Interstitial Telomeric Repeats Are Rare in Turtles
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Chromosome Preparation and Staining
2.3. Giemsa Staining and Karyotype Reconstruction
2.4. Fluorescence In Situ Hybridization with Probes for Telomeric Repeats
2.5. Distribution of ITRs across the Turtle Phylogeny
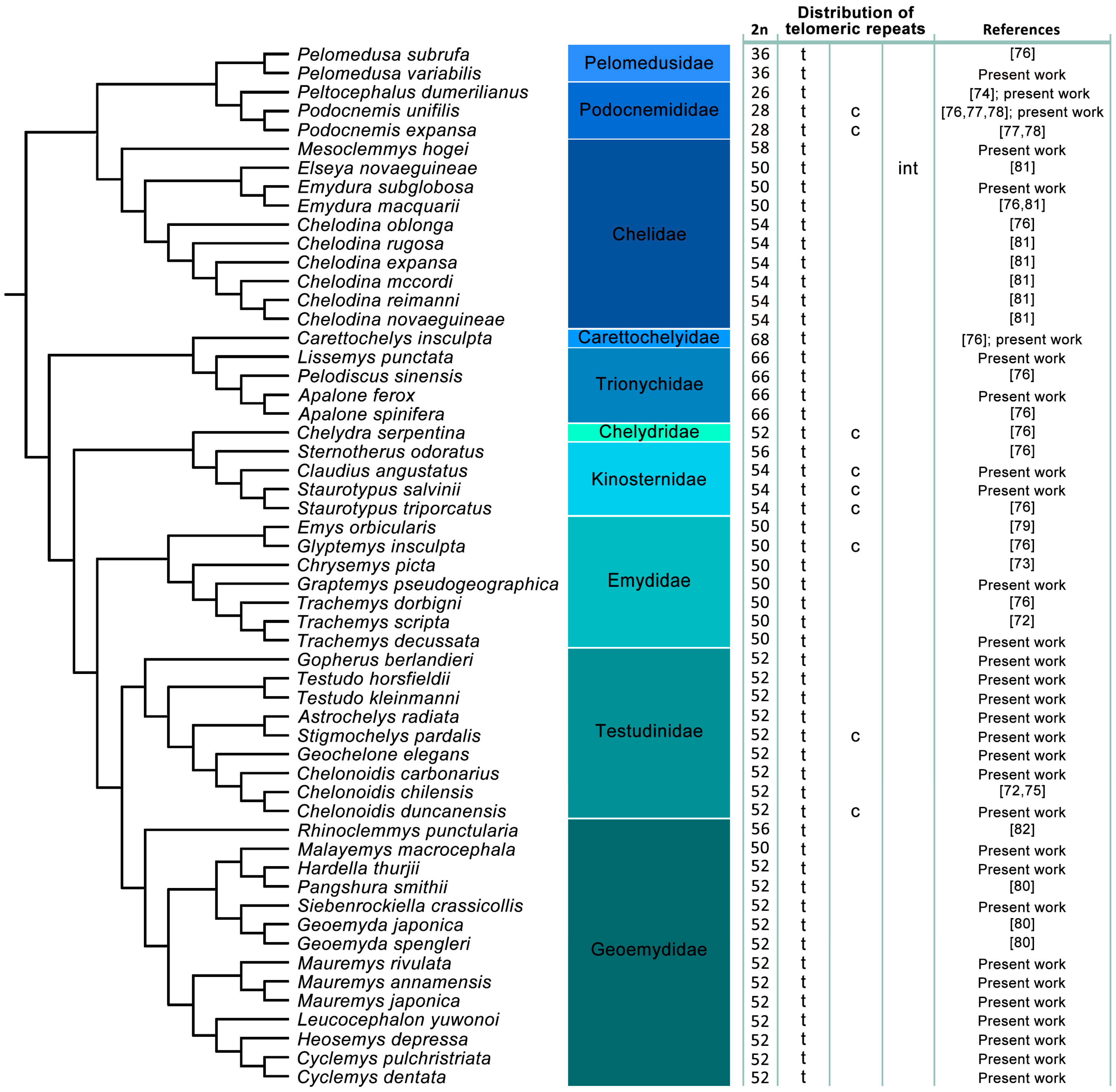
3. Results
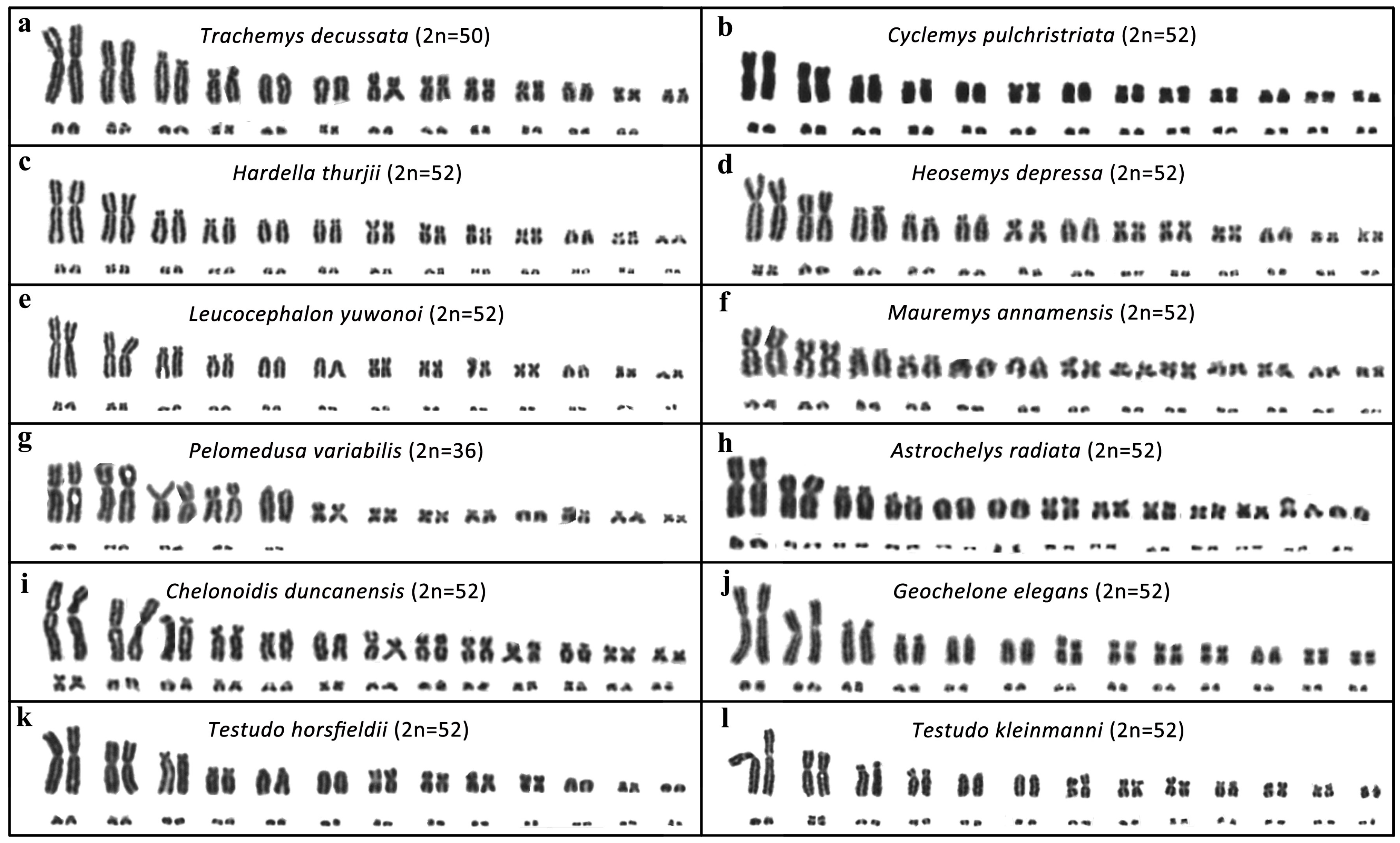
3.1. Karyotype Description
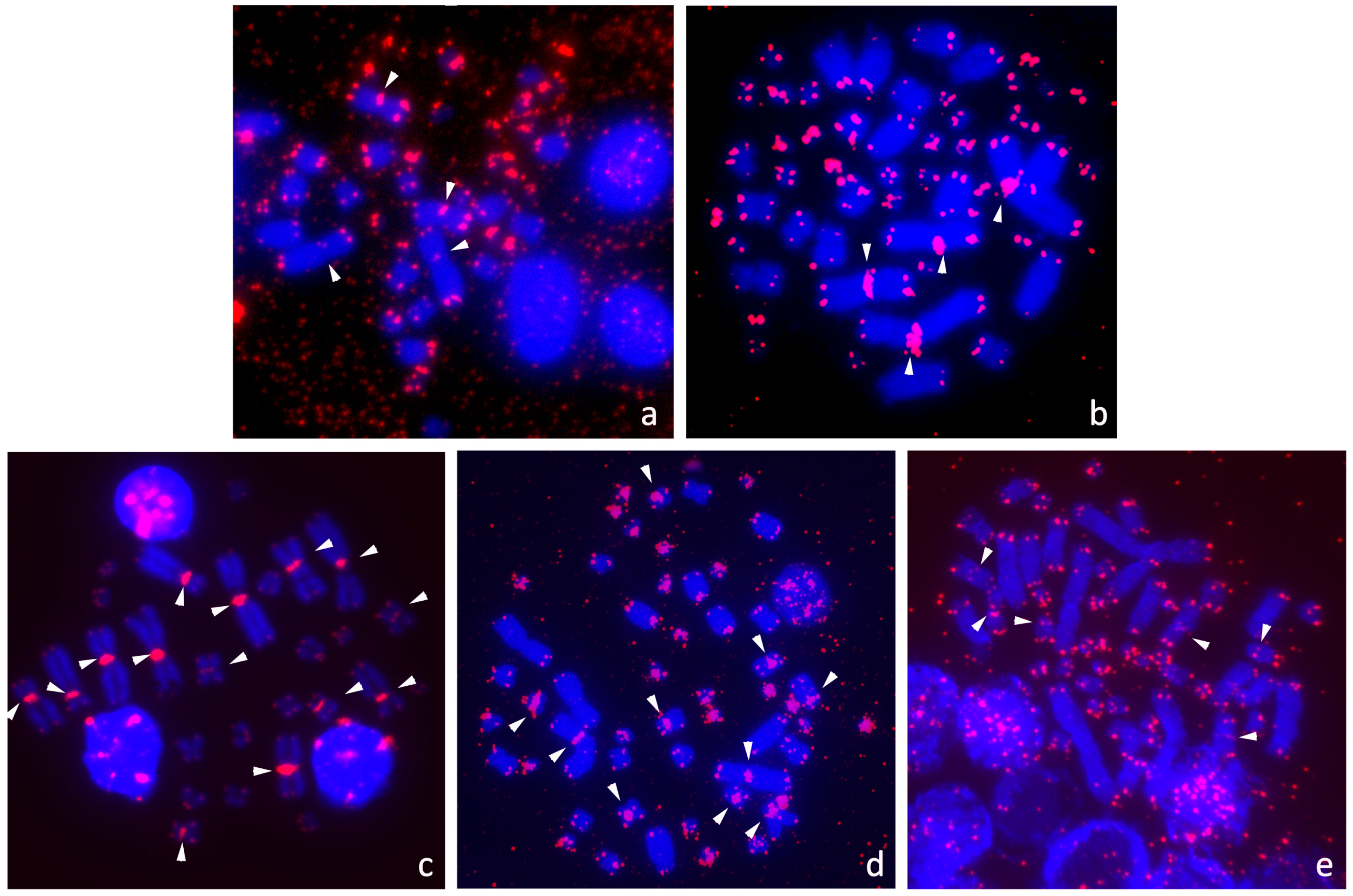
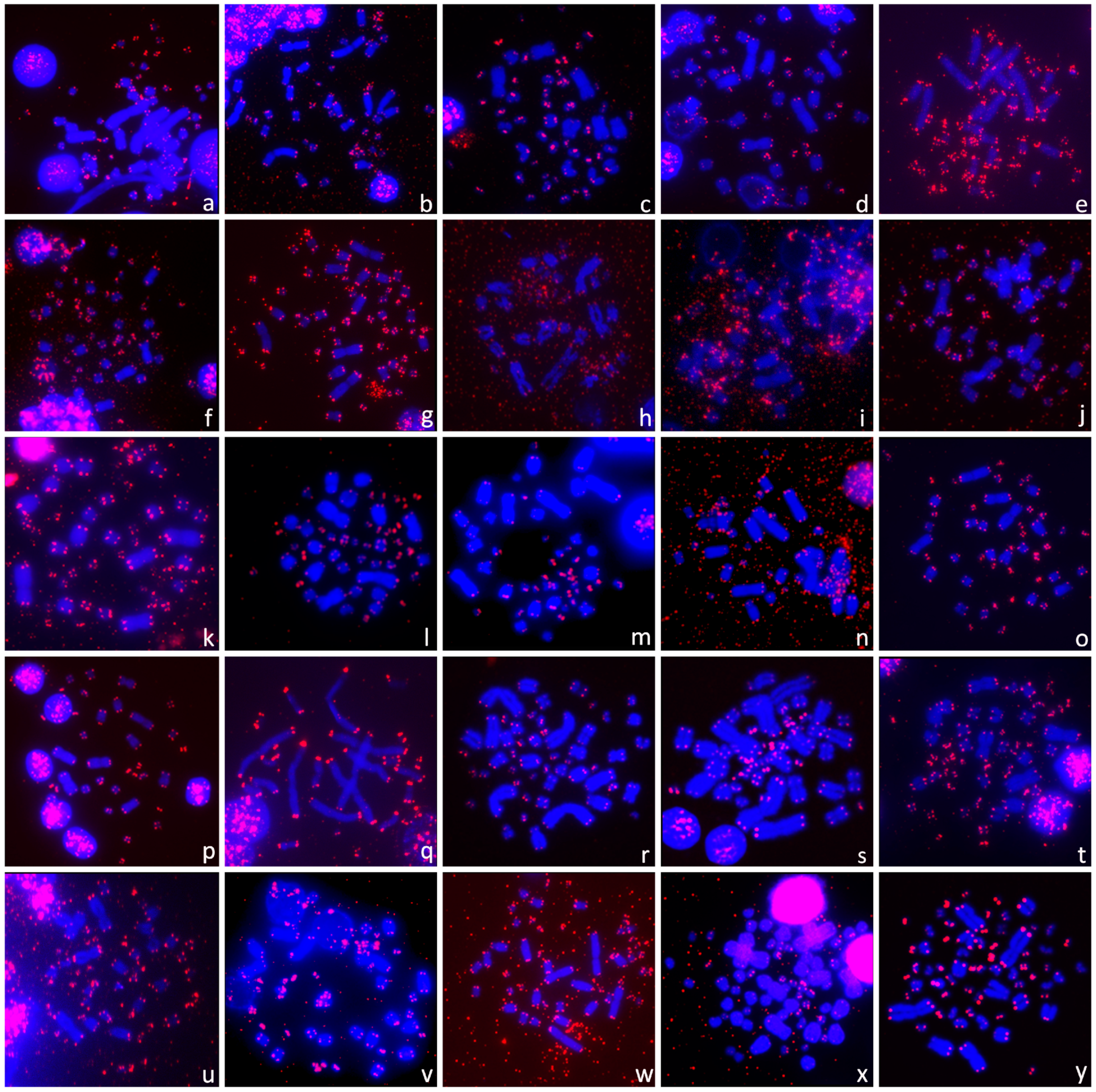
3.2. Presence of ITRs
3.3. Distribution of ITRs across the Turtle Phylogeny
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blackburn, E.H. Switching and signaling at the telomere. Cell 2001, 106, 661–673. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Crams, L.S.; Dani, M.; Larry, L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Chiodi, I.; Belgiovine, C.; Mondello, C. Telomerase and telomeric proteins: A life beyond telomeres. In Telomerase: Composition, Functions and Clinical Implications; Gagnon, A.N., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2010; pp. 35–58. [Google Scholar]
- Zhao, Z.; Pan, X.; Liu, L.; Liu, N. Telomere length maintenance, shortening, and lengthening. J. Cell. Physiol. 2014, 229, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.S.; Wright, W.E.; Shay, J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002, 66, 407–425. [Google Scholar] [CrossRef]
- Harley, B.C.; Futcher, B.A.; Greider, W.C. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Hastie, N.D.; Dempster, M.; Dunlop, M.G.; Thompson, A.M.; Green, D.K.; Allshire, R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990, 346, 866–868. [Google Scholar] [CrossRef]
- Lindsey, J.; McGill, N.I.; Lindsey, L.A.; Green, D.K.; Cooke, H.J. In vivo loss of telomeric repeats with age in humans. Mutat. Res. 1991, 256, 45–48. [Google Scholar] [CrossRef]
- Hayashi, M.T.; Cesare, A.J.; Rivera, T.; Karlseder, J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015, 522, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D.; Bianchi, M.S. Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat. Res. 2006, 612, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.W.; Yan, J. Endings in the middle: Current knowledge of interstitial telomeric sequences. Mutat. Res. 2008, 658, 95–110. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Nergadze, S.G.; Santagostino, M.; Giulotto, E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2008, 122, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Mucciolo, E.; Bertoni, L.; Giuliotto, E. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Genome Res. 1997, 78, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Santagostino, M.; Piras, F.M.; Cappelletti, E.; Del Giudice, S.; Semino, O.; Nergadze, S.G.; Giulotto, E. Insertion of telomeric repeats in the human and horse genomes: An evolutionary perspective. Int. J. Mol. Sci. 2020, 21, 2838. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Nergadze, S.G.; Giulotto, E. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 2001, 110, 75–82. [Google Scholar] [CrossRef]
- Faravelli, M.; Moralli, D.; Bertoni, L.; Attolini, C.; Chernova, O.; Raimondi, E.; Giulotto, E. Two extended arrays of a satellite DNA sequence at the centromere and at the short-arm telomere of Chinese hamster chromosome. Cytogenet. Genome Res. 1998, 83, 281–286. [Google Scholar] [CrossRef]
- Faravelli, M.; Azzalin, C.M.; Bertoni, L.; Chernova, O.; Attolini, C.; Mondello, C.; Giulotto, E. Molecular organization of internal telomeric sequences in Chinese hamster chromosomes. Gene 2002, 283, 11–16. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; García, F.; Azzalin, C.; Giulotto, E.; Egozcue, J.; Ponsà, M.; Garcia, M. Distribution of intrachromosomal telomeric sequences (ITS) on Macaca fascicularis (Primates) chromosomes and their implication for chromosome evolution. Hum. Genet. 2002, 110, 578–586. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; García, F.; Giulotto, E.; Attolini, C.; Egozcue, J.; Ponsà, M.; Garcia, M. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet. Genome Res. 2005, 108, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Santagostino, M.A.; Salzano, A.; Mondello, C.; Giulotto, E. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol. 2007, 8, R260. [Google Scholar] [CrossRef] [PubMed]
- Camats, N.; Ruiz-Herrera, A.; Parrilla, J.J.; Acien, M.; Payá, P.; Giulotto, E.; Egozcue, J.; García, F.; Garcia, M. Genomic instability in rat: Breakpoints induced by ionising radiation and interstitial telomeric-like sequences. Mutat. Res. 2006, 595, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.T.; Marchal, J.A.; Romero-Fernández, I.; Fernández, F.J.; Giagia-Athanosopoulou, E.B.; Sánchez, A. Rapid, independent, and extensive amplification of telomeric repeats in pericentromeric regions in karyotypes of arvicoline rodents. Chromosome Res. 2011, 19, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Johnson Pokorná, M. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Schillaci, O.; Sineo, L.; Dumas, F. Distribution of interstitial telomeric sequences in primates and the pygmy tree shrew (Scandentia). Cytogenet. Genome Res. 2017, 151, 141–150. [Google Scholar] [CrossRef]
- Milioto, V.; Vlah, S.; Mazzoleni, S.; Rovatsos, M.; Dumas, F. Chromosomal localization of 18S-28S rDNA and (TTAGGG)n sequences in two South African dormice of the genus Graphiurus (Rodentia: Gliridae). Cytogenet. Genome Res. 2019, 158, 145–151. [Google Scholar] [CrossRef]
- Swier, V.J.; Anwarali Khan, F.A.; Baker, R.J. Do time, heterochromatin, NORs, or chromosomal rearrangements correlate with distribution of interstitial telomeric repeats in Sigmodon (cotton rats)? J. Hered. 2012, 103, 493–502. [Google Scholar] [CrossRef]
- Nagamachi, C.Y.; Pieczarka, J.C.; O’Brien, P.C.M.; Pinto, J.A.; Malcher, S.M.; Pereira, A.L.; das Dores Rissino, J.; Mendes-Oliveira, A.C.; Rossi, R.V.; Ferguson-Smith, M.A. FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotype. Chromosome Res. 2013, 21, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Wiley, J.E.; Meyne, J.; Little, M.L.; Stout, J.C. Interstitial hybridization sites of the (TTAGGG)n telomeric sequence on the chromosomes of some North American hylid frogs. Cytogenet. Cell Genet. 1992, 61, 55–57. [Google Scholar] [CrossRef]
- Park, V.M.; Gustashaw, K.M.; Wathen, T.M. The presence of interstitial telomeric sequences in constitutional chromosome abnormalities. Am. J. Hum. Genet. 1992, 50, 914–923. [Google Scholar] [PubMed]
- Rossi, E.; Floridia, G.; Casali, M.; Danesino, C.; Chiumello, G.; Bernardi, F.; Magnani, I.; Papi, L.; Mura, M.; Zuffardi, O. Types, stability, and phenotypic consequences of chromosome rearrangements leading to interstitial telomeric sequences. J. Med. Genet. 1993, 30, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Vermeesch, J.R.; Petit, P.; Speleman, F.; Devriendt, K.; Fryns, J.P.; Marynen, P. Interstitial telomeric sequences at the junction site of a jumping translocation. Hum. Genet. 1997, 99, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Melek, M.; Shippen, D.E. Chromosome healing: Spontaneous and programmed de novo telomere formation by telomerase. BioEssays 1996, 18, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.R.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Nanda, I.; Schrama, D.; Feichtinger, W.; Haaf, T.; Schartl, M.; Schmid, M. Distribution of telomeric (TTAGGG)n sequences in avian chromosomes. Chromosoma 2002, 111, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Marchal, J.A.; Romero-Fernández, I.; Cano-Linares, M.; Fernández, F.J.; Giagia-Athanasopoulou, E.B.; Sánchez, A. Molecular and physical characterization of the complex pericentromeric heterochromatin of the vole species Microtus thomasi. Cytogenet. Genome Res. 2014, 144, 131–141. [Google Scholar] [CrossRef]
- Ocalewicz, K. Telomeres in fishes. Cytogenet. Genome Res. 2013, 141, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, D.P.; Rivera, M.; Lima, A.P.; Zúñiga, A.B.; Recco-Pimentel, S.M. Interstitial telomeric sequences (ITS) and major rDNA mapping reveal insights into the karyotypical evolution of Neotropical leaf frogs species (Phyllomedusa, Hylidae, Anura). Mol. Cytogenet. 2014, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Steinlein, C. Chromosome Banding in Amphibia. XXXIV. Intrachromosomal telomeric DNA sequences in Anura. Cytogenet. Genome Res. 2016, 148, 211–226. [Google Scholar] [CrossRef]
- De Oliveira, T.D.; Kretschmer, R.; Bertocchi, N.A.; Degrandi, T.M.; de Oliveira, E.H.C.; Cioffi, M.B.; Garnero, A.D.V.; Gunski, R.J. Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): Implications for karyotype and ZW sex chromosome differentiation. PLoS ONE 2017, 12, e0169987. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Azad, B.; Singchat, W.; Ezaz, T. Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS ONE 2019, 14, e0212683. [Google Scholar] [CrossRef] [PubMed]
- Zattera, M.L.; Lima, L.; Duarte, I.; de Sousa, D.Y. Cytogenetics chromosome spreading of the (TTAGGG)n repeats in the Pipa carvalhoi Miranda-Ribeiro, 1937 (Pipidae, Anura) karyotype. Comp. Cytogenet. 2019, 13, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Johnson Pokorná, M.; Altmanová, M.; Kratochvíl, L. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): Differentiation of sex and neo-sex chromosomes. Sci. Rep. 2015, 5, 13196. [Google Scholar] [CrossRef]
- Rovatsos, M.; Johnson Pokorná, M.; Kratochvíl, L. Differentiation of sex chromosomes and karyotype characterisation in the dragonsnake Xenodermus javanicus (Squamata: Xenodermatidae). Cytogenet. Genome Res. 2015, 147, 48–54. [Google Scholar] [CrossRef]
- Rovatsos, M.; Johnson Pokorná, M.; Altmanová, M.; Kratochvíl, L. Mixed-up sex chromosomes: Identification of sex chromosomes in the X1X1X2X2/X1X2Y system of the legless lizards of the genus Lialis (Squamata: Gekkota: Pygopodidae). Cytogenet. Genome Res. 2016, 149, 282–289. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Johnson Pokorná, M.; Velenský, P.; Sánchez Baca, A.; Kratochvíl, L. Evolution of karyotypes in chameleons. Genes 2017, 8, 382. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Johnson Pokorná, M.; Augstenová, B.; Kratochvíl, L. Cytogenetics of the Javan file snake (Acrochordus javanicus) and the evolution of snake sex chromosomes. J. Zool. Syst. Evol. Res. 2018, 56, 117–125. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Augstenová, B.; Mazzoleni, S.; Velenský, P.; Kratochvíl, L. ZZ/ZW sex determination with multiple neo-sex chromosomes is common in Madagascan chameleons of the genus Furcifer. Genes 2019, 10, 1020. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Matsuda, Y.; Miller, E.; Olsson, M. No interstitial telomeres on autosomes but remarkable amplification of telomeric repeats on the W sex chromosome in the sand lizard (Lacerta agilis). J. Hered. 2015, 106, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Ota, H.; Matsuda, Y. Karyotype reorganization in the Hokou gecko (Gekko hokouensis, Gekkonidae): The process of microchromosome disappearance in Gekkota. PloS ONE 2015, 10, e0134829. [Google Scholar] [CrossRef]
- Viana, P.F.; Ribeiro, L.B.; Souza, G.M.; Chalkidis, H.D.M.; Gross, M.C.; Feldberg, E. Is the karyotype of neotropical boid snakes really conserved? Cytotaxonomy, chromosomal rearrangements and karyotype organization in the Boidae family. PLoS ONE 2016, 11, e0160274. [Google Scholar] [CrossRef][Green Version]
- Viana, P.F.; Ezaz, T.; Cioffi, M.D.B.; Almeida, B.J.; Feldberg, E. Evolutionary insights of the ZW sex chromosomes in snakes: A new chapter added by the Amazonian puffing snakes of the genus Spilotes. Genes 2019, 10, 288. [Google Scholar] [CrossRef]
- Altmanová, M.; Rovatsos, M.; Kratochvíl, L.; Johnson Pokorná, M. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae). Biol. J. Linn. Soc. 2016, 118, 618–633. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Slimani, T.; Splendiani, A.; Paoletti, A.; Fawzi, A.; Olmo, E.; Caputo Barucchi, V. Cytogenetic characterization of a population of Acanthodactylus lineomaculatus Duméril and Bibron, 1839 (Reptilia, Lacertidae), from Southwestern Morocco and insights into sex chromosome evolution. Cytogenet. Genome Res. 2017, 153, 86–95. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Rojo, V.; Olmo, E.; Slimani, T.; Splendiani, A.; Caputo Barucchi, V. Characterization of a satellite DNA in the genera Lacerta and Timon (Reptilia, Lacertidae) and its role in the differentiation of the W chromosome. J. Exp. Zool. B Mol. Dev. Evol. 2018, 330, 83–95. [Google Scholar] [CrossRef]
- Augstenová, B.; Mazzoleni, S.; Kratochvíl, L.; Rovatsos, M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes 2018, 9, 5. [Google Scholar] [CrossRef]
- Augstenová, B.; Mazzoleni, S.; Kostmann, A.; Altmanová, M.; Frynta, D.; Kratochvíl, L.; Rovatsos, M. Cytogenetic analysis did not reveal differentiated sex chromosomes in ten species of boas and pythons (Reptilia: Serpentes). Genes 2019, 10, 934. [Google Scholar] [CrossRef]
- Singchat, W.; O’Connor, R.E.; Tawichasri, P.; Suntronpong, A.; Sillapaprayoon, S.; Suntrarachun, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genom. 2018, 19, 939. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.J.; de Araújo Vieira, A.P.; Galvão Cipriano, F.M.; dos Santos Cândido, M.R.; de Oliveira, E.H.C.; Gimenez Pinheiro, T.; da Silva, E.L. The karyotype of Salvator merianae (Squamata, Teiidae): Analyses by classical and molecular cytogenetic techniques. Cytogenet. Genome Res. 2020, 160, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Sidhom, M.; Said, K.; Chatti, N.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O.; Mezzasalma, M. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology 2020, 125738, in press. [Google Scholar] [CrossRef] [PubMed]
- Kawagoshi, T.; Nishida, C.; Ota, H.; Kumazawa, Y.; Endo, H.; Matsuda, Y. Molecular structures of centromeric heterochromatin and karyotypic evolution in the Siamese crocodile (Crocodylus siamensis) (Crocodylidae, Crocodylia). Chromosome Res. 2008, 16, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Nishida, C.; Ishijima, J.; Kosaka, A.; Tanabe, H.; Habermann, F.A.; Griffin, D.K.; Matsuda, Y. Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res. 2008, 16, 171–181. [Google Scholar] [CrossRef]
- Seibold-Torres, C.; Owens, E.; Chowdhary, R.; Ferguson-Smith, M.A.; Tizard, I.; Raudsepp, T. Comparative cytogenetics of the Congo African grey parrot (Psittacus erithacus). Cytogenet. Genome Res. 2015, 147, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Liangouzov, I.A.; Derjusheva, S.E.; Saifitdinova, A.F.; Malykh, A.G.; Gaginskaya, E.R. Monomers of a satellite DNA sequence of chaffinch (Fringilla coelebs L., Aves: Passeriformes) contain short clusters of the TTAGGG repeat. Russ. J. Genet. 2002, 38, 1359–1364. [Google Scholar] [CrossRef]
- Derjusheva, S.; Kurganova, A.; Habermann, F.; Gaginskaya, E. High chromosome conservation detected by comparative chromosome painting in chicken, pigeon and passerine birds. Chromosome Res. 2004, 12, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Freed, P.; Hošek, J. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 13 April 2020).
- Vargas-Ramírez, M.; Caballero, S.; Morales-Betancourt, M.A.; Lasso, C.A.; Amaya, L.; Martínez, J.G.; das Neves Silvia Viana, M.; Vogt, R.C.; Farias, I.P.; Hrbek, T.; et al. Genomic analyses reveal two species of the matamata (Testudines: Chelidae: Chelus spp.) and clarify their phylogeography. Mol. Phylogenet. Evol. 2020, 148, 106823. [Google Scholar]
- Martinez, P.A.; Boeris, J.M.; Sánchez, J.; Pastori, M.C.; Bolzán, A.D.; Ledesma, M.A. Karyotypic characterization of Trachemys dorbigni (Testudines: Emydidae) and Chelonoidis (Geochelone) donosobarrosi (Testudines: Testudinidae), two species of cryptodiran turtles from Argentina. Genetica 2009, 137, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Badenhorst, D.; Hillier, L.D.; Literman, R.; Montiel, E.E.; Radhakrishnan, S.; Shen, Y.; Minx, P.; Janes, D.E.; Warren, W.C.; Edwards, S.V.; et al. Physical mapping and refinement of the painted turtle genome (Chrysemys picta) inform amniote genome evolution and challenge turtle-bird chromosomal conservation. Genome Biol. Evol. 2013, 7, 2038–2050. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ventura, K.; Moreira, C.N.; Moretti, R.; Yonenaga-Yassuda, Y.; Rodrigues, M.T. The lowest diploid number in Testudines: Banding patterns, telomeric and 45s rDNA FISH in Peltocephalus dumerilianus, 2n = 26 and FN = 52 (Pleurodira, Podocnemididae). Genet. Mol. Biol. 2014, 37, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.; Alcalde, L.; Bolzán, A.D. First evidence of chromosomal variation within Chelonoidis chilensis (Testudines: Testudinidae). Herpetol. J. 2015, 25, 83–89. [Google Scholar]
- Montiel, E.E.; Badenhorst, D.; Lee, L.S.; Literman, R.; Trifonov, V.; Valenzuela, N. Cytogenetic insights into the evolution of chromosomes and sex determination reveal striking homology of turtle sex chromosomes to amphibian autosomes. Cytogenet. Genome Res. 2016, 148, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Noronha, R.C.R.; Barros, L.M.R.; Araújo, R.E.F.; Marques, D.F.; Nagamachi, C.Y.; Martins, C.; Pieczarka, J.C. New insights of karyoevolution in the Amazonian turtles Podocnemis expansa and Podocnemis unifilis (Testudines, Podocnemidae). Mol. Cytogenet. 2016, 9, 73. [Google Scholar] [CrossRef]
- Cavalcante, M.G.; Bastos, C.E.M.C.; Nagamachi, C.Y.; Pieczarka, J.C.; Vicari, M.R.; Noronha, R.C.R. Physical mapping of repetitive DNA suggests 2n reduction in Amazon turtles Podocnemis (Testudines: Podocnemididae). PLoS ONE 2018, 13, e0197536. [Google Scholar] [CrossRef] [PubMed]
- Iannucci, A.; Svartman, M.; Bellavita, M.; Chelazzi, G.; Stanyon, R.; Ciofi, C. Insights into Emydid turtle cytogenetics: The European pond turtle as a model species. Cytogenet. Genome Res. 2019, 157, 166–171. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Turtles of the genera Geoemyda and Pangshura (Testudines: Geoemydidae) lack differentiated sex chromosomes: The end of a 40-year error cascade for Pangshura. PeerJ 2019, 7, e6241. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 4276. [Google Scholar] [CrossRef]
- Cavalcante, M.G.; Souza, L.F.; Vicari, M.R.; de Bastos, C.E.M.; de Sousa, J.V.; Nagamachi, C.Y.; Pieczarka, J.C.; Martins, C.; Noronha, R.C.R. Molecular cytogenetics characterization of Rhinoclemmys punctularia (Testudines, Geoemydidae) and description of a Gypsy-H3 association in its genome. Gene 2020, 738, 144477. [Google Scholar] [CrossRef]
- Olmo, E.; Signorino, G.G. Chromorep: A reptile chromosomes database. Available online: http://chromorep.univpm.it (accessed on 14 April 2020).
- Bickham, J.W. Two hundred million year old chromosomes: Deceleration of the rate of karyotypic evolution in turtles. Science 1981, 212, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Fritz, U.; Guicking, D.; Auer, M.; Sommer, R.S.; Wink, M.; Hundsdörfer, A.K. Diversity of the Southeast Asian leaf turtle genus Cyclemys: How many leaves on its tree of life? Zool. Scr. 2008, 37, 367–390. [Google Scholar] [CrossRef]
- Petzold, A.; Vargas-Ramírez, M.; Kehlmaier, C.; Vamberger, M.; Branch, W.R.; Du Preez, L.; Hofmeyr, M.D.; Schleicher, A.; Široký, P.; Fritz, U. A revision of African helmeted terrapins (Testudines: Pelomedusidae: Pelomedusa), with descriptions of six new species. Zootaxa 2014, 3795, 523–548. [Google Scholar] [CrossRef]
- Ihlow, F.; Vamberger, M.; Flecks, M.; Hartmann, T.; Cota, M.; Makchai, S.; Meewattana, P.; Dawson, J.E.; Kheng, L.; Rödder, D. Integrative taxonomy of Southeast Asian snail-eating turtles (Geoemydidae: Malayemys) reveals a new species and mitochondrial introgression. PLoS ONE 2016, 11, e0153108. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef]
- Pereira, A.G.; Sterli, J.; Moreira, F.R.R.; Schrago, C.G. Multilocus phylogeny and statistical biogeography clarify the evolutionary history of major lineages of turtles. Mol. Phylogenet. Evol. 2017, 113, 59–66. [Google Scholar] [CrossRef]
- Kehlmaier, C.; Zhang, X.; Georges, A.; Campbell, P.D.; Thomson, S.; Fritz, U. Mitogenomics of historical type specimens of Australasian turtles: Clarification of taxonomic confusion and old mitochondrial introgression. Sci. Rep. 2019, 9, 5841. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A modular system for evolutionary analysis. Version 3.61. Available online: http://mesquiteproject.org (accessed on 15 March 2020).
- Delany, M.E.; Krupkin, A.B.; Miller, M.M. Organization of telomere sequences in birds: Evidence for arrays of extreme length and for in vivo shortening. Cytogenet. Cell Genet. 2000, 90, 139–145. [Google Scholar] [CrossRef]
- Raudsepp, T.; Houck, M.L.; O’Brien, P.C.; Ferguson-Smith, M.A.; Ryder, O.A.; Chowdhary, B.P. Cytogenetic analysis of California condor (Gymnogyps californianus) chromosomes: Comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet. Genome Res. 2002, 98, 54–60. [Google Scholar] [CrossRef]
- Swanberg, S.E.; Delany, M.E. Telomeres in aging: Birds. In Handbook of Models for Human Aging; Conn, P.M., Ed.; Elsevier Academic Press: Cambridge, MA, USA, 2006; pp. 339–349. [Google Scholar]
- Dos Santos, M.S.; Kretschmer, R.; Silva, F.A.O.; Ledesma, M.A.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Del Valle Garnero, A.; de Oliveira, E.H.C.; Gunski, R.J. Intrachromosomal rearrangements in two representatives of the genus Saltator (Thraupidae, Passeriformes) and the occurrence of heteromorphic Z chromosomes. Genetica 2015, 143, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Ferguson-Smith, M.A.; de Oliveira, E.H.C. Karyotype evolution in birds: From conventional staining to chromosome painting. Genes 2018, 9, 181. [Google Scholar] [CrossRef]
- Johnson Pokorná, M.; Rovatsos, M.; Kratochvíl, L. Sex chromosomes and karyotype of the (nearly) mythical creature, the Gila monster, Heloderma suspectum (Squamata: Helodermatidae). PLoS ONE 2014, 9, e104716. [Google Scholar]
- Stock, A.D. Karyological relationships in turtles (Reptilia: Chelonia). Can. J. Genet. Cytol. 1972, 14, 859–868. [Google Scholar] [CrossRef]
- Killebrew, F.C. Mitotic chromosomes of turtles. III. The Kinosternidae. Herpetologica 1975, 31, 398–403. [Google Scholar]
- Rhodin, A.G.J.; Iverson, J.B.; Bour, R.; Fritz, U.; Georges, A.; Shaffer, H.B. Turtles of the World: Annotated Checklist and Atlas of Taxonomy. Synon. Distrib. Conserv. Status 2017, 8, 9–14. [Google Scholar]
- Rovatsos, M.; Praschag, P.; Fritz, U.; Kratochvíl, L. Stable Cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae). Sci. Rep. 2017, 7, 42150. [Google Scholar] [CrossRef] [PubMed]
- Literman, R.; Radhakrishnan, S.; Tamplin, J.; Burke, R.; Dresser, C.; Valenzuela, N. Development of sexing primers in Glyptemys insculpta and Apalone spinifera turtles uncovers an XX/XY sex-determining system in the critically-endangered bog turtle Glyptemys muhlenbergii. Conserv. Genet. Res. 2017, 9, 651–658. [Google Scholar] [CrossRef]
- Bull, J.J.; Moon, R.G.; Legler, J.M. Male heterogamety in kinosternid turtles (genus Staurotypus). Cytogenet. Genome Res. 1974, 13, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Kawagoshi, T.; Uno, Y.; Nishida, C.; Matsuda, Y. The Staurotypus turtles and Aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS ONE 2014, 9, e105315. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kawagoshi, T.; Nishida, C.; Matsuda, Y. The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res. 2012, 20, 95–110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: Berlin, Germany, 1967; Volume 1. [Google Scholar]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef]
- Vicoso, B. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 2019, 3, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Kejnovsky, E.; Bertollo, L.A.C. The chromosomal distribution of microsatellite repeats in the genome of the wolf fish Hoplias malabaricus, focusing on the sex chromosomes. Cytogenet. Genome Res. 2011, 132, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Knopp, T.; Sarre, S.D.; Georges, A.; Ezaz, T. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink-tailed worm-lizard (Aprasia parapulchella, Pygopodidae, Squamata). Mol. Cytogenet. 2013, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Scacchetti, P.C.; Utsunomia, R.; Pansonato-Alves, J.C.; da Costa Silva, G.J.; Vicari, M.R.; Artoni, F.R.; Oliveira, C.; Foresti, F. Repetitive DNA sequences and evolution of ZZ/ZW sex chromosomes in Characidium (Teleostei: Characiformes). PLoS ONE 2015, 10, e0137231. [Google Scholar] [CrossRef] [PubMed]
- Pokorná, M.; Rens, W.; Rovatsos, M.; Kratochvíl, L. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 2014, 142, 190–196. [Google Scholar] [CrossRef]
- Meles, S.; Adega, F.; Guedes-Pinto, H.; Chaves, R. The karyotype and sex chromosomes of Praomys tullbergi (Muridae, Rodentia): A detailed characterization. Micron 2008, 39, 559–568. [Google Scholar] [CrossRef]
- Cioffi, M.B.; Bertollo, L.A.C. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this fish group. Heredity 2010, 105, 554–561. [Google Scholar] [CrossRef]
- Mank, J.E. Small but mighty: The evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 2012, 20, 21–33. [Google Scholar] [CrossRef]
- Deakin, J.E.; Potter, S.; O’ Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.B.; Fukui, K.; Marshall Graves, J.A.; Griffin, D.; Grutzner, F. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Presting, G.G. Retrotransposon insertion targeting: A mechanism for homogenization of centromere sequences on nonhomologous chromosomes. Genes. Dev. 2012, 26, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef]
- Hooper, D.M.; Price, T.D. Chromosomal inversion differences correlate with range overlap in passerine birds. Nat. Ecol. Evol. 2017, 1, 1526–1534. [Google Scholar] [CrossRef]
- Hooper, D.M.; Griffith, S.C.; Price, T.D. Sex chromosome inversions enforce reproductive isolation across an avian hybrid zone. Mol. Ecol. 2018, 28, 1246–1262. [Google Scholar] [CrossRef]
- Fuller, Z.L.; Leonard, C.J.; Young, R.E.; Schaeffer, W.; Phadnis, N. Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 2018, 14, e1007526. [Google Scholar] [CrossRef]
- Kirkpatrick, M.; Barton, N. Chromosome inversion, local adaptation and speciation. Genetics 2006, 173, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Wellenreuther, M.; Bernatchez, L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 2018, 33, 427–440. [Google Scholar] [CrossRef]
- Avise, J.C.; Bowen, B.W.; Lamb, T.; Meylan, A.B.; Bermingham, E. Mitochondrial DNA evolution at a turtle’s place: Evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Mol. Biol. Evol. 1992, 9, 457–473. [Google Scholar]
- Lourenço, J.M.; Glémin, S.; Chiari, Y.; Galtier, N. The determinants of molecular substitution process in turtles. J. Evol. Biol. 2013, 26, 38–50. [Google Scholar] [CrossRef]
- Shaffer, B.H.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K. The western painted turtle genome, a model for evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013, 14, R28. [Google Scholar] [CrossRef] [PubMed]
- Fritz, U. Schildkröten-Hybriden, 2. Halsberger-Schildkröten (Cryptodira). Herpetofauna 1995, 95, 19–34. [Google Scholar]
- Karl, S.A.; Bowen, B.W.; Avise, J.C. Hybridization among the ancient mariners: Characterization of marine turtle hybrids with molecular genetic assays. J. Hered. 1995, 86, 262–268. [Google Scholar] [CrossRef]
- Parham, J.F.; Simison, W.B.; Kozak, K.H.; Feldman, C.R.; Shi, H. New Chinese turtles: Endangered or invalid? A reassessment of two species using mitochondrial DNA, allozyme electrophoresis and known-locality specimens. Anim. Conserv. 2001, 4, 357–367. [Google Scholar] [CrossRef]
- Fritz, U.; Mendau, D. Ein Gattungsbastard zweier südostasiatischer Schildkröten: Cuora amboinensis kamaroma Rummler & Fritz, 1991 x Mauremys annamensis (Siebenrock, 1903). Salamandra 2002, 38, 129–134. [Google Scholar]
- Seminoff, J.A.; Karl, S.A.; Schwartz, T.; Resendiz, A. Hybridization of the green turtle (Chelonia mydas) and hawksbill turtle (Eretmochelys imbricata) in the Pacific Ocean: Indication of an absence of gender bias in the directionality of crosses. Bull. Mar. Sci. 2003, 73, 643–652. [Google Scholar]
- James, M.C.; Martin, K.; Dutton, P.H. Hybridization between a green turtle, Chelonia mydas, and loggerhead turtle, Caretta caretta, and the first record of a green turtle in Atlantic Canada. Can. Field-Nat. 2004, 118, 579–582. [Google Scholar] [CrossRef]
- Lara-Ruiz, P.; Lopez, G.G.; Santos, F.R.; Soares, L.S. Extensive hybridization in hawksbill turtles (Eretmochelys imbricata) nesting in Brazil revealed by mtDNA analyses. Conserv. Genet. 2006, 7, 773–781. [Google Scholar] [CrossRef]
- Stuart, B.L.; Parham, J.F. Recent hybrid origin of three rare Chinese turtles. Conserv. Genet. 2006, 8, 169–175. [Google Scholar] [CrossRef]
- Lee, Y.; Lin, J.W.; Tseng, S.P.; Chen, T.S.; Lin, S.M. Human disturbance as a possible cause of genetic introgression from exotic into native Mauremys turtles. Anim. Conserv. 2019, 22, 556–567. [Google Scholar] [CrossRef]
- Burgtorf, C.; Bünemann, H. A telomere-like satellite (GGGTCAT)n comprises 4% of genomic DNA of Drosophila hydei and is located mainly in centromeric heterochromatin of all large acrocentric autosomes. Gene 1993, 137, 287–291. [Google Scholar] [CrossRef]
| Suborder | Family | Species | Sex | ||
|---|---|---|---|---|---|
 |  | Unknown | |||
| Cryptodira | Carettochelyidae | Carettochelys insculpta | - | - | 1 |
| Emydidae | Graptemys pseudogeographica | 1 | |||
| Trachemys decussata | 1 | - | - | ||
| Geoemydidae | Cyclemys dentata | - | - | 1 | |
| Cyclemys pulchristriata | 2 | - | - | ||
| Hardella thurjii | 1 | ||||
| Heosemys depressa | - | 1 | - | ||
| Leucocephalon yuwonoi | 1 | 1 | - | ||
| Malayemys macrocephala | 1 | - | - | ||
| Mauremys annamensis | 2 | 2 | - | ||
| Mauremys japonica | - | - | 4 | ||
| Mauremys rivulata | - | 1 | - | ||
| Siebenrockiella crassicollis | - | 1 | - | ||
| Kinosternidae | Claudius angustatus | 1 | 1 | - | |
| Staurotypus salvinii | 1 | - | - | ||
| Testudinidae | Astrochelys radiata | - | - | 1 | |
| Chelonoidis carbonarius | - | - | 1 | ||
| Chelonoidis duncanensis | 1 | - | - | ||
| Geochelone elegans | 1 | 1 | - | ||
| Gopherus berlandieri | 1 | - | - | ||
| Stigmochelys pardalis | 1 | - | - | ||
| Testudo horsfieldii | - | 1 | - | ||
| Testudo kleinmanni | 1 | - | - | ||
| Trionychidae | Apalone ferox | - | 1 | 1 | |
| Lissemys punctata | - | 1 | - | ||
| Pleurodira | Chelidae | Emydura subglobosa | - | 1 | - |
| Mesoclemmys hogei | 1 | - | - | ||
| Pelomedusidae | Pelomedusa variabilis | 1 | 1 | - | |
| Podocnemididae | Peltocephalus dumerilianus | 1 | - | - | |
| Podocnemis unifilis | 1 | - | - | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clemente, L.; Mazzoleni, S.; Pensabene Bellavia, E.; Augstenová, B.; Auer, M.; Praschag, P.; Protiva, T.; Velenský, P.; Wagner, P.; Fritz, U.; et al. Interstitial Telomeric Repeats Are Rare in Turtles. Genes 2020, 11, 657. https://doi.org/10.3390/genes11060657
Clemente L, Mazzoleni S, Pensabene Bellavia E, Augstenová B, Auer M, Praschag P, Protiva T, Velenský P, Wagner P, Fritz U, et al. Interstitial Telomeric Repeats Are Rare in Turtles. Genes. 2020; 11(6):657. https://doi.org/10.3390/genes11060657
Chicago/Turabian StyleClemente, Lorenzo, Sofia Mazzoleni, Eleonora Pensabene Bellavia, Barbora Augstenová, Markus Auer, Peter Praschag, Tomáš Protiva, Petr Velenský, Philipp Wagner, Uwe Fritz, and et al. 2020. "Interstitial Telomeric Repeats Are Rare in Turtles" Genes 11, no. 6: 657. https://doi.org/10.3390/genes11060657
APA StyleClemente, L., Mazzoleni, S., Pensabene Bellavia, E., Augstenová, B., Auer, M., Praschag, P., Protiva, T., Velenský, P., Wagner, P., Fritz, U., Kratochvíl, L., & Rovatsos, M. (2020). Interstitial Telomeric Repeats Are Rare in Turtles. Genes, 11(6), 657. https://doi.org/10.3390/genes11060657







