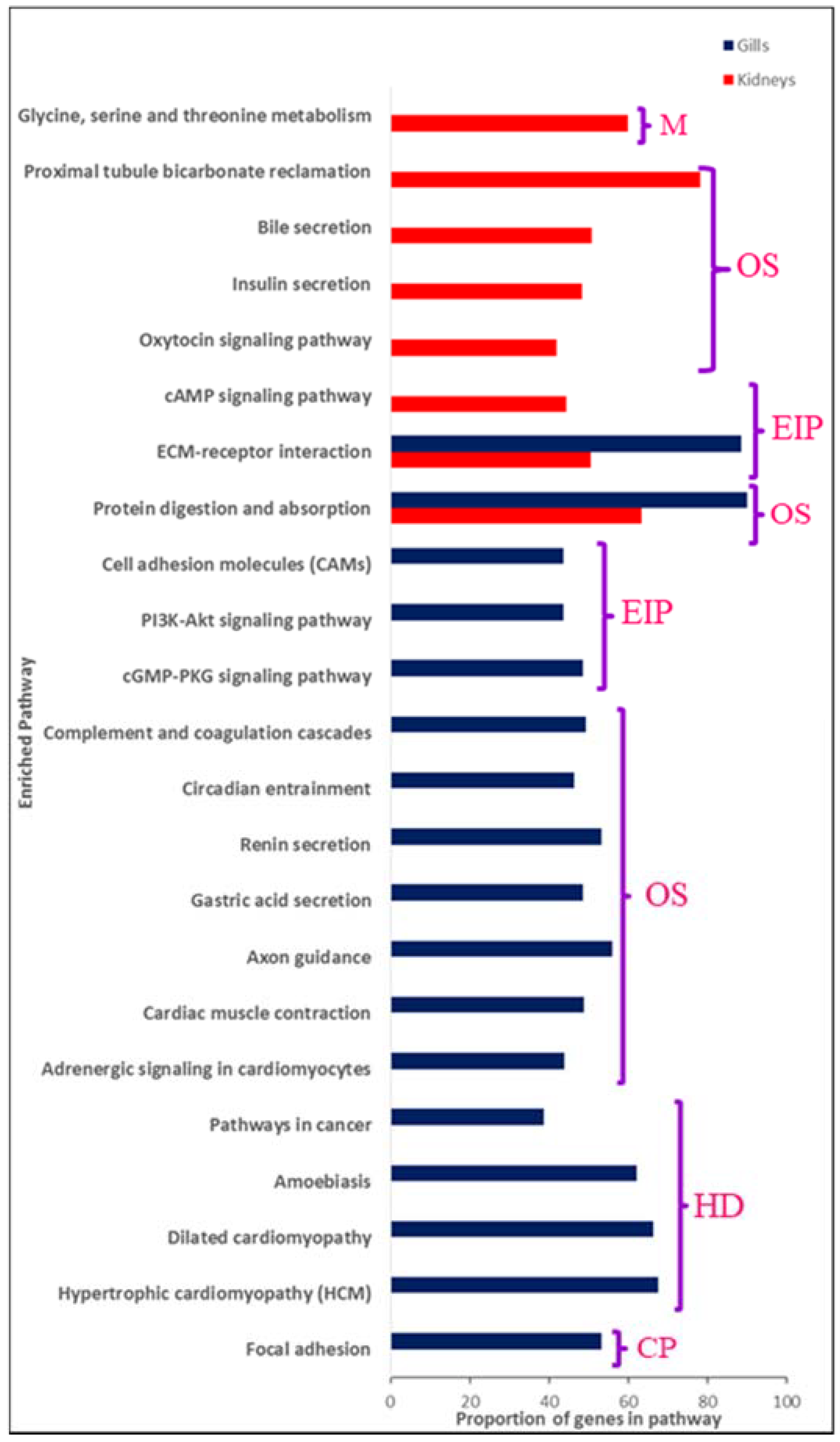
4. GO and Pathway Analyses
4.1. Genes Involved in Osmoregulation, Tissue/organ Morphogenesis and Regulation of Cell Volume Are Important for Salinity Acclimation
4.2. Ion Loss under Freshwater Conditions Seems to Be Restricted by Upregulation of Several Tight Junction Proteins
4.3. Genes Rrequired for Growth and Development Were Upregulated under Freshwater Conditions
4.4. Genes Coding for Mucins Were Upregulated under Saline Conditions
4.5. Hormones Play an Important Role in Regulating Euryhalinity
4.6. Circadian Entrainment Could Be Important for Establishing Euryhalinity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. Available online: www.fishbase.org (accessed on 27 March 2016).
- Kültz, D. Physiological mechanisms used by fish to cope with salinity stress. J. Exp. Biol. 2015, 218, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.T.; McCormick, D.S. Euryhalinity in an evolutionary context. Fish Physiol. 2012, 32, 477–533. [Google Scholar]
- Kirsch, R.; Humbert, W.; Rodeau, J.L. Control of the blood osmolarity in fishes with references to the functional anatomy of the gut. In Osmoregulation in Estuarine and Marine Animals: Proceedings of the Invited Lectures to a Symposium Organized within the 5th Conference of the European Society for Comparative Physiology and Biochemistry—Taormina, Sicily, Italy, 5–8 September 1983; Pequeux, A., Gilles, R., Bolis, L., Eds.; Springer: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- Beyenbach, K.W. Kidneys sans glomeruli. Am. J. Physiol. Ren. Physiol. 2004, 286, F811–F827. [Google Scholar] [CrossRef] [PubMed]
- Marshall, W.; Grosell, M. Ion transport, osmoregulation, and acid-base balance. Physiol. Fishes 2006, 3, 177–230. [Google Scholar]
- Takei, Y.; McCormick, S. Hormonal control of fish euryhalinity. Euryhaline Fishes. Fish. Physiol. 2013, 32, 69–123. [Google Scholar]
- Lam, S.H.; Lui, E.Y.; Li, Z.; Cai, S.; Sung, W.-K.; Mathavan, S.; Lam, T.J.; Ip, Y.K. Differential transcriptomic analyses revealed genes and signaling pathways involved in iono-osmoregulation and cellular remodeling in the gills of euryhaline Mozambique tilapia, Oreochromis mossambicus. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Genten, F.; Terwinghe, E.; Danguy, A. Atlas of fish histology. Sci. Publ. 2009. [Google Scholar] [CrossRef]
- Cutler, C.P.; Cramb, G. Two isoforms of the Na+/K+/2Cl- cotransporter are expressed in the European eel (Anguilla anguilla). Biochim. Biophys. Acta 2002, 1566, 92–103. [Google Scholar] [CrossRef]
- Kultz, D.; Chakravarty, D.; Adilakshmi, T. A novel 14-3-3 gene is osmoregulated in gill epithelium of the euryhaline teleost Fundulus heteroclitus. J. Exp. Biol. 2001, 204, 2975–2985. [Google Scholar]
- Scott, G.R.; Claiborne, J.B.; Edwards, S.L.; Schulte, P.M.; Wood, C.M. Gene expression after freshwater transfer in gills and opercular epithelia of killifish: Insight into divergent mechanisms of ion transport. J. Exp. Biol. 2005, 208, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.R.; Richards, J.G.; Forbush, B.; Isenring, P.; Schulte, P.M. Changes in gene expression in gills of the euryhaline killifish Fundulus heteroclitus after abrupt salinity transfer. Am. J. Physiol. Cell Physiol. 2004, 287, C300–C309. [Google Scholar] [CrossRef]
- Tang, C.H.; Lai, D.Y.; Lee, T.H. Effects of salinity acclimation on Na(+)/K(+)-ATPase responses and FXYD11 expression in the gills and kidneys of the Japanese eel (Anguilla japonica). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2012, 163, 302–310. [Google Scholar] [CrossRef]
- Brennan, R.S.; Galvez, F.; Whitehead, A. Reciprocal osmotic challenges reveal mechanisms of divergence in phenotypic plasticity in the killifish Fundulus heteroclitus. J. Exp. Biol. 2015, 218, 1212–1222. [Google Scholar] [CrossRef]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef]
- Gibbons, T.C.; Metzger, D.C.H.; Healy, T.M.; Schulte, P.M. Gene expression plasticity in response to salinity acclimation in threespine stickleback ecotypes from different salinity habitats. Mol. Ecol. 2017, 26, 2711–2725. [Google Scholar] [CrossRef]
- Jeffries, K.M.; Connon, R.E.; Verhille, C.E.; Dabruzzi, T.F.; Britton, M.T.; Durbin-Johnson, B.P.; Fangue, N.A. Divergent transcriptomic signatures in response to salinity exposure in two populations of an estuarine fish. Evol. Appl. 2019, 12, 1212–1226. [Google Scholar] [CrossRef]
- Velotta, J.P.; Wegrzyn, J.L.; Ginzburg, S.; Kang, L.; Czesny, S.; O’Neill, R.J.; McCormick, S.D.; Michalak, P.; Schultz, E.T. Transcriptomic imprints of adaptation to fresh water: Parallel evolution of osmoregulatory gene expression in the Alewife. Mol. Ecol. 2017, 26, 831–848. [Google Scholar] [CrossRef]
- Whitehead, A.; Zhang, S.; Roach, J.L.; Galvez, F. Common functional targets of adaptive micro-and macro-evolutionary divergence in killifish. Mol. Ecol. 2013, 22, 3780–3796. [Google Scholar] [CrossRef]
- Avella, M.; Ducoudret, O.; Pisani, D.F.; Poujeol, P. Swelling-activated transport of taurine in cultured gill cells of sea bass: Physiological adaptation and pavement cell plasticity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R1149–R1160. [Google Scholar] [CrossRef][Green Version]
- Hwang, P.-P.; Lee, T.-H. New insights into fish ion regulation and mitochondrion-rich cells. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 148, 479–497. [Google Scholar] [CrossRef]
- Kang, C.-K.; Chen, Y.-C.; Chang, C.-H.; Tsai, S.-C.; Lee, T.-H. Seawater-acclimation abates cold effects on Na+, K+- ATPase activity in gills of the juvenile milkfish, Chanos chanos. Aquaculture 2015, 446, 67–73. [Google Scholar] [CrossRef]
- Lu, X.-J.; Zhang, H.; Yang, G.-J.; Li, M.-Y.; Chen, J. Comparative transcriptome analysis on the alteration of gene expression in ayu (Plecoglossus altivelis) larvae associated with salinity change. Zool. Res. 2016, 37, 126–135. [Google Scholar]
- Peter, M.S. The role of thyroid hormones in stress response of fish. Gen. Comp. Endocrinol. 2011, 172, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; McCormick, S.D. Prolactin and growth hormone in fish osmoregulation. Gen. Comp. Endocrinol. 2006, 147, 24–30. [Google Scholar] [CrossRef]
- Tseng, Y.-C.; Hwang, P.-P. Some insights into energy metabolism for osmoregulation in fish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2008, 148, 419–429. [Google Scholar] [CrossRef]
- Xu, Z.; Gan, L.; Li, T.; Xu, C.; Chen, K.; Wang, X.; Qin, J.G.; Chen, L.; Li, E. Transcriptome profiling and molecular pathway analysis of genes in association with salinity adaptation in nile tilapia Oreochromis niloticus. PLoS ONE 2015, 10, e0136506. [Google Scholar] [CrossRef]
- Yada, T.; Tsuruta, T.; Sakano, H.; Yamamoto, S.; Abe, N.; Takasawa, T.; Yogo, S.; Suzuki, T.; Iguchi, K.I.; Uchida, K. Changes in prolactin mRNA levels during downstream migration of the amphidromous teleost, ayu Plecoglossus altivelis. Gen. Comp. Endocrinol. 2010, 167, 261–267. [Google Scholar] [CrossRef]
- Brennan, R.S.; Healy, T.M.; Bryant, H.J.; La, M.V.; Schulte, P.M.; Whitehead, A. Integrative population and physiological genomics reveals mechanisms of adaptation in killifish. Mol. Biol. Evol. 2018, 35, 2639–2653. [Google Scholar] [CrossRef]
- Dennenmoser, S.; Vamosi, S.M.; Nolte, A.W.; Rogers, S.M. Adaptive genomic divergence under high gene flow between freshwater and brackish-water ecotypes of prickly sculpin (Cottus asper) revealed by Pool-Seq. Mol. Ecol. 2017, 26, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; DeFaveri, J.; Sotelo, G.; Nair, A.; Merila, J. Population genomic evidence for adaptive differentiation in Baltic Sea three-spined sticklebacks. BMC Biol. 2015, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Li, Z.; Merila, J. Population genomic evidence for adaptive differentiation in the Baltic Sea herring. Mol. Ecol. 2016, 25, 2833–2852. [Google Scholar] [CrossRef]
- Komissarov, A.; Vij, S.; Yurchenko, A.; Trifonov, V.; Thevasagayam, N.; Saju, J.; Sridatta, P.; Purushothaman, K.; Graphodatsky, A.; Orbán, L. B chromosomes of the Asian seabass (Lates calcarifer) contribute to genome variations at the level of individuals and populations. Genes 2018, 9, 464. [Google Scholar] [CrossRef]
- Purushothaman, K.; Lau, D.; Saju, J.M.; Sk, S.M.; Lunny, D.P.; Vij, S.; Orbán, L. Morpho-histological characterisation of the alimentary canal of an important food fish, Asian seabass (Lates calcarifer). PeerJ 2016, 4, e2377. [Google Scholar] [CrossRef] [PubMed]
- Thevasagayam, N.M.; Sridatta, P.S.; Jiang, J.; Tong, A.; Saju, J.M.; Kathiresan, P.; Kwan, H.Y.; Ngoh, S.Y.; Liew, W.C.; Kuznetsova, I.S.; et al. Transcriptome survey of a marine food fish: Asian seabass (Lates calcarifer). J. Mar. Sci. Eng. 2015, 3, 382–400. [Google Scholar] [CrossRef]
- Vij, S.; Kuhl, H.; Kuznetsova, I.S.; Komissarov, A.; Yurchenko, A.A.; Heusden, P.V.; Singh, S.; Thevasagayam, N.M.; Prakki, S.R.S.; Purushothaman, K.; et al. Chromosomal-level assembly of the Asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet. 2016. [Google Scholar] [CrossRef]
- Vij, S.; Purushothaman, K.; Gopikrishna, G.; Lau, D.; Saju, J.M.; Shamsudheen, K.V.; Vinayumar, K.; Basheer, V.S.; Gopalakrishnan, A.; Mohommad, S.H.; et al. Barcoding of Asian seabass across its geographic range provides evidence for its bifurcation into two distinct species. Front. Mar. Sci. 2014, 1. [Google Scholar] [CrossRef]
- Jerry, D.R. Biology and Culture of Asian Seabass Lates Calcarifer; CRC Press: Boca, FL, USA, 2014. [Google Scholar]
- Russell, D.J. Lates calcarifer wildstocks: Their biology, ecology and fishery. In Biology and Culture of Asian Seabass Lates Calcarifer; Jerry, D.R., Ed.; CRC Press: Boca, FL, USA, 2014; pp. 273–292. [Google Scholar]
- Moore, R. Natural sex inversion in the giant perch (Lates calcarifer). Mar. Freshw. Res. 1979, 30, 803–813. [Google Scholar] [CrossRef]
- Moore, R.; Reynold, L. Migration patterns of barramundi, Lates calcarifer (Bloch), in Papua New Guinea. Mar. Freshw. Res. 1982, 33, 671–682. [Google Scholar] [CrossRef]
- Moore, R. Spawning and early life history of burramundi, Lates calcarifer (Bloch), in Papua New Guinea. Mar. Freshw. Res. 1982, 33, 647–661. [Google Scholar] [CrossRef]
- Ayson, F.G.S.K.; Yashiro, R.; de Jesus-Ayson, E.G. Nursery and grow-out culture of Asian seabass, Lates calcarifer, in selected countries in Southeast Asia. In Biology and Culture of Asian Seabass Lates Calcarifer; Jerry, D.R., Ed.; CRC Press: Boca, FL, USA, 2014; pp. 273–292. [Google Scholar]
- Domingos, J.A.; Smith-Keune, C.; Jerry, D.R. Fate of genetic diversity within and between generations and implications for DNA parentage analysis in selective breeding of mass spawners: A case study of commercially farmed barramundi, Lates calcarifer. Aquaculture 2014, 424, 174–182. [Google Scholar] [CrossRef]
- Frost, L.A.; Evans, B.S.; Jerry, D.R. Loss of genetic diversity due to hatchery culture practices in barramundi (Lates calcarifer). Aquaculture 2006, 261, 1056–1064. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Paulete, J.; Beçak, W. Técnicas de citologia e histologia. Livros Técnicos E CientíficosSão Paulo 1976, 2, 305. [Google Scholar]
- Pereira, B.F.; Caetano, F.H. Histochemical technique for the detection of chloride cells in fish. Micron 2009, 40, 783–786. [Google Scholar] [CrossRef]
- R Core Team. R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Dantzler, W.H. Comparative Physiology of the Vertebrate Kidney; Springer: Berlin, Germany, 1989. [Google Scholar]
- Greenwell, M.G.; Sherrill, J.; Clayton, L.A. Osmoregulation in fish. Mechanisms and clinical implications. Vet. Clin. N. Am. Exot. Anim. Pract. 2003, 6, 169–189. [Google Scholar] [CrossRef]
- Boron, W.F. Acid-base transport by the renal proximal tubule. J. Am. Soc. Nephrol. 2006, 17, 2368–2382. [Google Scholar] [CrossRef]
- Koeppen, B.M. The kidney and acid-base regulation. Adv. Physiol. Educ. 2009, 33, 275–281. [Google Scholar] [CrossRef]
- Nowik, M.; Lecca, M.R.; Velic, A.; Rehrauer, H.; Brandli, A.W.; Wagner, C.A. Genome-wide gene expression profiling reveals renal genes regulated during metabolic acidosis. Physiol. Genom. 2008, 32, 322–334. [Google Scholar] [CrossRef]
- Ivanis, G.; Braun, M.; Perry, S.F. Renal expression and localization of SLC9A3 sodium/hydrogen exchanger and its possible role in acid-base regulation in freshwater rainbow trout (Oncorhynchus mykiss). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R971–R978. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef] [PubMed]
- Deaton, L.E. Hyperosmotic volume regulation in the gills of the ribbed mussel, Geukensia demissa: Rapid accumulation of betaine and alanine. J. Exp. Mar. Biol. Ecol. 2001, 260, 185–197. [Google Scholar] [CrossRef]
- Abe, H.; Okuma, E.; Amano, H.; Noda, H.; Watanabe, K. Role of free D-and L-alanine in the Japanese mitten crab Eriocheir japonicus to intracellular osmoregulation during downstream spawning migration. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1999, 123, 55–59. [Google Scholar] [CrossRef]
- Abe, H.; Okuma, E.; Amano, H.; Noda, H.; Watanabe, K. Effects of seawater acclimation on the levels of free D-and L-alanine and other osmolytes in the japanese mitten crab eriocheir japonicus. Fish. Sci. 1999, 65, 949–954. [Google Scholar] [CrossRef]
- Edwards, H. Free amino acids as regulators of osmotic pressure in aquatic insect larvae. J. Exp. Biol. 1982, 101, 153–160. [Google Scholar]
- Bystriansky, J.S.; Kaplan, J.H. Sodium pump localization in epithelia. J. Bioenerg. Biomembr. 2007, 39, 373–378. [Google Scholar] [CrossRef]
- Fujimori, T.; Abe, H. Physiological roles of free D-and L-alanine in the crayfish Procambarus clarkii with special reference to osmotic and anoxic stress responses. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2002, 131, 893–900. [Google Scholar] [CrossRef]
- Ballantyne, J. Amino acid metabolism. Fish. Physiol. 2001, 20, 77–107. [Google Scholar]
- Berman, A.E.; Kozlova, N.I.; Morozevich, G.E. Integrins: Structure and signaling. Biochemistry 2003, 68, 1284–1299. [Google Scholar] [CrossRef]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Petit, V.; Thiery, J.P. Focal adhesions: Structure and dynamics. Biol. Cell 2000, 92, 477–494. [Google Scholar] [CrossRef]
- Elangbam, C.S.; Qualls, C.W., Jr.; Dahlgren, R.R. Cell adhesion molecules—Update. Vet. Pathol. 1997, 34, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Krauss, R.S.; Cole, F.; Gaio, U.; Takaesu, G.; Zhang, W.; Kang, J.S. Close encounters: Regulation of vertebrate skeletal myogenesis by cell-cell contact. J. Cell Sci. 2005, 118, 2355–2362. [Google Scholar] [CrossRef] [PubMed]
- Boeuf, G.; Payan, P. How should salinity influence fish growth? Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001, 130, 411–423. [Google Scholar] [CrossRef]
- Boivin, B.; Castonguay, M.; Audet, C.; Pavey, S.; Dionne, M.; Bernatchez, L. How does salinity influence habitat selection and growth in juvenile American eels Anguilla rostrata? J. Fish Biol. 2015, 86, 765–784. [Google Scholar] [CrossRef] [PubMed]
- Gillanders, B.M.; Izzo, C.; Doubleday, Z.A.; Ye, Q. Partial migration: Growth varies between resident and migratory fish. Biol. Lett. 2015, 11, 20140850. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.E.; Allen, P.J. Effects of salinity on growth and ion regulation of juvenile alligator gar Atractosteus spatula. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 169, 44–50. [Google Scholar] [CrossRef]
- Lee, C.-T.; Risom, T.; Strauss, W.M. Evolutionary conservation of microRNA regulatory circuits: An examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007, 26, 209–218. [Google Scholar] [CrossRef]
- Shoman, H.; Gaber, H.J.O.N. Histological and ultrastructural observations on Gills of Tilapia nilotica L. (Oreochromis niloticus) in lake Qarun, Fayoum province, Egypt. Egypt. J. Aquat. Biol. Fish. 2003, 7, 157–181. [Google Scholar] [CrossRef][Green Version]
- Roberts, S.D.; Powell, M.D. Comparative ionic flux and gill mucous cell histochemistry: Effects of salinity and disease status in Atlantic salmon (Salmo salar L.). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 134, 525–537. [Google Scholar] [CrossRef]
- Powell, M.D. Respiration in infectious and non-infectious gill diseases. Fish. Respir. Environ. 2007, 317–339. [Google Scholar] [CrossRef]
- Fernandes, M.N.; Mazon, A.d.F. Environmental pollution and fish gill morphology. Fish Adapt. Enfield Sci. Publ. 2003, 203–231. [Google Scholar]
- Parashar, R.S.; Banerjee, T.K. Toxic impact of lethal concentration of lead nitrate on the gills of air-breathing catfish Heteropneustes fossilis (Bloch). Vet. Arh. 2002, 72, 167–182. [Google Scholar]
- Banerjee, T. Histopathology of respiratory organs of certain air-breathing fishes of India. Fish Physiol. Biochem. 2007, 33, 441–454. [Google Scholar] [CrossRef]
- Chandra, S.; Banerjee, T.K. Histopathological analysis of the respiratory organs of the air-breathing catfish Clarias batrachus (Linn.) exposed to the air. Acta Zool. Taiwanica 2003, 14, 45–64. [Google Scholar]
- Chandra, S.; Banerjee, T.K. Histopathological analysis of the respiratory organs of Channa striata subjected to air exposure. Vet. Arh. 2004, 74, 37–52. [Google Scholar]
- Olson, K.R. Scanning electron microscopy of the fish gill. Fish Morphol. Horiz. New Res. 1996, 31–45. [Google Scholar] [CrossRef]
- Parashar, R.; Banerjee, T. Histopathological analysis of sublethal toxicity induced by lead nitrate to the accessory respiratory organs of the air-breathing teleost, Heteropneustes fossilis [Bloch]. Pol. Arch. Hydrobiol. 1999, 46, 194–205. [Google Scholar]
- Parashar, R.; Banerjee, T. Response of the gill of the air-breathing catfish Heteropneustes fossilis (Bloch) to acute stress of desiccation. J. Exp. Zool. 1999, 2, 169–174. [Google Scholar]
- Parashar, R.; Banerjee, T.K. Response of aerial respiratory organs of the air-breathing catfish Heteropneustes fossilis (Bloch) to extreme stress of desiccation Ram Sanehi Parashar and Tarun Kumar Banerjee. Vet. Arh. 1999, 69, 63–68. [Google Scholar]
- Kantham, K.; Richards, R. Effect of buffers on the gill structure of common carp, Cyprinus carpio L., and rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 1995, 18, 411–423. [Google Scholar] [CrossRef]
- Roberts, S.D.; Powell, M.D. The viscosity and glycoprotein biochemistry of salmonid mucus varies with species, salinity and the presence of amoebic gill disease. J. Comp. Physiol. B 2005, 175, 1–11. [Google Scholar] [CrossRef]
- Dezfuli, B.S.; Giari, L.; Konecny, R.; Jaeger, P.; Manera, M. Immunohistochemistry, ultrastructure and pathology of gills of Abramis brama from Lake Mondsee, Austria, infected with Ergasilus sieboldi (Copepoda). Dis. Aquat. Org. 2003, 53, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Mc Cahon, C.; Pascoe, D.; Mc Kavanagh, C. Histochemical observations on the salmonids Salmo salar L. and Salmo trutta L. and the ephemeropterans Baetis rhodani (Pict.) and Ecdyonurus venosus (Fabr.) following a simulated episode of acidity in an upland stream. Hydrobiologia 1987, 153, 3–12. [Google Scholar] [CrossRef]
- McCormick, S.D. Endocrine Control of Osmoregulation in Teleost Fish. Am. Zool. 2001, 41, 781–794. [Google Scholar] [CrossRef]
- Foulkes, N.S.; Borjigin, J.; Snyder, S.H. Rhythmic transcription: The molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997, 20, 487–492. [Google Scholar] [CrossRef]
- Falcón, J.; Bolliet, V.; Ravault, J.; Chesneau, D.; Ali, M.A.; Collin, J. Rhythmic secretion of melatonin by the superfused pike pineal organ: Thermo-and photoperiod interaction. Neuroendocrinology 1994, 60, 535–543. [Google Scholar] [CrossRef]
- López-Patiño, M.A.; Rodríguez-Illamola, A.; Gesto, M.; Soengas, J.L.; Míguez, J.M. Changes in plasma melatonin levels and pineal organ melatonin synthesis following acclimation of rainbow trout (Oncorhynchus mykiss) to different water salinities. J. Exp. Biol. 2011, 214, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Zachmann, A.; Ali, M.A.; Falcón, J. Melatonin and its effects in fishes: An overview. In Rhythms in Fishes; Springer: Berlin, Germany, 1992; pp. 149–165. [Google Scholar]
- Kleszczyńska, A.; Vargas-Chacoff, L.; Gozdowska, M.; Kalamarz, H.; Martínez-Rodríguez, G.; Mancera, J.M.; Kulczykowska, E. Arginine vasotocin, isotocin and melatonin responses following acclimation of gilthead sea bream (Sparus aurata) to different environmental salinities. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 145, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.; Randall, C.; Bromage, N.; Thorpe, J. The role of melatonin and the pineal gland on development and smoltification of Atlantic salmon (Salmo salar) parr. Aquaculture 1998, 168, 139–155. [Google Scholar] [CrossRef]
- Sangiao-Alvarellos, S.; Míguez, J.M.; Soengas, J.L. Melatonin treatment affects the osmoregulatory capacity of rainbow trout. Aquac. Res. 2007, 38, 325–330. [Google Scholar] [CrossRef]
- Golombek, D.A.; Ferreyra, G.A.; Agostino, P.V.; Murad, A.D.; Rubio, M.F.; Pizzio, G.A.; Katz, M.E.; Marpegan, L.; Bekinschtein, T.A. From light to genes: Moving the hands of the circadian clock. Front. Biosci. 2003, 8, s285–s293. [Google Scholar] [CrossRef][Green Version]
- Golombek, D.A.; Rosenstein, R.E. Physiology of circadian entrainment. Physiol. Rev. 2010, 90, 1063–1102. [Google Scholar] [CrossRef] [PubMed]
- Dubocovich, M.L.; Delagrange, P.; Krause, D.N.; Sugden, D.; Cardinali, D.P.; Olcese, J. International union of basic and clinical pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharm. Rev. 2010, 62, 343–380. [Google Scholar] [CrossRef]
- Dubocovich, M.L.; Markowska, M. Functional MT1 and MT2 melatonin receptors in mammals. Endocrine 2005, 27, 101–110. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin: Signaling mechanisms of a pleiotropic agent. Biofactors 2009, 35, 183–192. [Google Scholar] [CrossRef]
- López-Olmeda, J.; Oliveira, C.; Kalamarz, H.; Kulczykowska, E.; Delgado, M.; Sánchez-Vázquez, F. Effects of water salinity on melatonin levels in plasma and peripheral tissues and on melatonin binding sites in European sea bass (Dicentrarchus labrax). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 152, 486–490. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K.; Robinson, G.J.; Lundberg, A.E.; Bartlett, P.F.; Wray, N.R.; et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS ONE 2014, 9, e103207. [Google Scholar] [CrossRef]
- Seyednasrollah, F.; Laiho, A.; Elo, L.L. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief. Bioinform. 2015, 16, 59–70. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vij, S.; Purushothaman, K.; Sridatta, P.S.R.; Jerry, D.R. Transcriptomic Analysis of Gill and Kidney from Asian Seabass (Lates calcarifer) Acclimated to Different Salinities Reveals Pathways Involved with Euryhalinity. Genes 2020, 11, 733. https://doi.org/10.3390/genes11070733
Vij S, Purushothaman K, Sridatta PSR, Jerry DR. Transcriptomic Analysis of Gill and Kidney from Asian Seabass (Lates calcarifer) Acclimated to Different Salinities Reveals Pathways Involved with Euryhalinity. Genes. 2020; 11(7):733. https://doi.org/10.3390/genes11070733
Chicago/Turabian StyleVij, Shubha, Kathiresan Purushothaman, Prakki Sai Rama Sridatta, and Dean R. Jerry. 2020. "Transcriptomic Analysis of Gill and Kidney from Asian Seabass (Lates calcarifer) Acclimated to Different Salinities Reveals Pathways Involved with Euryhalinity" Genes 11, no. 7: 733. https://doi.org/10.3390/genes11070733
APA StyleVij, S., Purushothaman, K., Sridatta, P. S. R., & Jerry, D. R. (2020). Transcriptomic Analysis of Gill and Kidney from Asian Seabass (Lates calcarifer) Acclimated to Different Salinities Reveals Pathways Involved with Euryhalinity. Genes, 11(7), 733. https://doi.org/10.3390/genes11070733





