Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of MSTs in B. napus, B. rapa and B. oleracea
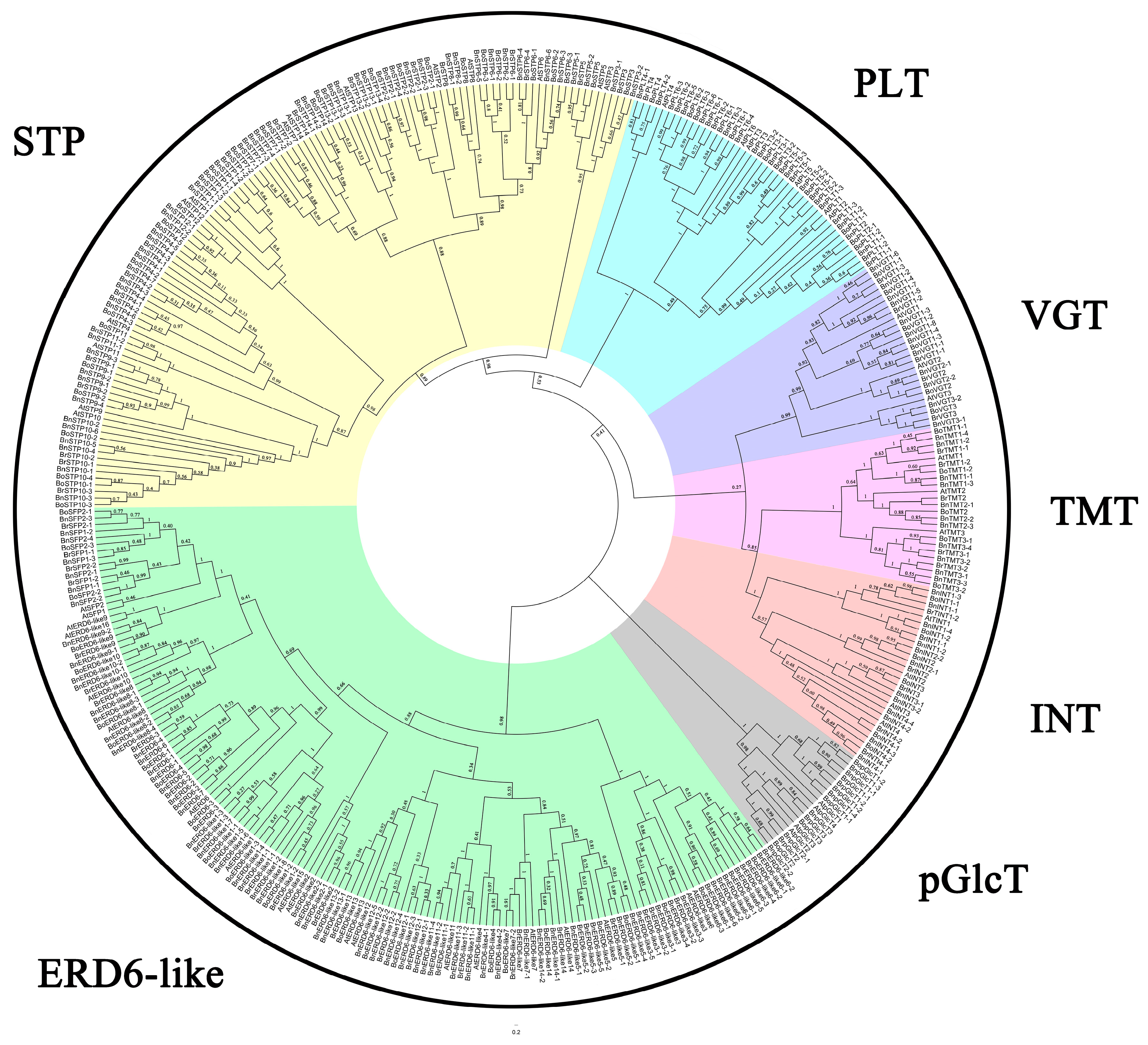
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
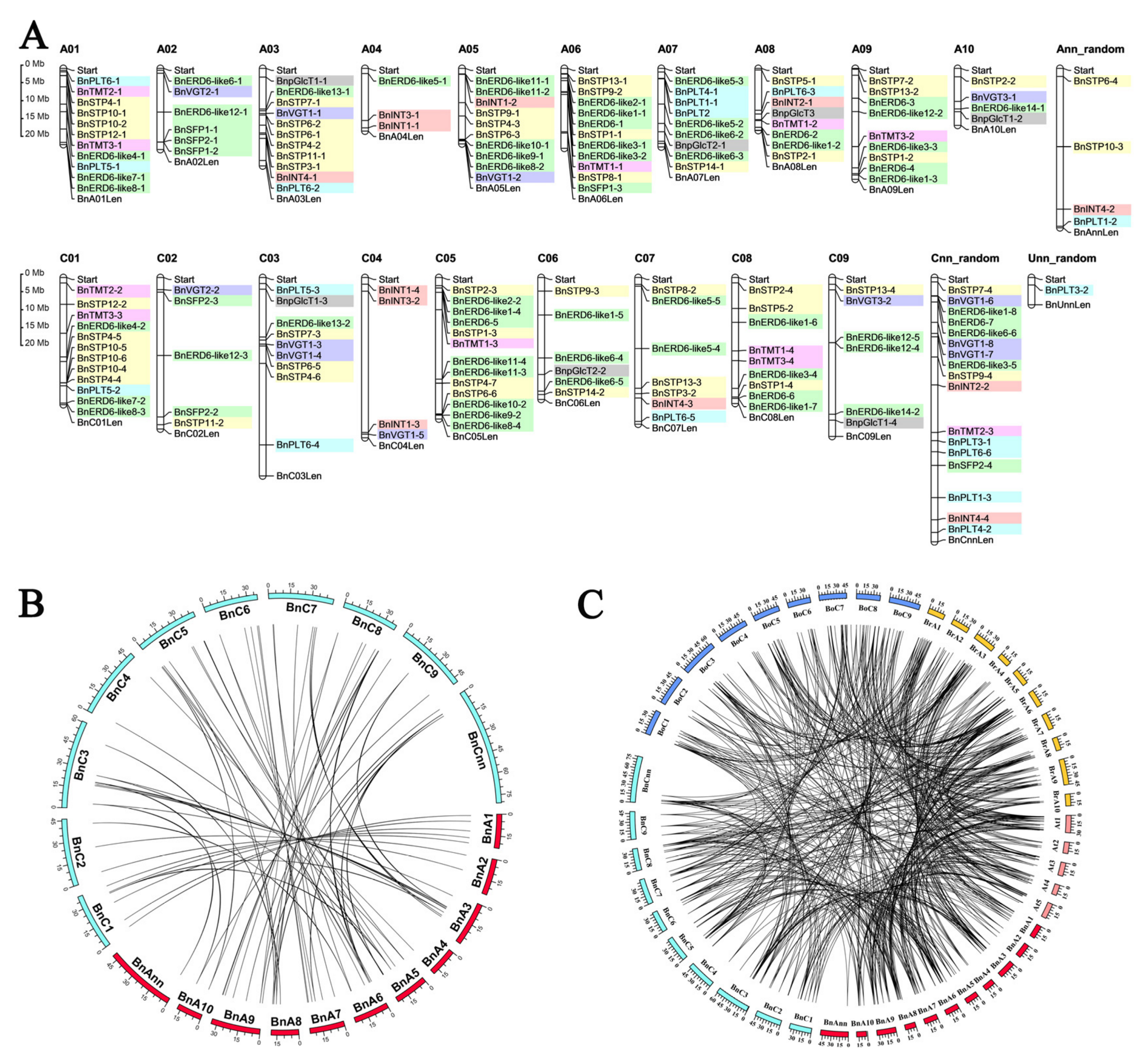
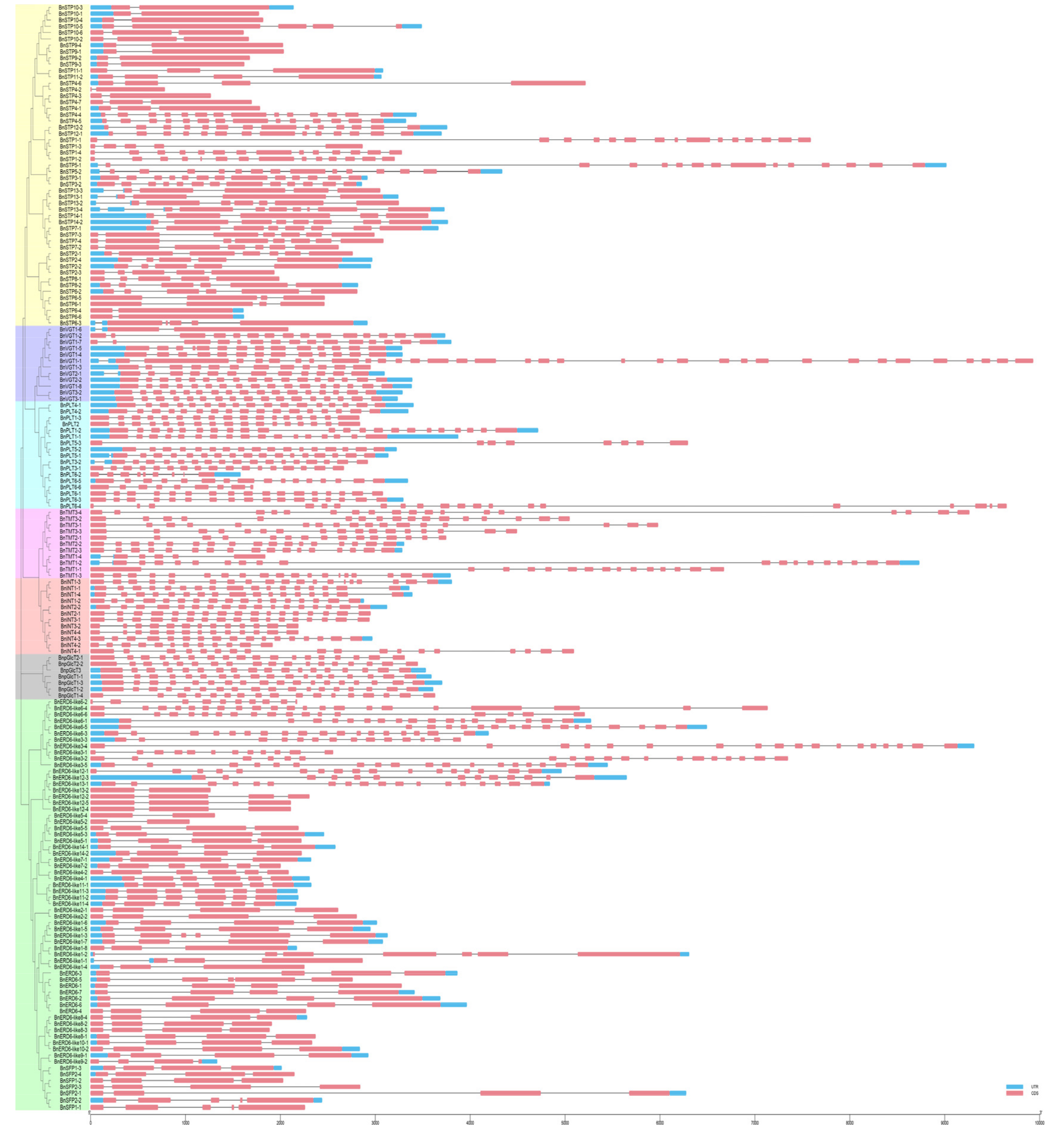
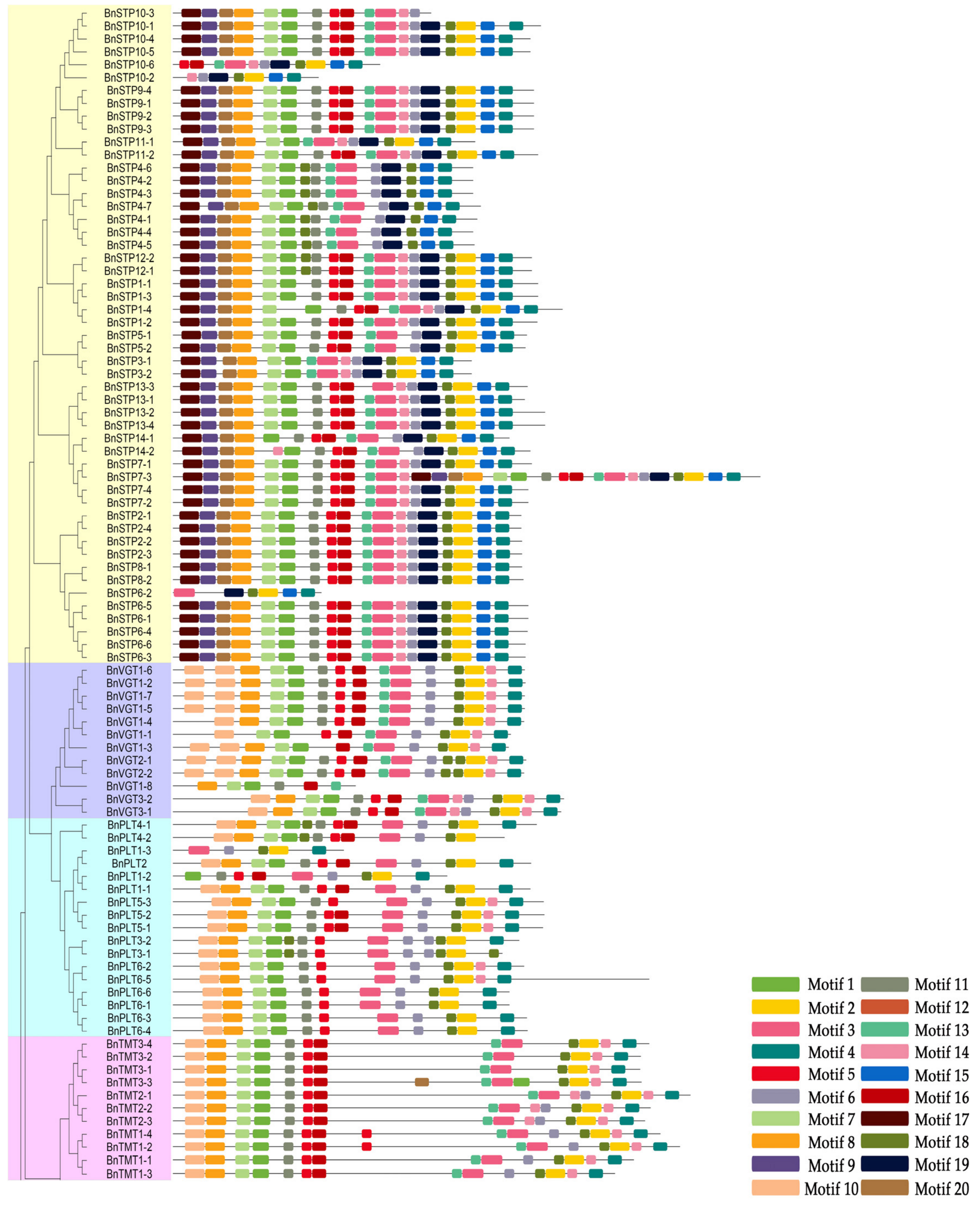
2.3. Analysis of the Chromosomal Locations, Gene Structures, and Conserved Motifs of the BnMST Genes
2.4. RNA-seq Analysis
2.5. Plant Materials
2.6. RNA Extraction and Validation of RNA-seq Data by qRT-PCR
3. Results
3.1. Identification and Phylogenetic Analysis of MSTs
3.2. Chromosome Locations of BnMST Genes and Duplication Analysis
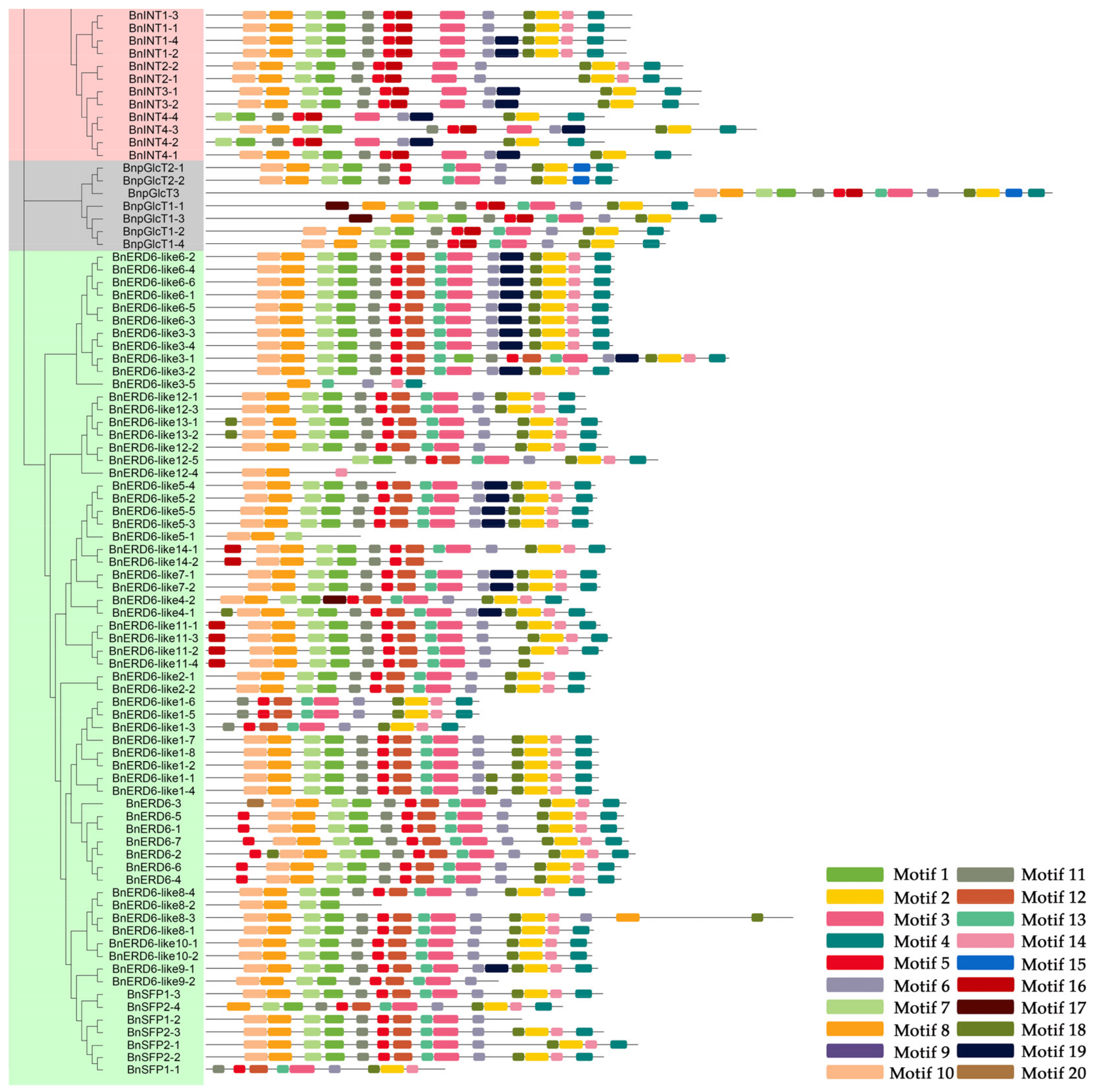
3.3. Exon-Intron Structures and Conserved Motif Analysis of the BnMSTs
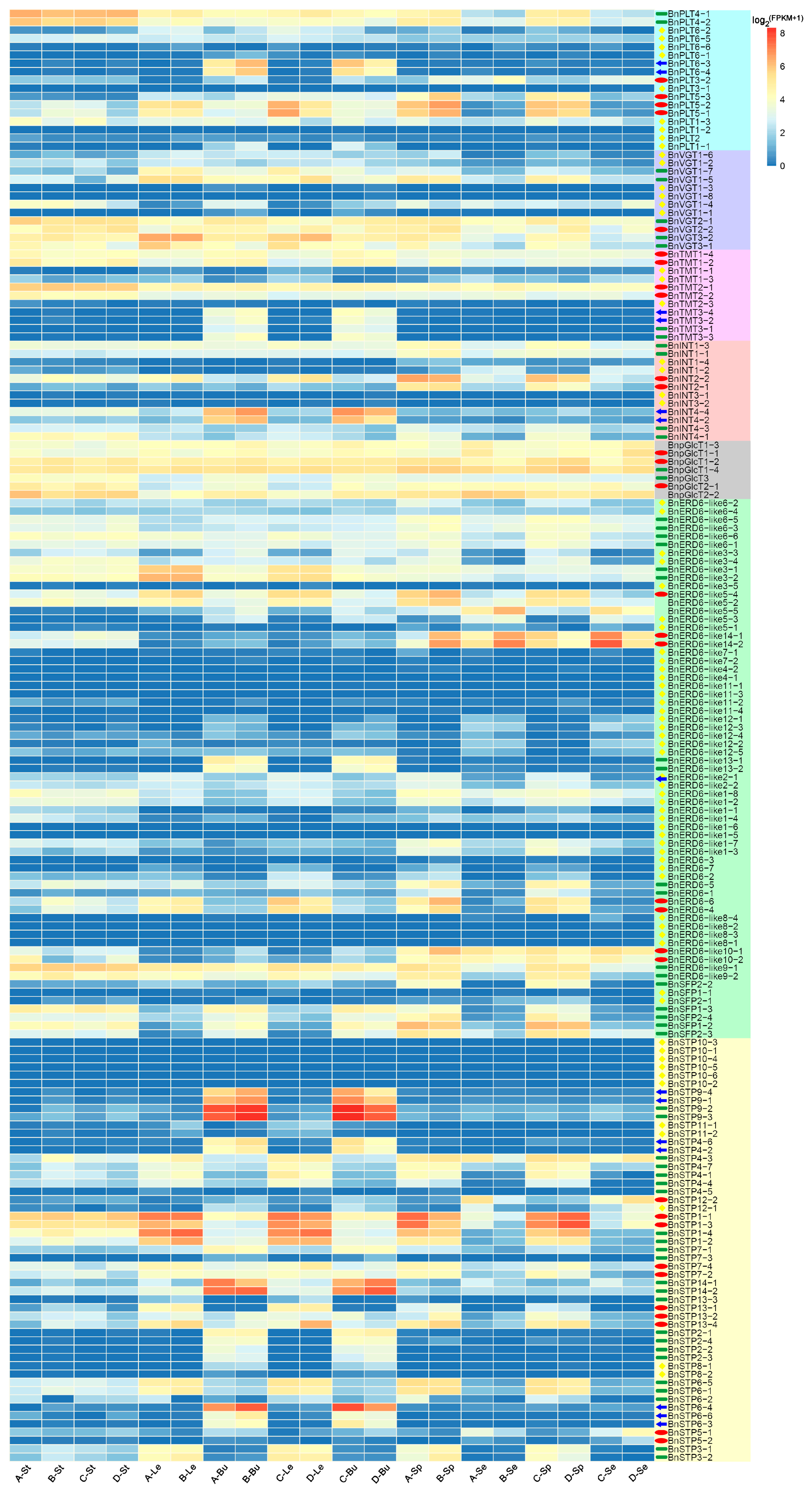
3.4. RNA-seq Analysis of the 175 BnMST Genes
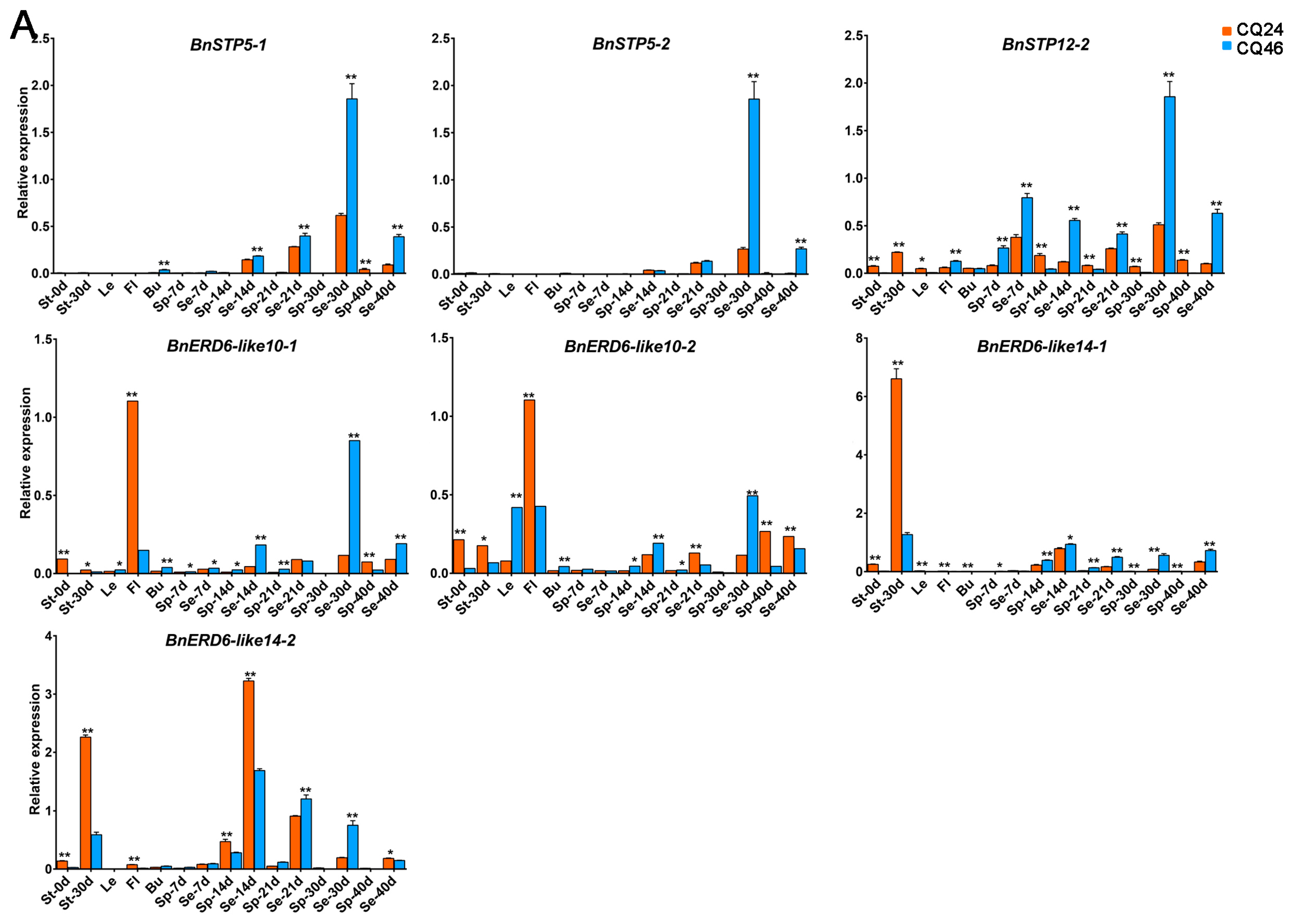
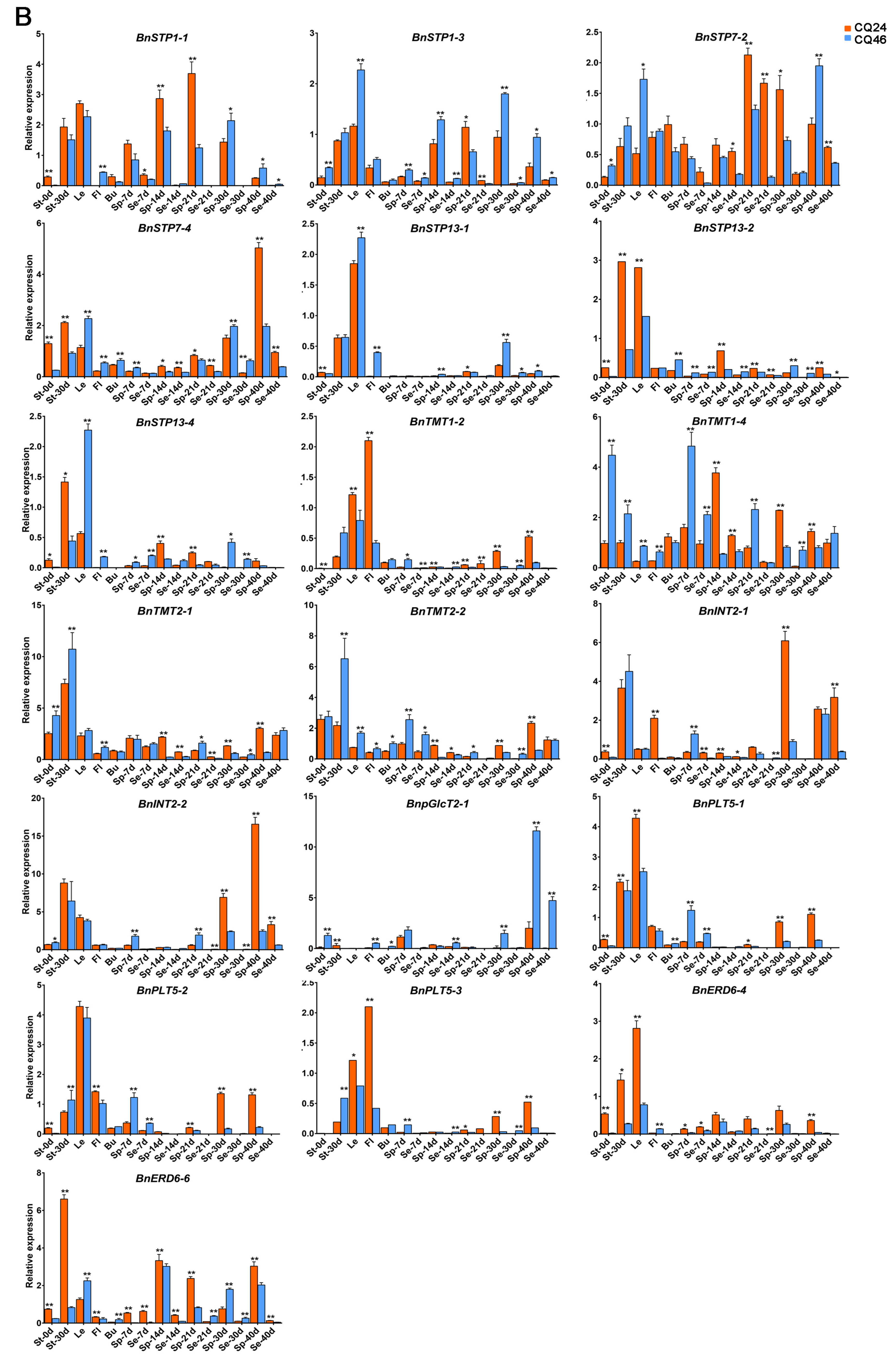
3.5. qRT-PCR Analysis of DEGs in Different Tissues between Diverse Materials
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Weise, S.E.; Schrader, S.M.; Kleinbeck, K.R.; Sharkey, T.D. Carbon balance and circadian regulation of hydrolytic and phosphorolytic breakdown of transitory starch. Plant Physiol. 2006, 141, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Gu, X.; Xue, L.J.; Leebens-Mack, J.H.; Tsai, C.J. Bayesian phylogeny of sucrose transporters: ancient origins, differential expansion and convergent evolution in monocots and dicots. Front. Plant. Sci 2014, 5, 615. [Google Scholar] [CrossRef] [PubMed]
- Bel, A.J.E.V. The phloem, a miracle of ingenuity. Plant. Cell Environ. 2003, 26, 125–149. [Google Scholar]
- Kuhn, C.; Franceschi, V.R.; Schulz, A.; Lemoine, R.; Frommer, W.B. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 1997, 275, 1298–1300. [Google Scholar] [CrossRef]
- Zheng, Q.M.; Tang, Z.; Xu, Q.; Deng, X.X. Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant. Cell Tiss. Org. 2014, 119, 609–624. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Peng, C.C.; Zou, K.Q.; Wang, X.L.; Fan, R.C.; Yu, X.C.; Zhang, X.Y.; Shen, Y.Y.; Zhang, D.P. A monosaccharide transporter is localized to sieve plate and plasmodesmal channel in developing apple fruit. Chinese Sci Bull. 2005, 50, 429–434. [Google Scholar] [CrossRef]
- Johnson, D.A.; Hill, J.P.; Thomas, M.A. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evol. Biol. 2006, 6, 64. [Google Scholar]
- Büttner, M. The monosaccharide transporter (-like) gene family in Arabidopsis. Febs Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C.; Smith, S.M. Starch degradation. Annu. Rev. Plant. Biol. 2005, 56, 73–98. [Google Scholar] [CrossRef]
- Patrick, J.W. PHLOEM UNLOADING: Sieve Element Unloading and Post-Sieve Element Transport. Front. Plant. Sci. 1997, 48, 191–222. [Google Scholar] [CrossRef]
- Patrick, J.W. Does Don Fisher’s high-pressure manifold model account for phloem transport and resource partitioning? Front. Plant. Sci. 2013, 4, 184. [Google Scholar] [CrossRef] [PubMed]
- Guo-Liang, W.; Xiao-Yan, Z.; Ling-Yun, Z.; Qiu-Hong, P.; Yuan-Yue, S.; Da-Peng, Z.J.P.; Physiology, C. Phloem unloading in developing walnut fruit is symplasmic in the seed pericarp and apoplasmic in the fleshy pericarp. Plant. Cell Physiol. 2004, 45, 1461–1470. [Google Scholar]
- Lalonde, S.; Tegeder, M.; Throne-Holst , M.; Frommer, W.B.; Patrick, J.W. Phloem loading and unloading of sugars and amino acids. Plant. Cell Env. 2010, 26, 37–56. [Google Scholar] [CrossRef]
- Haupt, S.; Duncan, G.H.; Holzberg, S.; Oparka, K. Evidence for symplastic phloem unloading in sink leaves of barley. Plant. Physiol. 2001, 125, 209–218. [Google Scholar] [CrossRef]
- Giaquinta, R.T.; Lin, W.; Sadler, N.L.; Franceschi, V.R. Pathway of Phloem unloading of sucrose in corn roots. Plant. Physiol. 1983, 72, 362–367. [Google Scholar] [CrossRef]
- Hackel, A.; Schauer, N.; Carrari, F.; Fernie, A.R.; Grimm, B.; Kuhn, C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant. J. 2006, 45, 180–192. [Google Scholar] [CrossRef]
- Rae, A.L.; Grof, C.P.L.; Casu, R.E.; Bonnett, G.D. Sucrose accumulation in the sugarcane stem: pathways and control points for transport and compartmentation. Field Crops Res. 2005, 92, 159–168. [Google Scholar] [CrossRef]
- Sherson, S.M.; Alford, H.L.; Forbes, S.M.; Wallace, G.; Smith, S.M. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 2003, 54, 525–531. [Google Scholar] [CrossRef]
- Weschke, W.; Panitz, R.; Gubatz, S.; Wang, Q.; Radchuk, R.; Weber, H.; Wobus, U. The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. Plant. J. 2003, 33, 395–411. [Google Scholar] [CrossRef]
- Sauer, N.; Tanner, W. The hexose carrier from Chlorella. cDNA cloning of a eucaryotic H+-cotransporter. FEBS Lett. 1989, 1, 43–46. [Google Scholar] [CrossRef]
- Sauer, N.; Friedländer, K.; Gräml-Wicke, U. Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 1990, 10, 3045–3050. [Google Scholar] [CrossRef]
- Sauer, N.; Stadler, R. A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker’s yeast. Plant. J. Cell Mol. Biol. 1993, 4, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Deborah, A.J.; Michael, A.H. The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Mol. Biol. Evol. 2007, 11, 2412–2423. [Google Scholar]
- Hayes, M.A.; Davies, C.; Dry, I.B. Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J. Exp. Bot. 2007, 58, 1985–1997. [Google Scholar] [CrossRef]
- Marger, M.D.; Saier, M.H. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 1993, 18, 13–20. [Google Scholar] [CrossRef]
- Hediger, M.A. Structure, function and evolution of solute transporters in prokaryotes and eukaryotes. J. Exp. Biol. 1994, 196, 15–49. [Google Scholar] [PubMed]
- Wingenter, K.; Schulz, A.; Wormit, A.; Wic, S.; Trentmann, O.; Hoermiller, I.I.; Heyer, A.G.; Marten, I.; Hedrich, R.; Neuhaus, H.E. Increased Activity of the Vacuolar Monosaccharide Transporter TMT1 Alters Cellular Sugar Partitioning, Sugar Signaling, and Seed Yield in Arabidopsis. Plant. Physiol. 2010, 154, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z. Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant. Mol. Biol. 2007, 65, 439–451. [Google Scholar]
- Yamaguchi, J. Characterization of Rice Functional Monosaccharide Transporter, OsMST5. Biosci. Biotechnol. Biochem. 2014, 67, 556–562. [Google Scholar]
- Jian, H.; Lu, K.; Bo, Y.; Wang, T.; Li, Z.; Zhang, A.; Jia, W.; Liu, L.; Qu, C.; Li, J. Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.). Front. Plant. Sci. 2016, 7, 1464. [Google Scholar] [CrossRef]
- Hay, R.K.M. Harvest index: A review of its use in plant breeding and crop physiology. Ann. Appl. Biol. 2008, 126, 197–216. [Google Scholar] [CrossRef]
- Lu, K.; Xiao, Z.; Jian, H.; Peng, L.; Qu, C.; Fu, M.; He, B.; Tie, L.; Liang, Y.; Xu, X. A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci. Rep. 2016, 6, 36452. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ma, C.; Yue, Y.; Hu, K.; Li, Y.; Duan, Z.; Wu, M.; Tu, J.; Shen, J.; Yi, B. Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genom. 2015, 16, 379. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.X.; Fu, T.D.; Yang, G.S.; Ma, C.Z.; Tu, J. Genetic analysis of rapeseed self-incompatibility lines reveals significant heterosis of different patterns for yield and oil content traits. Plant. Breed. 2010, 124, 111–116. [Google Scholar] [CrossRef]
- Chhabra, M.L. Translocation pattern of assimilates in Indian mustard. Int. Rapeseed Congr. 1988, 25, 102. [Google Scholar]
- Lemonnier, P.; Gaillard, C.; Veillet, F.; Verbeke, J.; Lemoine, R.; CoutosThévenot, P.; La Camera, S. Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant. Mol. Biol. 2011, 85, 473–484. [Google Scholar] [CrossRef]
- Schofield, R.A.; Bi, Y.M.; Kant, S.; Rothstein, S.J. Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant. Cell Environ. 2008, 32, 271–285. [Google Scholar] [CrossRef]
- Sherson, S.M.; Hemmann, G.; Wallace, G.; Forbes, S.; Smith, S.M. Monosaccharide/proton symporter AtSTP1 plays a major role in uptake and response of Arabidopsis seeds and seedlings to sugars. Plant. J. 2000, 24, 849–857. [Google Scholar] [CrossRef]
- Afoufa-Bastien, D.; Medici, A.; Jeauffre, J.; CoutosThévenot, P.; Lemoine, R.; Atanassova, R.; Laloi, M. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant. Biol. 2010, 10, 245. [Google Scholar] [CrossRef]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Li, Y. A Novel Insight into Functional Divergence of the MST Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Dang, H.; Chen, Z.; Wu, J.; Chen, Y.; Chen, S.; Luo, L. Genome-Wide Identification, Expression, and Functional Analysis of the Sugar Transporter Gene Family in Cassava (Manihot esculenta). Int. J. Mol. Sci. 2018, 19, 987. [Google Scholar]
- Jia-ming, L.; Dan-man, Z.; Lei-ting, L.; Qiao, X.; Shu-wei, W.; Bai, B.; Shao-ling, Z.; Wu, J. Genome-Wide Function, Evolutionary Characterization and Expression Analysis of Sugar Transporter Family Genes in Pear (Pyrus bretschneideri Rehd). Plant. Cell Physiol. 2015, 9, 1721–1737. [Google Scholar]
- Szenthe, A.; Schfer, H.; Hauf, J.; Schwend, T.; Wink, M. Characterisation and expression of monosaccharide transporters in lupins, Lupinus polyphyllus and L. albus. J. Plant. Res. 2007, 120, 697–705. [Google Scholar] [CrossRef] [PubMed]
- ALTSCHUL, S.F. Gapped BLAST and PSI-BLAST: a new generation of protein detabase search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant. Biol. 2011, 11, 136. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar]
- Voorrips, E.R. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 13, 3784–3788. [Google Scholar]
- Hoglund, A.; Donnes, P.; Blum, T.; Adolph, H.-W.; Kohlbacher, O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 2006, 22, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Fuyou, F.; Miao, L.; Huiyan, Z.; Chuan, L.; Jiana, L.; Zhanglin, T.; Xinfu, X.; Xiao, Q.; Rui, W. Comparative Transcriptome Analysis of Recessive Male Sterility (RGMS) in Sterile and Fertile Brassica napus Lines. PLoS ONE 2015, 10, e0144118. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer Premier: Program for Design of Degenerate Primers from a Protein Sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, L.; Wu, Y.; Cao, Y.; Lu, C. Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Brassica napus. J. Agric. Food Chem. 2010, 58, 2812–2817. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-△△Ct Method. Methods 2000, 25, 402–408. [Google Scholar] [CrossRef]
- Steland, A. Book Review: Fitting Models to Biological Data Using Linear and Nonlinear Regression. By Harvey Motulsky und Arthur Christopoulos. Biom. J. 2006, 48, 327. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Modeling 1997, 37, 411–412. [Google Scholar]
- Cheng, F.; Wu, J.; Wang, X. Genome Triplication Drove the Diversification of Brassica Plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef]
- Lysak, M.A.; Cheung, K.; Kitschke, M.; Bures, P. Ancestral Chromosomal Blocks Are Triplicated in Brassiceae Species with Varying Chromosome Number and Genome Size. Plant. Physiol. 2007, 145, 402–410. [Google Scholar] [CrossRef]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Arias, T.; Beilstein, M.A.; Tang, M.; Mckain, M.R.; Pires, J.C. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 2014, 1, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Yang, T.J.; Seol, Y.J.; Jin, M. Genome-wide comparative analysis of theBrassica rapagene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R11. [Google Scholar] [CrossRef] [PubMed]
- Moghe, G.D.; Hufnagel, D.E.; Tang, H.; Xiao, Y.; Dworkin, I.; Town, C.D.; Conner, J.K.; Shiu, S.H. Consequences of Whole-Genome Triplication as Revealed by Comparative Genomic Analyses of the Wild Radish Raphanus raphanistrum and Three Other Brassicaceae Species. Plant. Cell 2014, 26, 1925–1937. [Google Scholar] [CrossRef]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- NRholm, M.H.H.; NourEldin, H.H.; Brodersen, P.; Mundy, J.; Halkier, B.A. Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett. 2006, 580, 2381–2387. [Google Scholar] [CrossRef]
- Rottmann, T.M.; Klebl, F.; Schneider, S.; Kischka, D.; Rüscher, D.; Sauer, N.; Stadler, R. Characterization of three sugar transporters, STP7, STP8 and STP12. Plant. Physiol. 2018, 3. [Google Scholar] [CrossRef]
- Reinders, A.; Panshyshyn, J.A.; Ward, J.M. Analysis of Transport Activity of Arabidopsis Sugar Alcohol Permease Homolog AtPLT5. J. Biol. Chem. 2005, 280, 1594–1602. [Google Scholar] [CrossRef]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S.; Schmidt, U.; Martinoia, E.; Neuhaus, H.E. Molecular Identification and Physiological Characterization of a Novel Monosaccharide Transporter from Involved in Vacuolar Sugar Transport. Plant. Cell 2006, 18, 3476–3490. [Google Scholar] [CrossRef]
- Cho, M.H.; Lim, H.; Shin, D.H.; Jeon, J.S.; Bhoo, S.H.; Park, Y.I.; Hahn, T. Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 2011, 190, 101–112. [Google Scholar] [CrossRef] [PubMed]
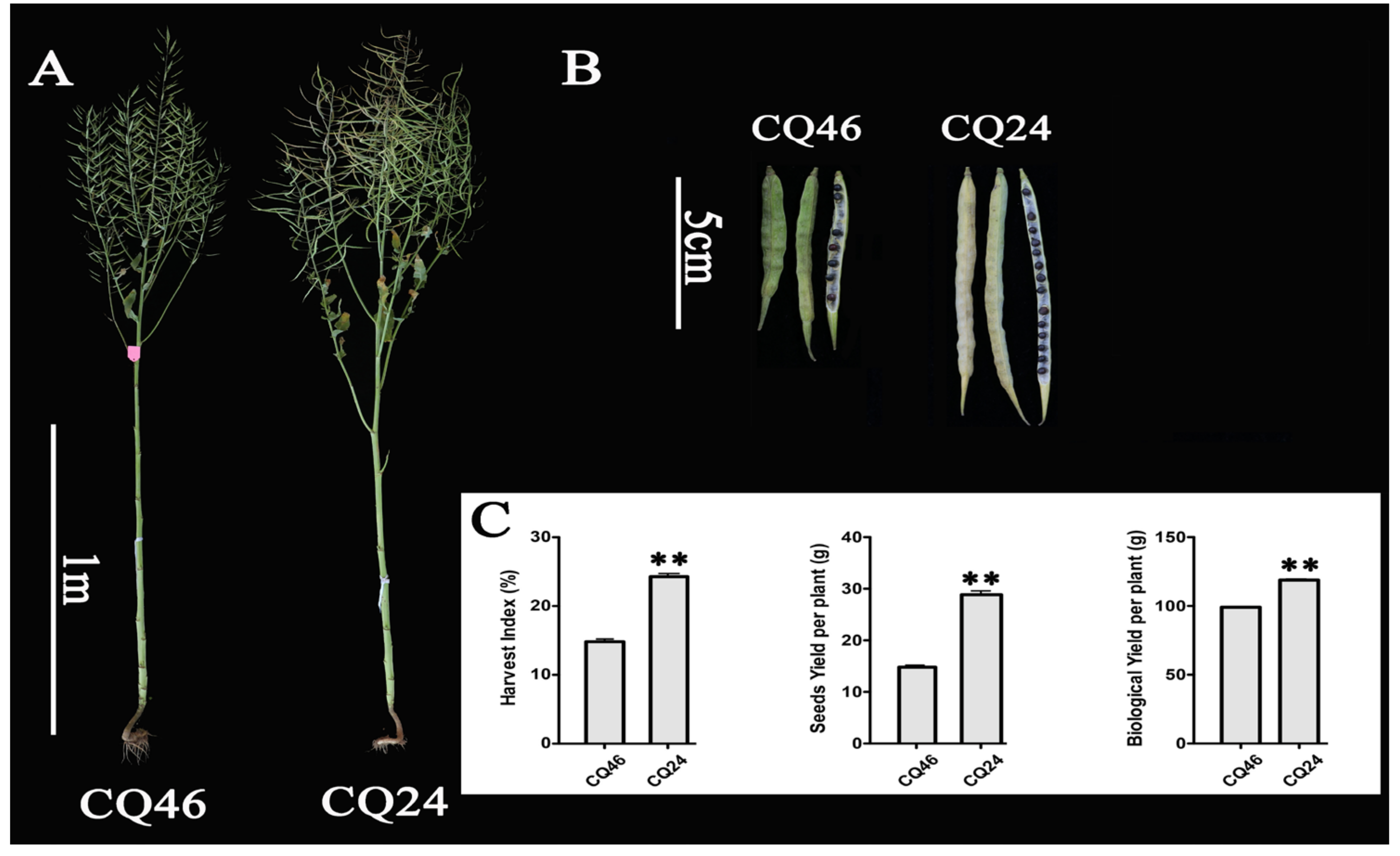
| Trait | Material | 2016 | 2017 | Mean Value | SEM | P-Value |
|---|---|---|---|---|---|---|
| Harvest Index /% (HI) | CQ24 | 24.75 | 24.33 | 24.28 | 0.47 | 0.004 |
| CQ46 | 15.21 | 14.43 | 14.82 | 0.39 | ||
| Seed Yield per Plant /g (SY) | CQ24 | 29.60 | 28.13 | 28.86 | 0.73 | 0.004 |
| CQ46 | 15.21 | 14.37 | 14.79 | 0.42 | ||
| Biological Yield per Plant /g (BY) | CQ24 | 119.60 | 118.15 | 118.87 | 0.73 | 0.002 |
| CQ46 | 98.60 | 99.55 | 99.08 | 0.48 |
| Gene Name | A. thaliana | B. rapa | B. oleracea | B. napus (A) | B. napus (C) |
|---|---|---|---|---|---|
| Zero-Copy in B. napus | |||||
| ERD6-like15 | 1 | 0 | 0 | 0 | 0 |
| ERD6-like16 | 1 | 0 | 0 | 0 | 0 |
| pGlcT4 | 1 | 0 | 0 | 0 | 0 |
| One-Copy in B. napus | |||||
| PLT2 | 1 | 0 | 1 | 1 | 0 |
| pGlcT3 | 1 | 1 | 1 | 1 | 0 |
| Two-Copy in B. napus | |||||
| ERD6-like2 | 1 | 1 | 1 | 1 | 1 |
| ERD6-like4 | 1 | 0 | 1 | 1 | 1 |
| ERD6-like7 | 1 | 1 | 1 | 1 | 1 |
| ERD6-like9 | 1 | 1 | 1 | 1 | 1 |
| ERD6-like10 | 1 | 1 | 1 | 1 | 1 |
| ERD6-like13 | 1 | 1 | 1 | 1 | 1 |
| ERD6-like14 | 1 | 1 | 1 | 1 | 1 |
| pGlcT2 | 1 | 1 | 1 | 1 | 1 |
| STP3 | 1 | 1 | 1 | 1 | 1 |
| STP5 | 1 | 1 | 1 | 1 | 1 |
| STP8 | 1 | 1 | 1 | 1 | 1 |
| STP11 | 1 | 0 | 1 | 1 | 1 |
| STP12 | 1 | 1 | 1 | 1 | 1 |
| STP14 | 1 | 1 | 1 | 1 | 1 |
| VGT2 | 1 | 1 | 1 | 1 | 1 |
| VGT3 | 1 | 1 | 1 | 1 | 1 |
| INT2 | 1 | 1 | 1 | 1 | 1 |
| INT3 | 1 | 1 | 0 | 1 | 1 |
| PLT3 | 1 | 1 | 1 | 1 | 1 |
| PLT4 | 1 | 0 | 0 | 1 | 1 |
| Three-Copy in B. napus | |||||
| SFP1 | 1 | 2 | 0 | 3 | 0 |
| TMT2 | 1 | 1 | 1 | 1 | 2 |
| PLT1 | 1 | 3 | 2 | 2 | 1 |
| PLT5 | 1 | 2 | 2 | 1 | 2 |
| Four-Copy in B. napus | |||||
| ERD6-like8 | 1 | 1 | 2 | 2 | 2 |
| ERD6-like11 | 1 | 2 | 0 | 2 | 2 |
| SFP2 | 1 | 2 | 3 | 1 | 3 |
| INT1 | 1 | 2 | 2 | 2 | 2 |
| INT4 | 1 | 2 | 2 | 2 | 2 |
| pGlcT1 | 1 | 2 | 2 | 2 | 2 |
| TMT1 | 1 | 2 | 2 | 2 | 2 |
| TMT3 | 1 | 2 | 2 | 2 | 2 |
| STP1 | 1 | 2 | 2 | 2 | 2 |
| STP2 | 1 | 2 | 2 | 2 | 2 |
| STP7 | 1 | 2 | 2 | 2 | 2 |
| STP9 | 1 | 2 | 2 | 2 | 2 |
| STP13 | 1 | 2 | 2 | 2 | 2 |
| Five-Copy in B. napus | |||||
| ERD6-like3 | 1 | 2 | 1 | 3 | 2 |
| ERD6-like5 | 1 | 2 | 2 | 3 | 3 |
| ERD6-like12 | 1 | 2 | 2 | 2 | 3 |
| Six-Copy in B. napus | |||||
| ERD6-like6 | 1 | 3 | 3 | 3 | 3 |
| STP6 | 1 | 4 | 3 | 4 | 3 |
| STP10 | 1 | 3 | 4 | 3 | 3 |
| PLT6 | 1 | 3 | 3 | 3 | 3 |
| Seven-Copy in B. napus | |||||
| ERD6 | 1 | 3 | 4 | 4 | 3 |
| STP4 | 1 | 3 | 5 | 2 | 3 |
| Eight-Copy in B. napus | |||||
| ERD6-like1 | 1 | 3 | 3 | 4 | 4 |
| VGT1 | 1 | 3 | 4 | 2 | 6 |
| TOTAL | 53 | 83 | 87 | 87 | 88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, C.; Yang, B.; Xiao, Z.; Ma, J.; Liu, J.; Jian, H.; Qu, C.; Lu, K.; Li, J. Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.). Genes 2020, 11, 653. https://doi.org/10.3390/genes11060653
Zhang L, Zhang C, Yang B, Xiao Z, Ma J, Liu J, Jian H, Qu C, Lu K, Li J. Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.). Genes. 2020; 11(6):653. https://doi.org/10.3390/genes11060653
Chicago/Turabian StyleZhang, Liyuan, Chao Zhang, Bo Yang, Zhongchun Xiao, Jinqi Ma, Jingsen Liu, Hongju Jian, Cunmin Qu, Kun Lu, and Jiana Li. 2020. "Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.)" Genes 11, no. 6: 653. https://doi.org/10.3390/genes11060653
APA StyleZhang, L., Zhang, C., Yang, B., Xiao, Z., Ma, J., Liu, J., Jian, H., Qu, C., Lu, K., & Li, J. (2020). Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.). Genes, 11(6), 653. https://doi.org/10.3390/genes11060653








