Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Wine Yeast Strains, Growth Conditions, and Study Design
2.2. Kinetics of Growth Assay
2.3. Cell Viability
2.4. Pulsed-Field Gel Electrophoresis (PFGE)
2.5. Spot Test Assay
2.6. Iodine-Staining of Yeast Colonies
2.7. Laboratory-Scale Fermentations and Measurements of Glucose, Fructose, Glycerol, and Resveratrol Concentrations
2.8. HPLC
2.9. Semi-Quantitative PCR
3. Results and Discussion
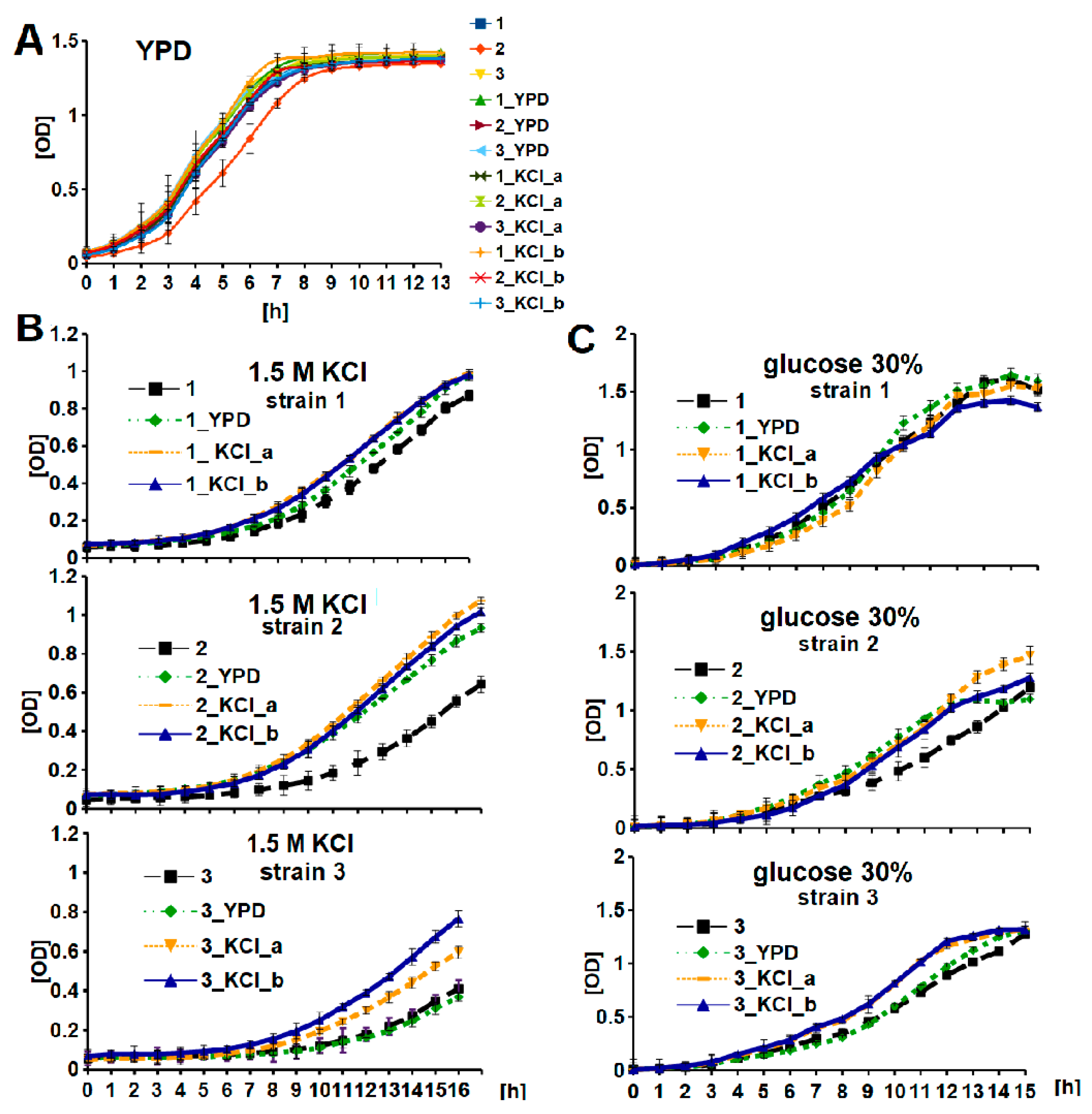
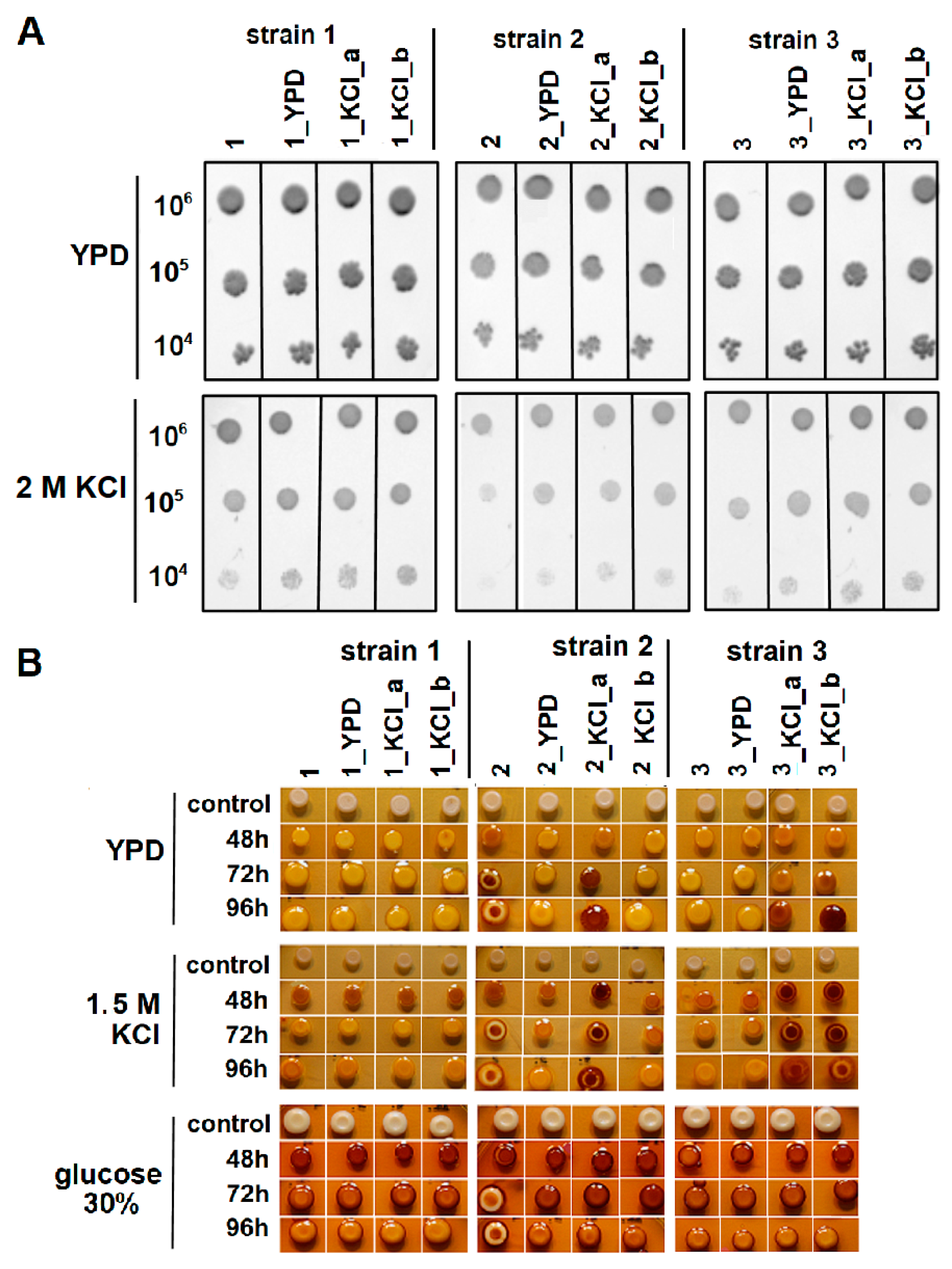
3.1. Wine Yeast Strain Screening and Characterization
3.2. KCl-ALE-Based Improvement of Wine Yeasts’ Tolerance to Osmotic Stress and Glycogen Accumulation
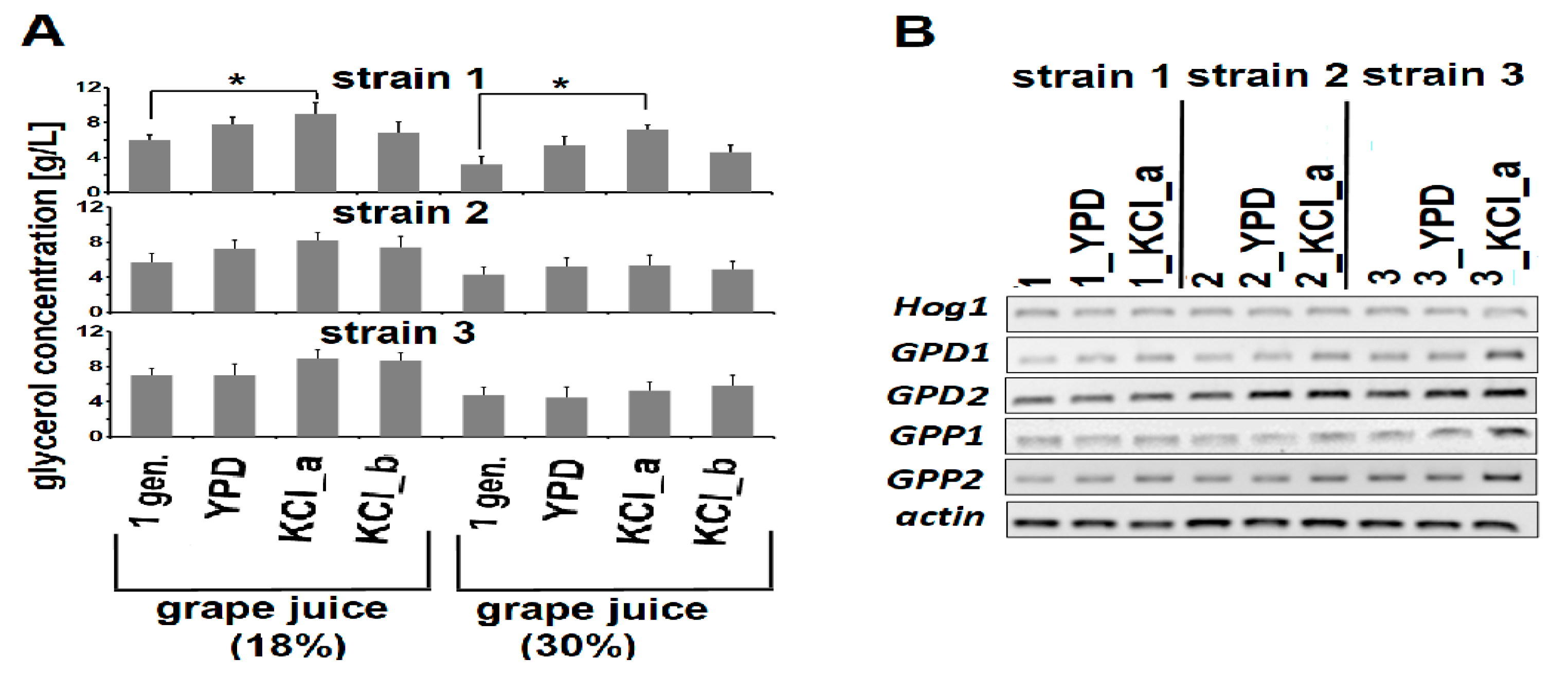
3.3. Adaptive Evolution-Mediated Changes in Glycerol Concentrations
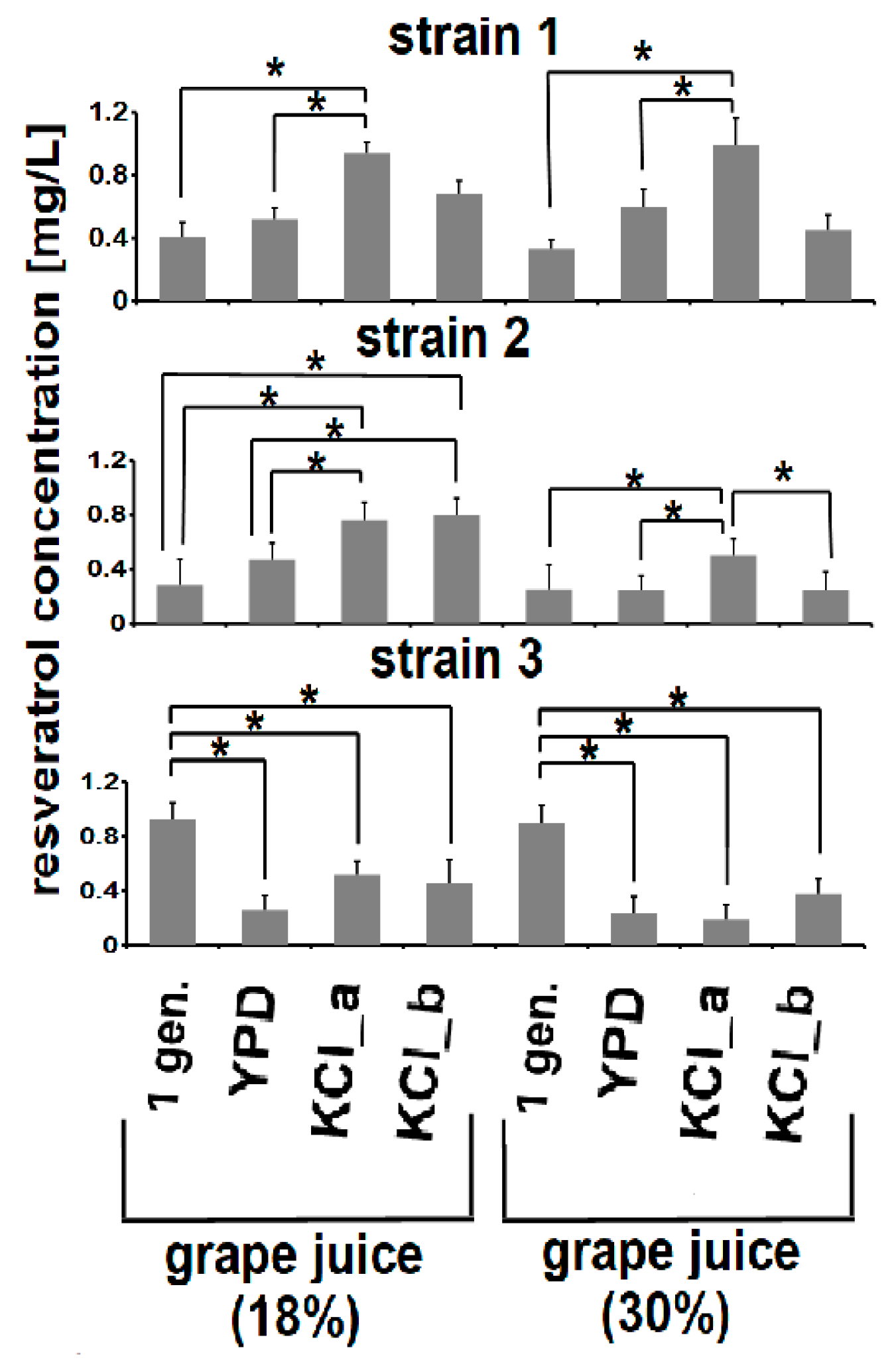
3.4. Adaptive Evolution-Mediated Changes in Resveratrol Concentrations
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pretorius, I.S. Tasting the terroir of wine yeast innovation. FEMS Yeast Res. 2020, 20. [Google Scholar] [CrossRef]
- van Wyk, N.; Grossmann, M.; Wendland, J.; von Wallbrunn, C.; Pretorius, I.S. The whiff of wine yeast innovation: Strategies for enhancing aroma production by yeast during wine fermentation. J. Agric. Food Chem. 2019, 67, 13496–13505. [Google Scholar] [CrossRef]
- Marsit, S.; Dequin, S. Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Dequin, S.; Casaregola, S. The genomes of fermentative Saccharomyces. Comptes Rendus Boil. 2011, 334, 687–693. [Google Scholar] [CrossRef]
- Cray, J.A.; Stevenson, A.; Ball, P.; Bankar, S.B.; Eleutherio, E.C.; Ezeji, T.C.; Singhal, R.S.; Thevelein, J.M.; Timson, D.J.; Hallsworth, J.E. Chaotropicity: A key factor in product tolerance of biofuel-producing microorganisms. Curr. Opin. Biotechnol. 2015, 33, 228–259. [Google Scholar] [CrossRef]
- Hallsworth, J.E.; Nomura, Y. A simple method to determine the water activity of ethanol-containing samples. Biotechnol. Bioeng. 1999, 62, 242–245. [Google Scholar] [CrossRef]
- Hallsworth, J.E. Ethanol-induced water stress in yeast. J. Ferment Bioeng. 1998, 85, 125–137. [Google Scholar] [CrossRef]
- Cray, J.A.; Bell, A.N.; Bhaganna, P.; Mswaka, A.Y.; Timson, D.J.; Hallsworth, J.E. The biology of habitat dominance; can microbes behave as weeds? Microb Biotechnol. 2013, 6, 453–492. [Google Scholar] [CrossRef]
- Kutyna, D.R.; Varela, C.; Stanley, G.A.; Borneman, A.R.; Henschke, P.A.; Chambers, P.J. Adaptive evolution of Saccharomyces cerevisiae to generate strains with enhanced glycerol production. Appl. Microbiol. Biotechnol. 2012, 93, 1175–1184. [Google Scholar] [CrossRef]
- Tilloy, V.; Ortiz-Julien, A.; Dequin, S. Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl. Environ. Microbiol. 2014, 80, 2623–2632. [Google Scholar] [CrossRef]
- Caspeta, L.; Chen, Y.; Nielsen, J. Thermotolerant yeasts selected by adaptive evolution express heat stress response at 30 °C. Sci. Rep. 2016, 6, 27003. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Thevelein, J.M.; Gibson, B. Multimodal Microorganism Development: Integrating Top-Down Biological Engineering with Bottom-Up Rational Design. Trends Biotechnol. 2020, 38, 241–253. [Google Scholar] [CrossRef]
- Cambon, B.; Monteil, V.; Remize, F.; Camarasa, C.; Dequin, S. Effects of GPD1 overexpression in Saccharomyces cerevisiae commercial wine yeast strains lacking ALD6 genes. Appl. Environ. Microbiol. 2006, 72, 4688–4694. [Google Scholar] [CrossRef]
- Doğan, A.; Demirci, S.; Aytekin, A.Ö.; Şahin, F. Improvements of tolerance to stress conditions by genetic engineering in Saccharomyces cerevisiae during ethanol production. Appl. Biochem. Biotechnol. 2014, 174, 28–42. [Google Scholar] [CrossRef]
- Adamo, G.M.; Brocca, S.; Passolunghi, S.; Salvato, B.; Lotti, M. Laboratory evolution of copper tolerant yeast strains. Microbial Cell Factories 2012, 11, 1. [Google Scholar] [CrossRef]
- Dhar, R.; Sagesser, R.; Weikert, C.; Yuan, J.; Wagner, A. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol. 2011, 24, 1135–1153. [Google Scholar] [CrossRef]
- Zhao, X.; Procopio, S.; Becker, T. Flavor impacts of glycerol in the processing of yeast fermented beverages: A review. J. Food Sci. Technol. 2015, 52, 7588–7598. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- de Lima Alves, F.; Stevenson, A.; Baxter, E.; Gillion, J.L.; Hejazi, F.; Hayes, S.; Morrison, I.E.; Prior, B.A.; McGenity, T.J.; Rangel, D.E.; et al. Concomitant osmotic and chaotropicity-induced stresses in Aspergillus wentii: Compatible solutes determine the biotic window. Curr. Genet. 2015, 61, 457–477. [Google Scholar] [CrossRef]
- Pérez-Torrado, R.; Gimeno-alcañiz, J.V.; Matallana, E. Wine Yeast Strains Engineered for Glycogen Overproduction Display Enhanced Viability under Glucose Deprivation Conditions. Appl. Environ. Microbiol. 2002, 68, 3339–3344. [Google Scholar] [CrossRef][Green Version]
- Claus, H.; Mojsov, K. Enzymes for wine fermentation: Current and perspective applications. Fermentation 2018, 4, 52. [Google Scholar]
- Xia, N.; Daiber, A.; Förstermann, U.; Li, H. Antioxidant effects of resveratrol in the cardiovascular system. Br. J. Pharmacol. 2017, 174, 1633–1646. [Google Scholar]
- D’Amore, T.; Panchal, C.J.; Russeil, I.; Stewart, G.G. Osmotic pressure effects and intracellular accumulation of ethanol in yeast during fermentation. J. Ind. Microbiol. 1988, 6, 365–372. [Google Scholar]
- Adamczyk, J.; Deregowska, A.; Skoneczny, M.; Skoneczna, A.; Kwiatkowska, A.; Potocki, L.; Rawska, E.; Pabian, S.; Kaplan, J.; Lewinska, A.; et al. Adaptive response to chronic mild ethanol stress involves ROS, sirtuins and changes in chromosome dosage in wine yeasts. Oncotarget 2016, 7, 29958–29976. [Google Scholar] [CrossRef]
- Deregowska, A.; Adamczyk, J.; Kwiatkowska, A.; Gurgul, A.; Skoneczny, M.; Skoneczna, A.; Szmatola, T.; Jasielczuk, I.; Magda, M.; Rawska, E.; et al. Shifts in rDNA levels act as a genome buffer promoting chromosome homeostasis. Cell Cycle 2015, 14, 3475–3487. [Google Scholar]
- Lewinska, A.; Miedziak, B.; Wnuk, M. Assessment of yeast chromosome XII instability: Single chromosome comet assay. Fungal Genet. Biol. 2014, 63, 9–16. [Google Scholar] [CrossRef]
- Chester, V.E. Heritable glycogen-storage deficiency in yeast and its induction by ultra-violet light. J. Gen. Microbiol. 1968, 51, 49–56. [Google Scholar] [CrossRef]
- Harsch, M.J.; Lee, S.A.; Goddard, M.R.; Gardner, R.C. Optimized fermentation of grape juice by laboratory strains of Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 10, 72–82. [Google Scholar]
- Warringer, J.; Zörgö, E.; Cubillos, F.A.; Zia, A.; Gjuvsland, A.; Simpson, J.T.; Forsmark, A.; Durbin, R.; Omholt, S.W.; Louis, E.J.; et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011, 7, e1002111. [Google Scholar]
- Almeida, P.; Barbosa, R.; Zalar, P.; Imanishi, Y.; Shimizu, K.; Turchetti, B.; Legras, J.L.; Serra, M.; Dequin, S.; Couloux, A.; et al. A Population genomics insight into the mediterranean origins of wine yeast domestication. Mol. Ecol. 2015, 24, 5412–5427. [Google Scholar]
- Borneman, A.R.; Desany, B.A.; Riches, D.; Affourtit, J.P.; Forgan, A.H.; Pretorius, I.S.; Egholm, M.; Chambers, P.J. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011, 7, e1001287. [Google Scholar] [CrossRef]
- Hallsworth, J.E.; Magan, N. Effects of KCl concentration on accumulation of acyclic sugar alcohols and trehalose in conidia of three entomopathogenic fungi. Lett. Appl. Microbiol. 1994, 18, 8–11. [Google Scholar]
- Araújo, C.A.S.; Ferreira, P.C.; Pupin, B.; Dias, L.P.; Avalos, J.; Edwards, J.; Hallsworth, J.E.; Rangel, D.E.N. Osmotolerance as a determinant of microbial ecology: A study of phylogenetically diverse fungi. Fungal Biol. 2020, 124, 273–288. [Google Scholar]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar]
- Toh, T.H.; Kayingo, G.; van der Merwe, M.J.; Kilian, S.G.; Hallsworth, J.E.; Hohmann, S.; Prior, B.A. Implications of FPS1 deletion and membrane ergosterol content for glycerol efflux from Saccharomyces cerevisiae. FEMS Yeast Res. 2001, 1, 205–211. [Google Scholar]
- Hamill, P.G.; Stevenson, A.; McMullan, P.E.; Williams, J.P.; Lewis, A.D.R.; Stevenson, K.E.; Farnsworth, K.D.; Khroustalyova, G.; Takemoto, J.Y.; Quinn, J.P.; et al. Microbial lag phase can be indicative of, or independent from, cellular stress. Sci. Rep. 2020, 10, 5948. [Google Scholar] [CrossRef]
- Chang, Y.C.; Khanal Lamichhane, A.; Garraffo, H.M.; Walter, P.J.; Leerkes, M.; Kwon-Chung, K.J. Molecular mechanisms of hypoxic responses via unique roles of Ras1, Cdc24 and Ptp3 in a human fungal pathogen Cryptococcus neoformans. PLoS Genet. 2014, 10, e1004292. [Google Scholar] [CrossRef]
- Anderson, C.; Tatchell, K. Hyperactive glycogen synthase mutants of Saccharomyces cerevisiae suppress the glc7-1 protein phosphatase mutant. J. Bacteriol. 2001, 183, 821–829. [Google Scholar]
- Wilson, W.A.; Roach, P.J.; Montero, M.; Baroja-Fernández, E.; Muñoz, F.J.; Eydallin, G.; Viale, A.M.; Pozueta-Romero, J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010, 34, 952–985. [Google Scholar]
- Albertyn, J.; Hohmann, S.; Prior, B.A. Characterization of the osmotic-stress response in Saccharomyces cerevisiae: Osmotic stress and glucose repression regulate glycerol-3-phosphate dehydrogenase independently. Curr. Genet. 1994, 25, 12–18. [Google Scholar]
- Babazadeh, R.; Furukawa, T.; Hohmann, S.; Furukawa, K. Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci. Rep. 2014, 4, 4697. [Google Scholar]
- Ansell, R.; Granath, K.; Hohmann, S.; Thevelein, J.M.; Adler, L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997, 16, 2179–2187. [Google Scholar]
- Pahlman, A.K.; Granath, K.; Ansell, R.; Hohmann, S.; Adler, L. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 2001, 276, 3555–3563. [Google Scholar]
- Valadi, A.; Granath, K.; Gustafsson, L.; Adler, L. Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J. Biol. Chem. 2004, 279, 39677–39685. [Google Scholar]
- Lee, Y.J.; Jeschke, G.R.; Roelants, F.M.; Thorner, J.; Turk, B.E. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 2012, 32, 4705–4717. [Google Scholar]
- Remize, F.; Cambon, B.; Barnavon, L.; Dequin, S. Glycerol formation during wine fermentation is mainly linked to Gpd1p and is only partially controlled by the HOG pathway. Yeast 2003, 20, 1243–1253. [Google Scholar]
- Dhar, R.; Sagesser, R.; Weikert, C.; Wagner, A. Yeast Adapts to a Changing Stressful Environment by Evolving Cross-Protection and Anticipatory Gene Regulation. Mol. Biol. Evol. 2012, 30, 573–588. [Google Scholar]
- Berry, D.B.; Guan, Q.; Hose, J.; Haroon, S.; Gebbia, M.; Heisler, L.E.; Nislow, C.; Giaever, G.; Gasch, A.P. Multiple means to the same end: The genetic basis of acquired stress resistance in yeast. PLoS Genet. 2011, 7, e1002353. [Google Scholar] [CrossRef]
- Mattenberger, F.; Sabater-Muñoz, B.; Hallsworth, J.E.; Fares, M.A. Glycerol stress in Saccharomyces cerevisiae: Cellular responses and evolved adaptations. Environ. Microbiol. 2017, 19, 990–1007. [Google Scholar]
- Romero-Santacreu, L.; Moreno, J.; Pérez-Ortín, J.E.; Alepuz, P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 2009, 15, 1110–1120. [Google Scholar]
- Miller, C.; Schwalb, B.; Maier, K.; Schulz, D.; Dümcke, S.; Zacher, B.; Mayer, A.; Sydow, J.; Marcinowski, L.; Dölken, L.; et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 2011, 7, 458. [Google Scholar]
- Gaensly, F.; Agustini, B.C.; da Silva, A.G.; Picheth, G.; Bonfim, T.M.B. Autochthonous yeasts with β-glucosidase activity increase resveratrol concentration during the alcoholic fermentation of Vitis labrusca grape must. J. Funct. Foods 2015, 19, 288–295. [Google Scholar] [CrossRef]
- Rosi, I.; Vinella, M.; Domizio, P. Characterization of beta-glucosidase activity in yeasts of oenological origin. J. Appl. Bacteriol. 1994, 77, 19–27. [Google Scholar] [CrossRef]
- Becker, J.V.B.; Armstrong, G.O.; van der Merwe, M.J.; Lambrechts, M.G.; Vivier, M.A.; Pretorius, I.S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. JFEMS Yeast Res. 2003, 4, 79–85. [Google Scholar] [CrossRef]
- Kuo, H.P.; Wang, R.; Huang, C.-H.; Lai, J.-T.; Lo, Y.-C.; Huang, S.-T. Characterization of an extracellular b-glucosidase from Dekkera bruxellensis for resveratrol production. J. Food Drug Anal. 2018, 26, 163–171. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betlej, G.; Bator, E.; Oklejewicz, B.; Potocki, L.; Górka, A.; Slowik-Borowiec, M.; Czarny, W.; Domka, W.; Kwiatkowska, A. Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast. Genes 2020, 11, 576. https://doi.org/10.3390/genes11050576
Betlej G, Bator E, Oklejewicz B, Potocki L, Górka A, Slowik-Borowiec M, Czarny W, Domka W, Kwiatkowska A. Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast. Genes. 2020; 11(5):576. https://doi.org/10.3390/genes11050576
Chicago/Turabian StyleBetlej, Gabriela, Ewelina Bator, Bernadetta Oklejewicz, Leszek Potocki, Anna Górka, Magdalena Slowik-Borowiec, Wojciech Czarny, Wojciech Domka, and Aleksandra Kwiatkowska. 2020. "Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast" Genes 11, no. 5: 576. https://doi.org/10.3390/genes11050576
APA StyleBetlej, G., Bator, E., Oklejewicz, B., Potocki, L., Górka, A., Slowik-Borowiec, M., Czarny, W., Domka, W., & Kwiatkowska, A. (2020). Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast. Genes, 11(5), 576. https://doi.org/10.3390/genes11050576




