Chasing the Apomictic Factors in the Ranunculus auricomus Complex: Exploring Gene Expression Patterns in Microdissected Sexual and Apomictic Ovules
Abstract
1. Introduction
2. Materials and Methods
2.1. Custom Microarray Development
2.2. Transcriptomal Profiling of Sexual and Apomictic Ovules
2.2.1. Sample Selection, Ovule Microdissection, and RNA Extraction
2.2.2. cDNA Synthesis and Amplification
2.2.3. Microarray Hybridization and Data Processing
2.2.4. Analyses of Gene Expression throughout Development
2.3. Analyses for Signatures of Ploidy, Parent of Origin Effects or Hybridization
2.3.1. Ploidy
2.3.2. Parent of Origin Effects
2.3.3. Hybridization
2.4. Microarray Validation Using qRT-PCR
2.5. blastx, tblastx, Gene Ontology
3. Results
3.1. Gene Expression Differences
3.1.1. Principal Component Analysis (PCA)
3.1.2. Stage Specific Differential Expression Analysis
3.2. Transcriptome Wide Signatures of Hybridization, Ploidy Variation and Parent of Origin Effects
3.3. qPCR Validation
4. Discussion
4.1. Transcriptional Variation Reflect Contrasted Sexual vs. Apomictic Developments in the Ovule
4.2. Apomixis Inheritance in Allopolyploid Ranunculus and Partitioned Gene-Expression Effects
4.2.1. Homologous Parent of Origin Effects in the Apomictic Hybrid
4.2.2. Ploidy Effect and Apomixis Expressivity in the Hybrid
4.2.3. Transgressive Gene Expression in the Apomictic Hybrid
5. Conclusions and Prospects
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barton, N.; Charlesworth, D. Why sex and recombination? Science 1998, 281, 1986–1990. [Google Scholar] [CrossRef]
- Mittwoch, U. Parthenogenesis. J. Med. Genet. 1978, 15, 165–181. [Google Scholar] [CrossRef]
- Suomalainen, E. Parthenogenesis in animals. In Advances in Genetics; Academic Press Inc.: New York, NY, USA, 1950; pp. 193–253. [Google Scholar]
- Hörand, E.; Grossniklaus, U.; van Dijk, P.J.; Sharbel, T.S. Apomixis: Evolution, Mechanisms and Perspectives, 1st ed.; Regnum Vegetabile; Hörand, E., Grossniklaus, U., van Dijk, P.J., Sharbel, T.S., Eds.; A.R.G. Gantner Verlag: Rugell, Liechtenstein, 2007; Volume 147, ISBN 978-3-906166-60-5. [Google Scholar]
- Schön, I.; Van Dijk, P.; Martens, K. Lost Sex; Schön, I., Martens, K., Dijk, P., Eds.; Springer Science & Business Media: Berlin, Germany, 2009; ISBN 978-90-481-2769-6. [Google Scholar]
- Maynard Smith, J. The Evolution of Sex, 1st ed.; Cambridge University Press: Cambridge, NY, USA, 1978; ISBN 0521293022. [Google Scholar]
- Bell, G. The Masterpiece of Nature—The Evolution and Genetics of Sexuality, 1st ed.; University of California Press: Berkeley, CA, USA, 1982; ISBN 10: 0520045831, ISBN 13: 9780520045835. [Google Scholar]
- Crow, J.F.; Kimura, M. Evolution in sexual and asexual populations. Am. Nat. 1965, 99, 439–450. [Google Scholar] [CrossRef]
- Vrijenhoek, R.C. Ecological differentiation among clones: The Frozen Niche Variation model. In Population Biology and Evolution; Woehrmann, K., Loeschcke, V., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 217–231. [Google Scholar]
- Hörandl, E. A combinational theory for maintenance of sex. Heredity (Edinb.) 2009, 103, 445–457. [Google Scholar] [CrossRef]
- Lovell, J.T.; Williamson, R.J.; Wright, S.I.; McKay, J.K.; Sharbel, T.F. Mutation accumulation in an asexual relative of Arabidopsis. PLoS Genet. 2017, 13, 1006550. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. Mol. Mech. Mutagen. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Lynch, M.; Butcher, D.; Bürger, R.; Gabriel, W. The mutational meltdown in asexual populations. J. Hered. 1993, 84, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Kondrashov, A.S. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 1982, 40, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The Genetical Theory of Natural Selection; The Clarendon Press: Oxford, UK, 1930. [Google Scholar]
- Lewontin, R.C. The Genetic Basis of Evolutionary Change; Columbia University Press: New York, NY, USA, 1974; ISBN 0-231-03392-3. [Google Scholar]
- Asker, S.E.; Jerling, L. Apomixis in Plants, 1st ed.; CRC Press: Boca Raton, FL, USA, 1992; ISBN 0-8493-4545-6. [Google Scholar]
- Carman, J.G. Asynchonous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Klatt, S.; Baier, R.; Carman, J.G.; Hörandl, E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. CRC Crit. Rev. Plant Sci. 2014, 33, 414–427. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [PubMed]
- Carman, J.G. Do duplicate genes cause apomixis? In Apomixis: Evolution, Mechanisms and Perspectives; Regnum Vegetabile; Hörandl, E., Grossniklaus, U., van Dijk, P., Sharbel, T.S., Eds.; A.R.G. Gantner Verlag: Rugell, Liechtenstein, 2007; ISBN 978-3-906166-60-5. [Google Scholar]
- Hand, M.L.; Koltunow, A.M.G.G. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A. Controlling apomixis: Shared features and distinct characteristics of gene regulation. Genes (Basel) 2020, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Kantama, L.; Sharbel, T.F.; Schranz, M.E.; Mitchell-Olds, T.; de Vries, S.; de Jong, H. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 14026–14031. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Morán, E.; Benavente, E.; Orellana, J. Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 2001, 110, 371–377. [Google Scholar] [CrossRef]
- Adams, K.L.; Percifield, R.; Wendel, J.F. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 2004, 168, 2217–2226. [Google Scholar] [CrossRef]
- Pikaard, C.S. Genomic change and gene silencing in polyploids. Trends Genet. 2001, 17, 675–677. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S. Polyploidy: Recurrent formation and genome evolution. Trends Ecol. Evol. 1999, 14, 348–352. [Google Scholar] [CrossRef]
- Adams, K.L. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 2007, 98, 136–141. [Google Scholar] [CrossRef]
- Grimanelli, D.; García, M.; Kaszas, E.; Perotti, E.; Leblanc, O. Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 2003, 165, 1521–1531. [Google Scholar] [PubMed]
- Sharbel, T.F.; Voigt, M.-L.; Corral, J.M.; Galla, G.; Kumlehn, J.; Klukas, C.; Schreiber, F.; Vogel, H.; Rotter, B. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 2010, 22, 655–671. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef] [PubMed]
- Nogler, G.A. Genetik der Aposporie bei Ranunculus auricomus s.l. W. Koch. I. Embryologie. Ber. Schweiz. Bot. 1971, 81, 139–179. [Google Scholar]
- Nogler, G.A. Genetik der aposporie bei Ranunculus auricomus. III. F2-Rückkreuzungsbastarde. Ber. Schweiz. Bot. 1973, 83, 295–305. [Google Scholar]
- Nogler, G.A. Genetics of apospory in apomictic Ranunculus auricomus. V: Conclusion. Bot. Helv. 1984, 94, 411–422. [Google Scholar]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 475–518. ISBN 3-540-12739-9. [Google Scholar]
- Barke, B.H.; Daubert, M.; Hörandl, E. Establishment of apomixis in diploid F2 hybrids and inheritance of apospory from F1 to F2 hybrids of the Ranunculus auricomus complex. Front. Plant Sci. 2018, 9, 1111. [Google Scholar] [CrossRef]
- Hodač, L.; Scheben, A.P.; Hojsgaard, D.; Paun, O.; Hörand, E. ITS Polymorphisms shed light on hybrid evolution in apomictic plants: A case study on the Ranunculus auricomus complex. PLoS ONE 2014, 9, 28–30. [Google Scholar] [CrossRef]
- Hörandl, E.; Greilhuber, J.; Klímová, K.; Paun, O.; Temsch, E.; Emadzade, K.; Hodálová, I. Reticulate evolution and taxonomic concepts in the Ranunculus auricomus complex (Ranunculaceae): Insights from analysis of morphological, karyological and molecular data. Taxon 2009, 58, 1194–1215. [Google Scholar] [CrossRef]
- Paun, O.; Ho, E. Evolution of Hypervariable Microsatellites in Apomictic Polyploid Lineages of Ranunculus carpaticola: Directional Bias at Dinucleotide Loci. Genetics 2006, 174, 387–398. [Google Scholar] [CrossRef]
- Pellino, M.; Hojsgaard, D.; Schmutzer, T.; Scholz, U.; Hörandl, E.; Vogel, H.; Sharbel, T.F. Asexual genome evolution in the apomictic Ranunculus auricomus complex: Examining the effects of hybridization and mutation accumulation. Mol. Ecol. 2013, 22, 5908–5921. [Google Scholar] [CrossRef]
- Hörandl, E.; Cosendai, A.C.; Temsch, E.M. Understanding the geographic distributions of apomictic plants: A case for a pluralistic approach. Plant Ecol. Divers. 2008, 1, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Izmailow, R. Megasporogenesis in the apomictic species Ranunculus cassubicus. Acta Biol. Crac. 1965, 8, 183–195. [Google Scholar]
- Hojsgaard, D.; Greilhuber, J.; Pellino, M.; Paun, O.; Sharbel, T.F.; Elvira, H.; Hörandl, E. Emergence of apospory and bypass of meiosis via apomixis after sexual hybridisation and polyploidisation. New Phytol. 2014, 204, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Klatt, S.; Hadacek, F.; Hodač, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef]
- Ulum, F.B.; Costa Castro, C.; Hörandl, E. Ploidy-dependent effects of light stress on the mode of reproduction in the Ranunculus auricomus complex (Ranunculaceae). Front. Plant Sci. 2020, 11, 104. [Google Scholar] [CrossRef]
- Hörandl, E.; Greilhuber, J. Diploid and autotetraploid sexuals and their relationships to apomicts in the Ranunculus cassubicus group: Insights from DNA content and isozyme variation. Plant Syst. Evol. 2002, 234, 85–100. [Google Scholar] [CrossRef]
- Paun, O.; Stuessy, T.F.; Hörandl, E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006, 171, 223–236. [Google Scholar] [CrossRef]
- Vogel, H.; Heidel, A.J.; Heckel, D.G.; Groot, A.T. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genom. 2010, 11, 29. [Google Scholar] [CrossRef]
- Vogel, H.; Wheat, C.W. Accessing the transcriptome: How to normalize mRNA pools. In Molecular Methods for Evolutionary Genetics; Humana Press: Totowa, NJ, USA, 2011; Volume 772, pp. 105–128. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Rapp, R.A.; Udall, J.A.; Wendel, J.F. Genomic expression dominance in allopolyploids. BMC Biol. 2009, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity (Edinb.) 2013, 110, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Albertini, E. Apomixis in plant reproduction: A novel perspective on an old dilemma. Plant Reprod. 2013, 26, 159–179. [Google Scholar] [CrossRef] [PubMed]
- Pupilli, F.; Barcaccia, G. Cloning plants by seeds: Inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J. Biotechnol. 2012, 159, 291–311. [Google Scholar] [CrossRef]
- Polegri, L.; Calderini, O.; Arcioni, S.; Pupilli, F. Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. J. Exp. Bot. 2010, 61, 1869–1883. [Google Scholar] [CrossRef]
- Bräuning, S.; Catanach, A.; Lord, J.M.; Bicknell, R.; Macknight, R.C. Comparative transcriptome analysis of the wild-type model apomict Hieracium praealtum and its loss of parthenogenesis (lop) mutant. BMC Plant Biol. 2018, 18, 206. [Google Scholar] [CrossRef]
- Osborn, T.C.; Chris Pires, J.; Birchler, J.A.; Auger, D.L.; Jeffery Chen, Z.; Lee, H.-S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Mau, M.; Lovell, J.T.; Corral, J.M.; Kiefer, C.; Koch, M.A.; Aliyu, O.M.; Sharbel, T.F. Hybrid apomicts trapped in the ecological niches of their sexual ancestors. Proc. Natl. Acad. Sci. USA 2015, 112, E2357–E2365. [Google Scholar] [CrossRef]
- Grossniklaus, U. From sexuality to apomixis: Molecular and genetic approaches. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; CIMMYT: El Batán, Mexico, 2001; pp. 168–211. ISBN 970-648-074-9. [Google Scholar]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Jackson, S.; Chen, Z.J. Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 2010, 13, 153–159. [Google Scholar] [CrossRef]
- Grimanelli, D.; Leblanc, O.; Perotti, E.; Grossniklaus, U. Developmental genetics of gametophytic apomixis. Trends Genet. 2001, 17, 597–604. [Google Scholar] [CrossRef]
- Delgado, L.; Galdeano, F.; Sartor, M.E.; Quarin, C.L.; Espinoza, F.; Ortiz, J.P.A. Analysis of variation for apomictic reproduction in diploid Paspalum rufum. Ann. Bot. 2014, 113, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, C.C.F.; Kirchheimer, B.; Dellinger, A.S.; Klatt, S.; Winkler, M.; Dullinger, S.; Hörandl, E. Correlations of polyploidy and apomixis with elevation and associated environmental gradients in an alpine plant. AoB Plants 2016, 8, plw064. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D. Transient activation of apomixis in sexual neotriploids may retain genomically altered states and enhance polyploid establishment. Front. Plant Sci. 2018, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.; Hörandl, E. The rise of apomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef] [PubMed]
- Sharbel, T.F.; Voigt, M.L.; Corral, J.M.; Thiel, T.; Varshney, A.; Kumlehn, J.; Vogel, H.; Rotter, B. Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J. 2009, 58, 870–882. [Google Scholar] [CrossRef]
- Galla, G.; Zenoni, S.; Avesani, L.; Altschmied, L.; Rizzo, P.; Sharbel, T.F.; Barcaccia, G. Pistil transcriptome analysis to disclose genes and gene products related to aposporous apomixis in Hypericum perforatum L. Front. Plant Sci. 2017, 8, 79. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010, 186, 184–193. [Google Scholar] [CrossRef]
- Swanson-Wagner, R.A.; Jia, Y.; DeCook, R.; Borsuk, L.A.; Nettleton, D.; Schnable, P.S. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 2006, 103, 6805–6810. [Google Scholar] [CrossRef]
- He, G.; Zhu, X.; Elling, A.A.; Chen, L.; Wang, X.; Guo, L.; Liang, M.; He, H.; Zhang, H.; Chen, F.; et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 2010, 22, 17–33. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Barker, G.L.; Wilson, I.D.; Abbott, R.J.; Edwards, K.J.; Hiscock, S.J. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 2006, 16, 1652–1659. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.D.M.; Kane, N.C.; Rieseberg, L.H.; Adams, K.L. RNA-Seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol. Evol. 2013, 5, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Hojsgaard, D.H.; Martínez, E.J.; Quarin, C.L. Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytol. 2013, 197, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Hand, M.L.; Vít, P.; Krahulcová, A.; Johnson, S.D.; Oelkers, K.; Siddons, H.; Chrtek, J.; Fehrer, J.; Koltunow, A.M.G. Evolution of apomixis loci in Pilosella and Hieracium (Asteraceae) inferred from the conservation of apomixis-linked markers in natural and experimental populations. Heredity (Edinb.) 2015, 114, 17–26. [Google Scholar] [CrossRef]
- Washburn, J.D.; Birchler, J.A. Polyploids as a “model system” for the study of heterosis. Plant Reprod. 2014, 27, 1–5. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Fort, A.; Ryder, P.; Mckeown, P.C.; Wijnen, C.; Aarts, M.G.; Sulpice, R.; Spillane, C. Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol. 2016, 209, 590–599. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Schegg, E.; Valls, J.F.M.; Martínez, E.J.; Quarin, C.L. Sexuality, apomixis, ploidy levels, and genomic relationships among four Paspalum species of the subgenus Anachyris (Poaceae). Flora Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 535–547. [Google Scholar] [CrossRef]
- Ortiz, J.P.A.; Quarin, C.L.; Pessino, S.C.; Acuña, C.; Martínez, E.J.; Espinoza, F.; Hojsgaard, D.H.; Sartor, M.E.; Cáceres, M.E.; Pupilli, F. Harnessing apomictic reproduction in grasses: What we have learned from Paspalum. Ann. Bot. 2013, 112, 767–787. [Google Scholar] [CrossRef]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef]
- Chapman, H.; Houliston, G.J.; Robson, B.; Iline, I. A case of reversal: The evolution and maintenance of sexuals from parthenogenetic clones in Hieracium pilosella. Int. J. Plant Sci. 2003, 164, 719–728. [Google Scholar] [CrossRef]
- Hörandl, E.; Hojsgaard, D. The evolution of apomixis in angiosperms: A reappraisal. Plant Biosyst. 2012, 146, 681–693. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Hörandl, E. A little bit of sex matters for genome evolution in asexual plants. Front. Plant Sci. 2015, 6, 82. [Google Scholar] [CrossRef] [PubMed]
- Emadzade, K.; Hörandl, E. Northern Hemisphere origin, transoceanic dispersal, and diversification of Ranunculeae DC. (Ranunculaceae) in the Cenozoic. J. Biogeogr. 2011, 38, 517–530. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Widmer, A.; Arntz, A.M.; Burke, J.M.; Carr, D.E.; Abbott, R.J.; Meagher, T.R. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 1141–1147. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Hiscock, S.J. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 2008, 18, 435–444. [Google Scholar] [CrossRef]
- Mancini, M.; Permingeat, H.; Colono, C.; Siena, L.; Pupilli, F.; Azzaro, C.; de Alencar Dusi, D.M.; de Campos Carneiro, V.T.; Podio, M.; Seijo, J.G.; et al. The MAP3K-coding QUI-GON JINN (QGJ) gene is essential to the formation of unreduced embryo sacs in Paspalum. Front. Plant Sci. 2018, 9, 1547. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Yu, H.J.; Ngo, Q.A.; Rajani, S.; Mayalagu, S.; Johnson, C.S.; Capron, A.; Xie, L.F.; Ye, D.; Sundaresan, V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef]
- Ray, S.M.; Park, S.S.; Ray, A. Pollen tube guidance by the female gametophyte. Development 1997, 124, 2489–2498. [Google Scholar] [PubMed]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.-P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, C.C.F.; Syngelaki, E.; Kirchheimer, B.; Dullinger, S.; Klatt, S.; Hörandl, E. Epigenetic patterns and geographical parthenogenesis in the alpine plant species Ranunculus kuepferi (Ranunculaceae). Int. J. Mol. Sci. 2020, 21, 3318. [Google Scholar] [CrossRef] [PubMed]
- Corral, J.M.; Vogel, H.; Aliyu, O.M.; Hensel, G.; Thiel, T.; Kumlehn, J.; Sharbel, T.F. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013, 163, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Mau, M.; Corral, J.M.; Vogel, H.; Melzer, M.; Fuchs, J.; Kuhlmann, M.; de Storme, N.; Geelen, D.; Sharbel, T.F. The conserved chimeric transcript UPGRADE2 is associated with unreduced pollen formation and is exclusively found in apomictic Boechera species. Plant Physiol. 2013, 163, 1640–1659. [Google Scholar] [CrossRef] [PubMed]
- Schallau, A.; Arzenton, F.; Johnston, A.J.; Hähnel, U.; Koszegi, D.; Blattner, F.R.; Altschmied, L.; Haberer, G.; Barcaccia, G.; Bäumlein, H. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010, 62, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [PubMed]
- Albertini, E.; Marconi, G.; Reale, L.; Barcaccia, G.; Porceddu, A.; Ferranti, F.; Falcinelli, M. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiol. 2005, 138, 2185–2199. [Google Scholar] [CrossRef]
- Bocchini, M.; Galla, G.; Pupilli, F.; Bellucci, M.; Barcaccia, G.; Ortiz, J.P.A.; Pessino, S.C.; Albertini, E. The vesicle trafficking regulator PN-SCD1 is demethylated and overexpressed in florets of apomictic Paspalum notatum genotypes. Sci. Rep. 2018, 8, 3030. [Google Scholar] [CrossRef]
- Rodrigues, J.C.M.; Luo, M.; Berger, F.; Koltunow, A.M.G. Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex. Plant Reprod. 2010, 23, 123–133. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Barker, G.L.; Brennan, A.C.; Edwards, K.J.; Abbott, R.J.; Hiscock, S.J. Changes to gene expression associated with hybrid speciation in plants: Further insights from transcriptomic studies in Senecio. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3055–3069. [Google Scholar] [CrossRef]
- Schmidt, A.; Schmid, M.W.; Klostermeier, U.C.; Qi, W.; Guthörl, D.; Sailer, C.; Waller, M.; Rosenstiel, P.; Grossniklaus, U. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet. 2014, 10, e1004476. [Google Scholar] [CrossRef] [PubMed]
- Carman, J.G.; Mateo de Arias, M.; Gao, L.; Zhao, X.; Kowallis, B.M.; Sherwood, D.A.; Srivastava, M.K.; Dwivedi, K.K.; Price, B.J.; Watts, L.; et al. Apospory and diplospory in diploid Boechera (Brassicaceae) may facilitate speciation by recombination-driven apomixis-to-sex reversals. Front. Plant Sci. 2019, 10, 724. [Google Scholar] [CrossRef] [PubMed]
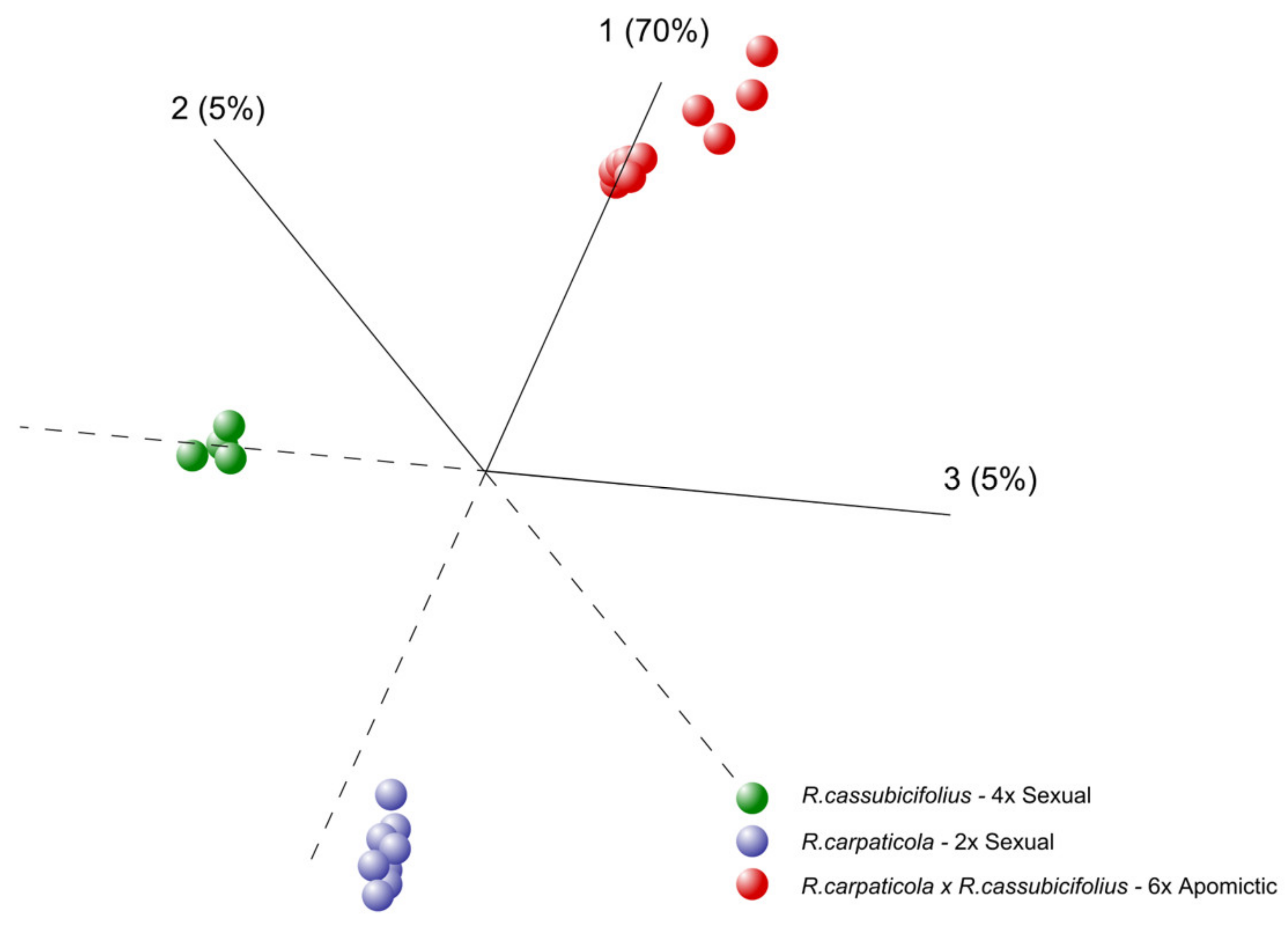
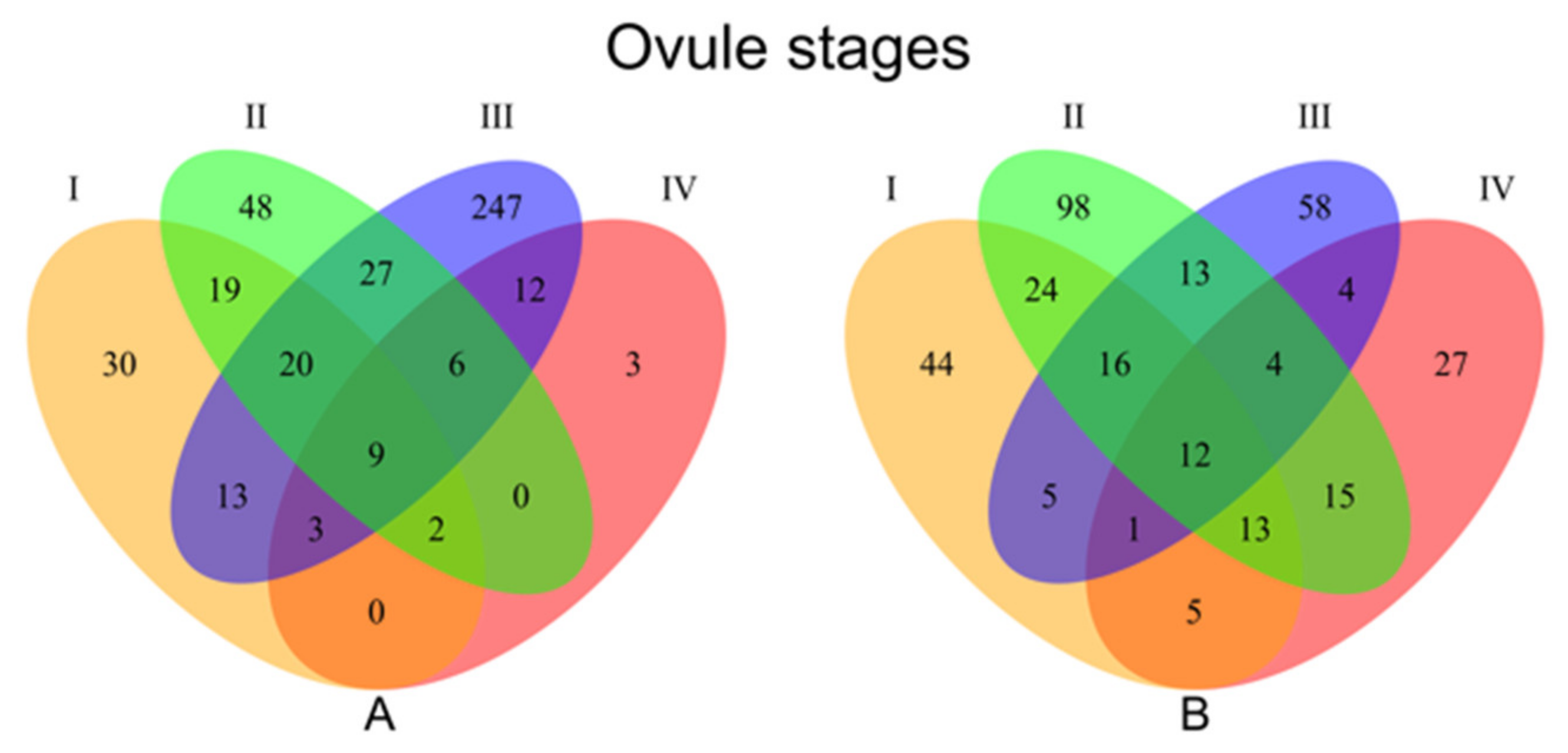
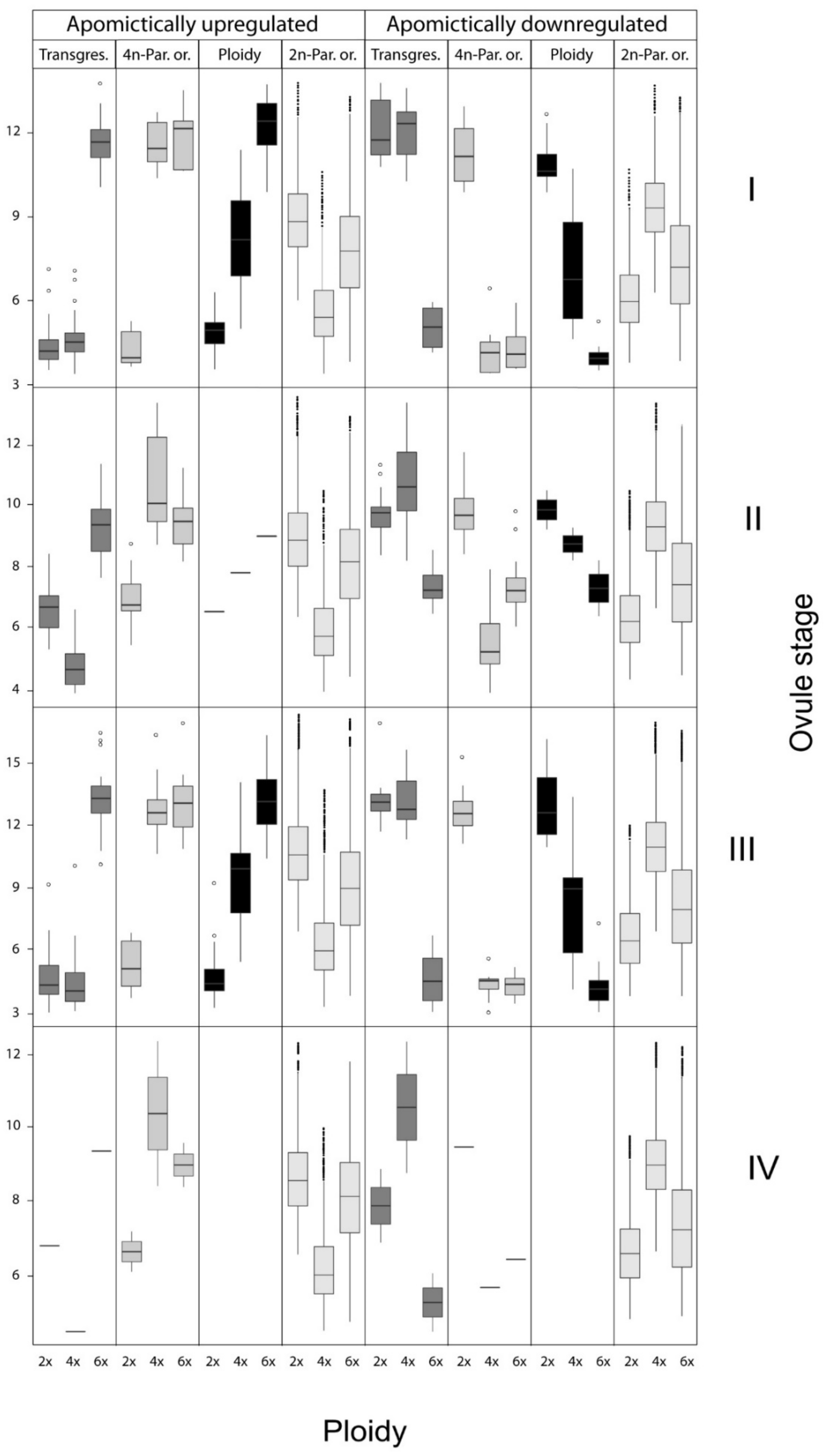
| Taxon (Code) | Nind | Ploidy 1 | Reprod Mode | Apospory Expressivity 2 | Collection Locality, Collector, Collection Number, and Vouchers |
|---|---|---|---|---|---|
| R. carpaticola REV1 | 3 | 2x | Sex | 0.0 | Slovakia, Slovenské rudohorie, Revúca, hill Skalka (forest) Hörandl 8483 (WU) |
| R. carpaticola × cassubicifolius TRE | 1 | 6x | Apo | 0.226 | Slovakia, Strázovské vrchy (near Trençín), between Kubra and Kubrica (margin of forest and meadow) Hörandl et al. C29 (SAV) |
| R. carpaticola × cassubicifolius VRU 2 | 2 | 6x | Apo | 0.339 | Slovakia, Turçianska kotlina, Vrútky-Piatrová (meadow) Hörandl et al. C35 (SAV) |
| R. cassubicifolius YBB 1 | 1 | 4x | Sex | 0.0 | Austria, Lower Austria, Wülfachgraben, SE Ybbsitz (forest) Hörandl 8472 (WU) |
| Number of Contigs | Number of Contigs, >300 bp | Differentially Expressed Transcripts | Annotated | ||
|---|---|---|---|---|---|
| 462102 | 62102 | ||||
| Develop. stage | Upregulated 1 | Downregulated 1 | |||
| I | 120 (44) | 96 (30) | 72 | ||
| II | 195 (98) | 131 (48) | 107 | ||
| III | 113 (58) | 337 (247) | 156 | ||
| IV | 81 (3) | 35 (3) | 30 | ||
| Expression Class 2 | Develop. Stage | Transgressive Effect | Parent of Origin Effect | Ploidy Effect | Total |
|---|---|---|---|---|---|
| Apomictic 6x upregulated 3 | I | 5 (0.1) | 27 (0.49) | 23 (0.41) | 55 |
| II | 15 (0.58) | 10 (0.38) | 1 (0.04) | 26 | |
| III | 16 (0.16) | 44 (0.46) | 36 (0.38) | 96 | |
| IV | 2 (0.4) | 1 (0.2) | 2 (0.4) | 5 | |
| Total up | 38 (0.21) | 82 (0.45) | 62 (0.34) | 182 | |
| Apomictic 6x downregulated 3 | I | 6 (0.26) | 7 (0.3) | 10 (0.44) | 23 |
| II | 18 (0.32) | 37 (0.65) | 2 (0.03) | 57 | |
| III | 9 (0.24) | 11 (0.3) | 17 (0.46) | 37 | |
| IV | 2 (0.4) | 1 (0.2) | 2 (0.4) | 5 | |
| Total down | 35 (0.29) | 56 (0.46) | 31 (0.25) | 122 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellino, M.; Hojsgaard, D.; Hörandl, E.; Sharbel, T.F. Chasing the Apomictic Factors in the Ranunculus auricomus Complex: Exploring Gene Expression Patterns in Microdissected Sexual and Apomictic Ovules. Genes 2020, 11, 728. https://doi.org/10.3390/genes11070728
Pellino M, Hojsgaard D, Hörandl E, Sharbel TF. Chasing the Apomictic Factors in the Ranunculus auricomus Complex: Exploring Gene Expression Patterns in Microdissected Sexual and Apomictic Ovules. Genes. 2020; 11(7):728. https://doi.org/10.3390/genes11070728
Chicago/Turabian StylePellino, Marco, Diego Hojsgaard, Elvira Hörandl, and Timothy F. Sharbel. 2020. "Chasing the Apomictic Factors in the Ranunculus auricomus Complex: Exploring Gene Expression Patterns in Microdissected Sexual and Apomictic Ovules" Genes 11, no. 7: 728. https://doi.org/10.3390/genes11070728
APA StylePellino, M., Hojsgaard, D., Hörandl, E., & Sharbel, T. F. (2020). Chasing the Apomictic Factors in the Ranunculus auricomus Complex: Exploring Gene Expression Patterns in Microdissected Sexual and Apomictic Ovules. Genes, 11(7), 728. https://doi.org/10.3390/genes11070728






