Evolution of the Human Chromosome 13 Synteny: Evolutionary Rearrangements, Plasticity, Human Disease Genes and Cancer Breakpoints
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
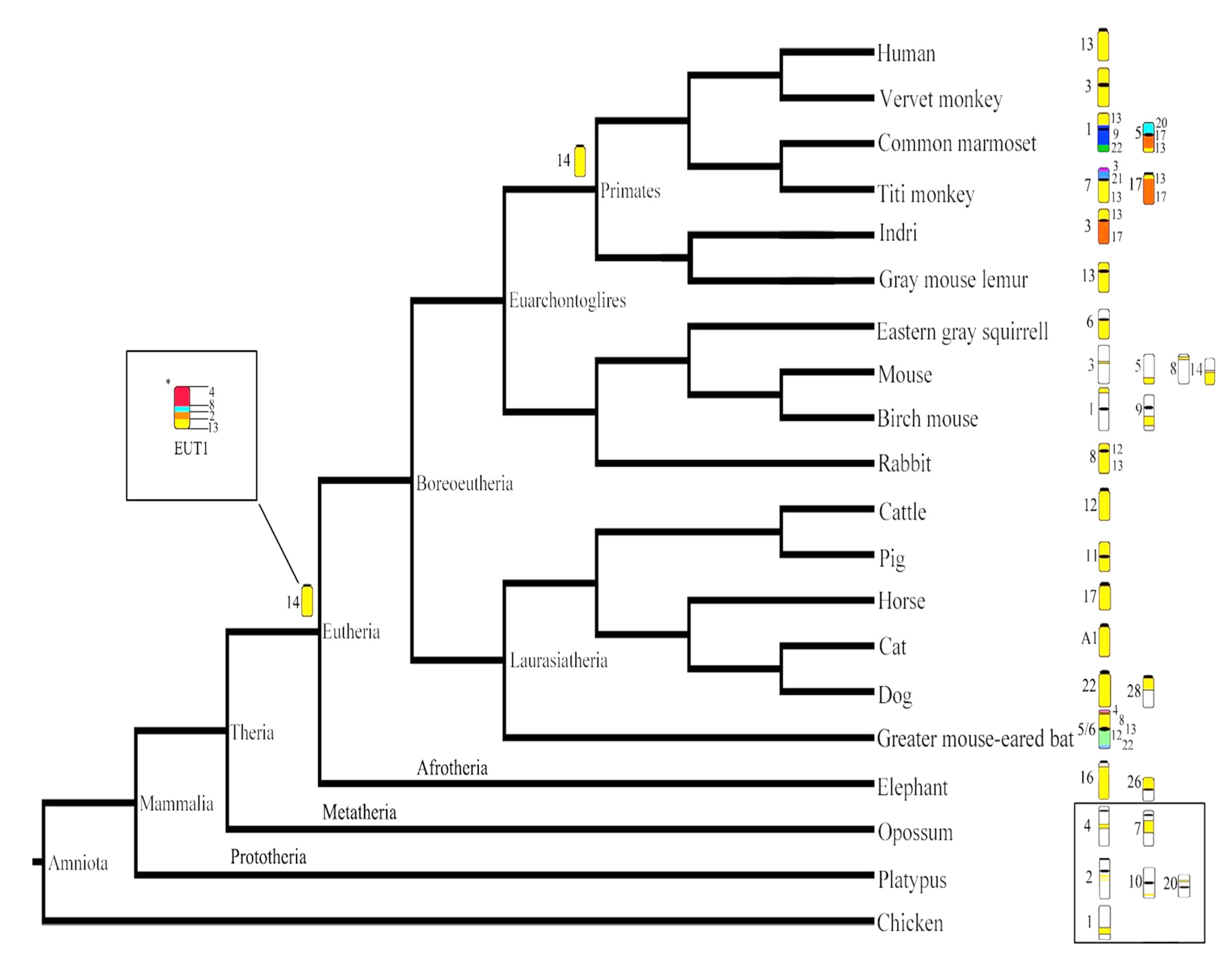
4.1. Evolutionary History of HSA13 Synteny in Eutherian Mammals
4.2. Evolutionary History of HSA 13 Synteny in Primates
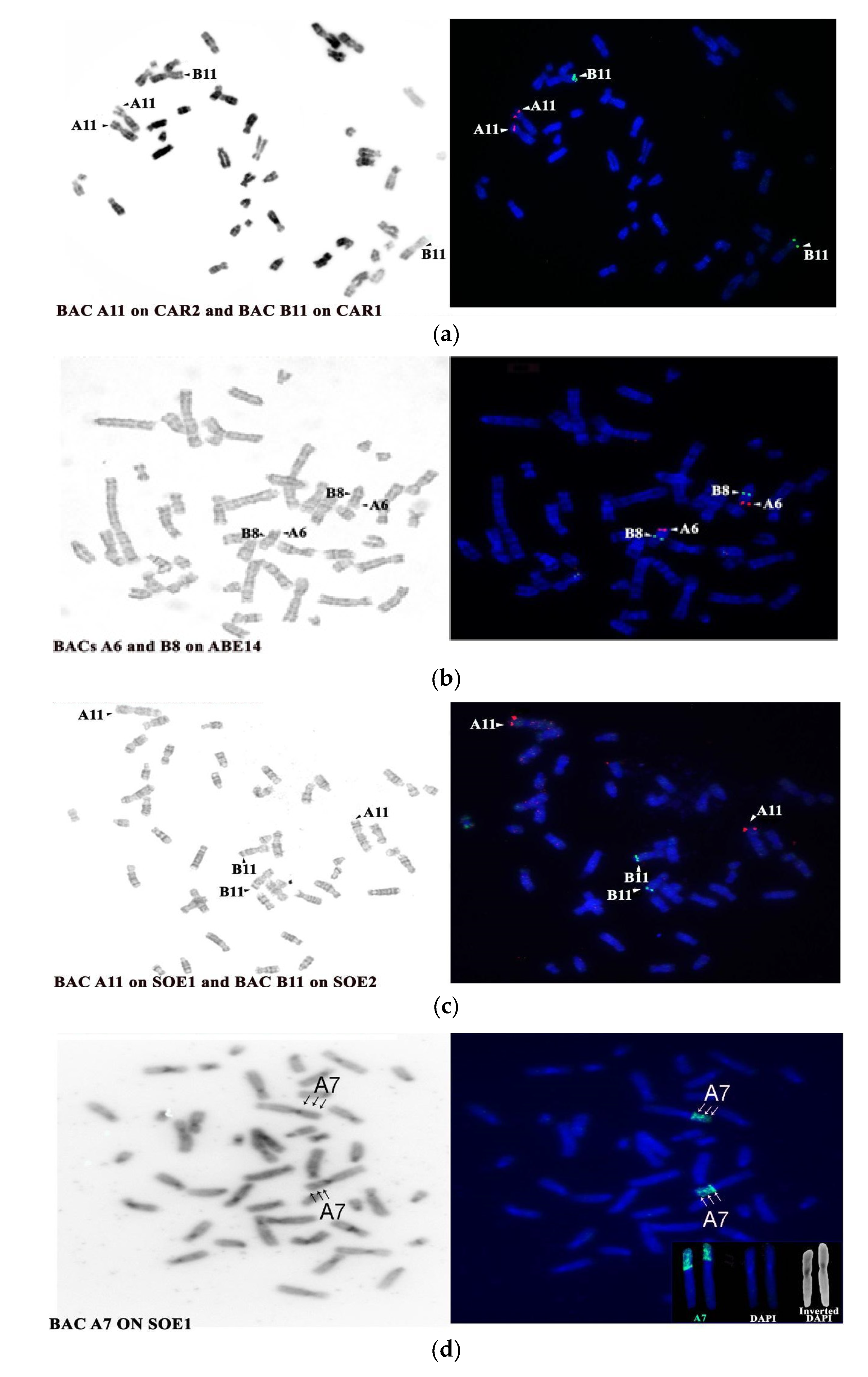
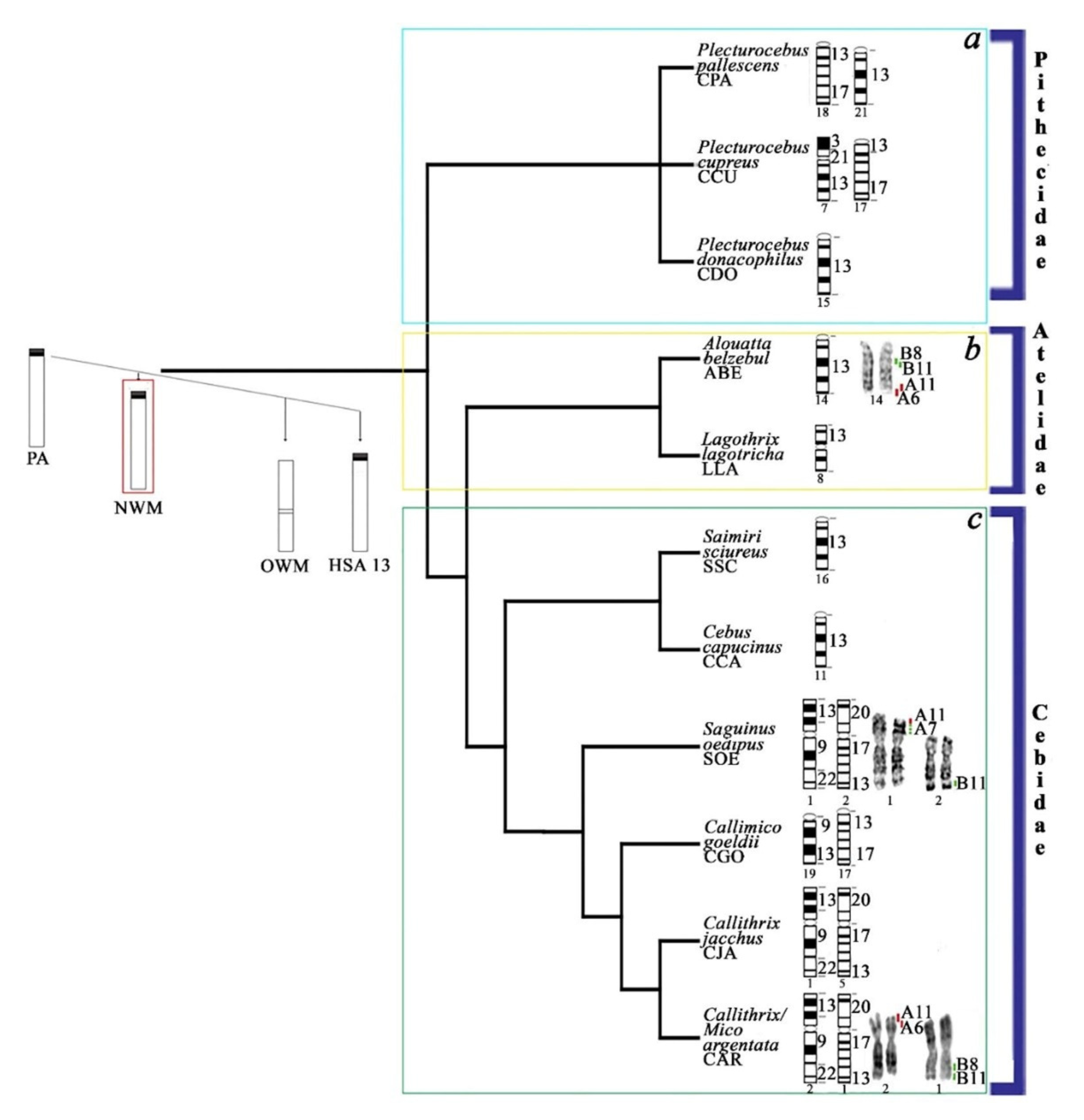
4.3. Evolutionary History of HSA13 Synteny in Platyrrhines
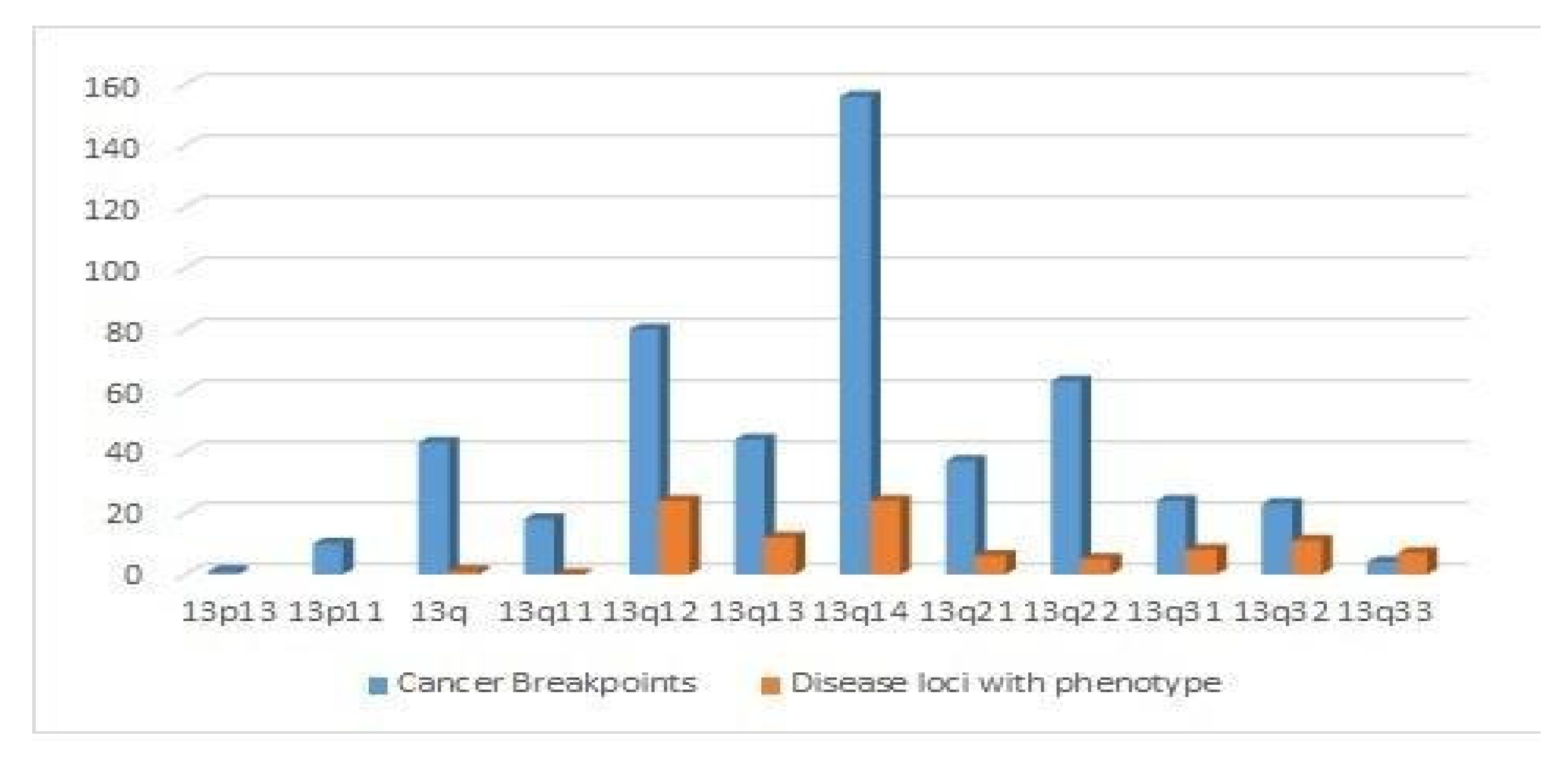
4.4. Distribution of Cancer Breakpoints and Loci Implicated in Human Diseases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dutrillaux, B. Chromosomal evolution in Primates: Tentative phylogeny from Microcebus murinus (Prosimian) to man. Qual. Life Res. 1979, 48, 251–314. [Google Scholar] [CrossRef]
- Stanyon, R.; Rocchi, M.; Capozzi, O.; Roberto, R.; Misceo, D.; Ventura, M.; Cardone, M.F.; Bigoni, F.; Archidiacono, N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosom. Res. 2008, 16, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2011, 108, 59–67. [Google Scholar] [CrossRef]
- Dumas, F.; Mazzoleni, S. Neotropical primate evolution and phylogenetic reconstruction using chromosomal data. Eur. Zool. J. 2017, 84, 1–18. [Google Scholar] [CrossRef][Green Version]
- Sineo, L.; Dumas, F.; Vitturi, R.; Picone, B.; Privitera, O.; Stanyon, R. Williams-Beuren mapping in Callithrix argentata, Callicebus cupreus and Alouatta caraya indicates different patterns of chromosomal rearrangements in neotropical primates. J. Zool. Syst. Evol. Res. 2007, 45, 366–371. [Google Scholar] [CrossRef]
- Picone, B.; Dumas, F.; Stanyon, R.; Lannino, A.; Bigoni, F.; Privitera, O.; Sineo, L. Exploring evolution in Ceboidea (Platyrrhini, Primates) by Williams-Beuren Probe (HSA 7q11.23) chromosome mapping. Folia Primatol. 2008, 79, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Jauch, A.; Wienberg, J.; Stanyon, R.; Arnold, N.; Tofanelli, S.; Ishida, T.; Cremer, T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. USA 1992, 89, 8611–8615. [Google Scholar] [CrossRef] [PubMed]
- Dumas, F.; Stanyon, R.; Sineo, L.; Stone, G.; Bigoni, F. Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting. BMC Evol. Boil. 2007, 7, S11. [Google Scholar] [CrossRef]
- Trifonov, V.; Stanyon, R.; Nesterenko, A.I.; Fu, B.; Perelman, P.L.; O’Brien, P.C.M.; Stone, G.; Rubtsova, N.V.; Houck, M.L.; Robinson, T.; et al. Multidirectional cross-species painting illuminates the history of karyotypic evolution in Perissodactyla. Chromosom. Res. 2008, 16, 89–107. [Google Scholar] [CrossRef]
- Dumas, F.; Sineo, L. Chromosomal dynamics in platyrrhinae by mapping BACs probes. S. Biol. Res 2012, LXXXV, 299–301. [Google Scholar]
- Dumas, F.; Sineo, L. The evolution of human synteny 4 by mapping sub-chromosomal specific probes in Primates. Caryologia 2014, 67, 281–291. [Google Scholar] [CrossRef]
- Dumas, F.; Sineo, L. Chromosomal dynamics in Cercopithecini studied by Williams-Beuren probe mapping. Caryologia 2010, 63, 435–442. [Google Scholar]
- Dumas, F.; Sineo, L.; Ishida, T. Taxonomic identification of Aotus (Platyrrhinae) through cytogenetics|Identificazione tassonomica di Aotus (Platyrrhinae) mediante la citogenetica. J. Biol. Res. 2015, 88, 65–66. [Google Scholar]
- Dumas, F.; Cuttaia, H.; Sineo, L. Chromosomal distribution of interstitial telomeric sequences in nine neotropical primates (Platyrrhini): Possible implications in evolution and phylogeny. J. Zool. Syst. Evol. Res. 2016, 54, 226–236. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Schillaci, O.; Sineo, L.; Dumas, F. Distribution of interstitial telomeric sequences in primates and the pygmy tree shrew (Scandentia). Cytogenet. Genome Res. 2017, 151, 141–150. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Rovatsos, M.; Schillaci, O.; Dumas, F. Evolutionary insight on localization of 18S, 28S rDNA genes on homologous chromosomes in Primates genomes. Comp. Cytogenet. 2018, 12, 27–40. [Google Scholar] [CrossRef]
- Milioto, V.; Vlah, S.; Mazzoleni, S.; Rovatsos, M.; Dumas, F. Chromosomal localization of 18S-28S rDNA and (TTAGGG)n sequences in two South African dormice of the genus Graphiurus (Rodentia: Gliridae). Cytogenet. Genome Res. 2019, 158, 145–151. [Google Scholar] [CrossRef]
- Hruba, M.; Dvorak, P.; Weberova, L.; Subrt, I. Independent coexistence of clones with 13q14 deletion at reciprocal translocation breakpoint and 13q14 interstitial deletion in chronic lymphocytic leukemia. Leuk. Lymphoma 2012, 53, 2054–2062. [Google Scholar] [CrossRef]
- Froenicke, L. Origins of primate chromosomes—As delineated by Zoo-FISH and alignments of human and mouse draft genome sequences. Cytogenet. Genome Res. 2005, 108, 122–138. [Google Scholar] [CrossRef]
- Murphy, W.J.; Larkin, D.M.; Der Wind, A.E.-V.; Bourque, G.; Tesler, G.; Auvil, L.; E Beever, J.; Chowdhary, B.P.; Galibert, F.; Gatzke, L.; et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative Maps. Science 2005, 309, 613–617. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, L.; Suh, B.B.; Raney, B.J.; Burhans, R.C.; Kent, W.J.; Blanchette, M.; Haussler, D.; Miller, W. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006, 16, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.J.; Ruiz-herrera, A.; Froenicke, L. Dissecting the mammalian genome—New insights into chromosomal evolution. Trends Genet. 2006, 22, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Farré, M.; Auvil, L.; Capitanu, B.; Larkin, D.M.; Ma, J.; Lewin, H.A. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E5379–E5388. [Google Scholar] [CrossRef] [PubMed]
- Dunham, A.; Matthews, L.H.; Burton, J.; Ashurst, J.L.; Howe, K.L.; Ashcroft, K.J.; Beare, D.M.; Burford, D.C.; Hunt, S.E.; Griffiths-Jones, S.; et al. The DNA sequence and analysis of human chromosome 13. Nature 2004, 428, 522–528. [Google Scholar] [CrossRef]
- Bailey, J.; Gu, Z.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef]
- Yunis, J.; Prakash, O. The origin of man: A chromosomal pictorial legacy. Science 1982, 215, 1525–1530. [Google Scholar] [CrossRef]
- Cardone, M.F.; Alonso, A.; Pazienza, M.; Ventura, M.; Montemurro, G.; Carbone, L.; De Jong, P.J.; Stanyon, R.; D’Addabbo, P.; Archidiacono, N.; et al. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Boil. 2006, 7, R91. [Google Scholar] [CrossRef]
- Alonso, A.; Mahmood, R.; Li, S.; Cheung, F.; Yoda, K.; Warburton, P.E. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003, 12, 2711–2721. [Google Scholar] [CrossRef]
- Scardino, R.; Milioto, V.; Dumas, F. Comparative Cytogenetics Allows the Reconstruction of Human Chromosome History: The Case of Human Chromosome 13. In Cytogenetics-Past, Present and Further Perspectives, 1st ed.; IntechOpen: Rijeka, Croatia, 2018; p. 79380. [Google Scholar]
- Small, M.F.; Stanyon, R.; Smith, D.G.; Sineo, L. High-resolution chromosomes of rhesus macaques (Macaca mulatta). Am. J. Primatol. 1985, 9, 63–67. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; Johnson, W.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R.V. Mesquite: A modular system for evolutionary analysis. Biology 2008, 11, 1103–1118. [Google Scholar]
- Koehler, U.; Bigoni, F.; Wienberg, J.; Stanyon, R. Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Am. J. Phys. Anthropol. 1995, 292, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Hollatz, M.; Wienberg, J.; Müller, S. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Qual. Life Res. 2003, 113, 493–501. [Google Scholar]
- Bigoni, F.; Houck, M.L.; Ryder, O.A.; Wienberg, J.; Stanyon, R. Chromosome painting shows that Pygathrix nemaeus has the most basal karyotype among Asian colobinae. Int. J. Primatol. 2004, 25, 679–688. [Google Scholar] [CrossRef]
- Bigoni, F.; Stanyon, R.; Wimmer, R.; Schempp, W. Chromosome painting shows that the proboscis monkey (Nasalis larvatus) has a derived karyotype and is phylogenetically nested within Asian colobines. Am. J. Primatol. 2003, 60, 85–93. [Google Scholar] [CrossRef]
- Nie, W.; Liu, R.; Chen, Y.; Wang, J.; Yang, F. Mapping chromosomal homologies between humans and two langurs (Semnopithecus francoisi and S. phayrei) by chromosome painting. Chromosom. Res. 1998, 6, 447–453. [Google Scholar] [CrossRef]
- Bigoni, F.; Koehler, U.; Stanyon, R.; Ishida, T.; Wienberg, J. Fluorescence in situ hybridization establishes homology between human and silvered leaf monkey chromosomes, reveals reciprocal translocations between chromosomes homologous to human Y/5, 1/9, and 6/16, and delineates an X1X2Y1Y2/X1X1X2X2 sex-chromosome system. Am. J. Phys. Anthr. 1997, 102, 315–327. [Google Scholar]
- Bigoni, F.; Stanyon, R.; Koehler, U.; Morescalchi, A.M.; Wienberg, J. Mapping homology between human and black and white colobine monkey chromosomes by fluorescent in situ hybridization. Am. J. Primatol. 1997, 42, 289–298. [Google Scholar] [CrossRef]
- Stanyon, R.; Bruening, R.; Stone, G.; Shearin, A.; Bigoni, F. Reciprocal painting between humans, De Brazza’s and patas monkeys reveals a major bifurcation in the Cercopithecini phylogenetic tree. Cytogenet. Genome Res. 2004, 108, 175–182. [Google Scholar] [CrossRef]
- Finelli, P.; Stanyon, R.; Plesker, R.; Ferguson-Smith, M.; O’Brien, P. Reciprocal chromosome painting shows that the great difference in diploid number between human and African green monkey is mostly due to non-Robertsonian fissions. Mamm. Genome 1999, 10, 713–718. [Google Scholar] [CrossRef]
- Moulin, S.; Gerbault-Seureau, M.; Dutrillaux, B.; Richard, F.A. Phylogenomics of African guenons. Chromosom. Res. 2008, 16, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Wienberg, J.; Stanyon, R.; Jauch, A.; Cremer, T. Homologies in human and Macasa fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma 1992, 101, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Neusser, M.; Stanyon, R.; Bigoni, F.; Wienberg, J.; Müller, S. Molecular cytotaxonomy of New World monkeys (Platyrrhini)—Comparative analysis of five species by multi-color chromosome painting gives evidence for a classification of Callimico goeldii within the family of Callitrichidae. Cytogenet. Cell Genet. 2001, 94, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Gerbault-Serreau, M.; Bonnet-Garnier, A.; Richard, F.; Dutrillaux, B. Chromosome painting comparison of Leontopithecus chrysomelas (Callitrichine, Platyrrhini) with man and its phylogenetic position. Chromosom. Res. 2004, 12, 691–701. [Google Scholar] [CrossRef]
- Stanyon, R.; Bigoni, F.; Slaby, T.; Müller, S.; Stone, G.; Bonvicino, C.R. Multi-directional chromosome painting maps homologies between species belonging to three genera of New World monkeys and humans. Chromosoma 2004, 113, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Araújo, N.P.; Stanyon, R.; Pereira, V.D.S.; Svartman, M. interspecific chromosome painting provides clues to the ancestral karyotype of the New World monkey genus aotus. J. Mamm. Evol. 2017, 26, 283–290. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Garcia, F.; Aguilera, M.; Garcia, M.; Fontanals, M.P. Comparative chromosome painting in Aotus reveals a highly derived evolution. Am. J. Primatol. 2005, 65, 73–85. [Google Scholar] [CrossRef]
- Stanyon, R.; Garofalo, F.; Steinberg, E.R.; Capozzi, O.; Di Marco, S.; Nieves, M.; Archidiacono, N.; Mudry, M. Chromosome painting in two genera of South American monkeys: Species identification, conservation, and management. Cytogenet. Genome Res. 2011, 134, 40–50. [Google Scholar] [CrossRef]
- Stanyon, R.; Consigliere, S.; Müller, S.; Morescalchi, A.; Neusser, M.; Wienberg, J. Fluorescence in situ hybridization (FISH) maps chromosomal homologies between the dusky titi and squirrel monkey. Am. J. Primatol. 2000, 50, 95–107. [Google Scholar] [CrossRef]
- Garcia, F.; Nogues, C.; Ponsa, M.; Ruiz-Herrera, A.; Egozcue, J.; García, M. Chromosomal homologies between humans and Cebus apella (Primates) revealed by ZOO-FISH. Mamm. Genome 2000, 11, 399–401. [Google Scholar] [CrossRef]
- Richard, F.; Lombard, M.; Dutrillaux, B. ZOO-FISH Suggests a complete homology between human and Capuchin Monkey (Platyrrhini) euchromatin. Chromosome Res. 1996, 36, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Garcia, F.; Ruiz-Herrera, A.; Egozcue, J.; Ponsa, M.; Garcia, M.; García, M. Chromosomal homologies between Cebusand Ateles (Primates) based on ZOO-FISH and G-banding comparisons. Am. J. Primatol. 2002, 57, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Stanyon, R.; Consigliere, S.; Bigoni, F.; Ferguson-Smith, M.; O’Brien, P.; Wienberg, J. Reciprocal chromosome painting between a New World primate, the woolly monkey, and humans. Chromosom. Res. 2001, 9, 97–106. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, E.H.C.; Neusser, M.; Pieczarka, J.C.; Nagamachi, C.Y.; Sbalqueiro, I.; Müller, S. Phylogenetic inferences of Atelinae (Platyrrhini) based on multi-directional chromosome painting in Brachyteles arachnoides, Ateles paniscus paniscus and Ateles b. marginatus. Cytogenet. Genome Res. 2004, 108, 183–190. [Google Scholar] [CrossRef]
- Morescalchi, M.A.; Schempp, W.; Consigliere, S.; Bigoni, F.; Wienberg, J.; Stanyon, R. Mapping chromosomal homology between humans and the black-handed spider monkey by fluorescence in situ hybridization. Chromosom. Res. 1997, 5, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Consigliere, S.; Stanyon, R.; Koehler, U.; Arnold, N.; Wienberg, J. In situ hybridization (FISH) maps chromosomal homologies between Alouatta belzebul (Platyrrhini, Cebidae) and other primates and reveals extensive interchromosomal rearrangements between howler monkey genomes. Am. J. Primatol. 1999, 46, 119–133. [Google Scholar] [CrossRef]
- De Oliveira, E.H.C.; Neusser, M.; Figueiredo, W.B.; Nagamachi, C.Y.; Pieczarka, J.C.; Sbalqueiro, I.J.; Wienberg, J.; Müller, S. The phylogeny of howler monkeys (Alouatta, Platyrrhini): Reconstruction by multicolor cross-species chromosome painting. Chromosom. Res. 2002, 10, 669–683. [Google Scholar] [CrossRef]
- Finotelo, L.; Amaral, P.; Pieczarka, J.C.; De Oliveira, E.H.C.; Pissinati, A.; Neusser, M.; Müller, S.; Nagamachi, C.Y. Chromosome phylogeny of the subfamily Pitheciinae (Platyrrhini, Primates) by classic cytogenetics and chromosome painting. BMC Evol. Boil. 2010, 10, 189. [Google Scholar] [CrossRef]
- Barros, R.M.S.; Nagamachi, C.Y.; Pieczarka, J.C.; Rodrigues, L.R.R.; Neusser, M.; De Oliveira, E.H.C.; Wienberg, J.; Muniz, J.A.P.C.; Rissino, J.D.; Müller, S. Chromosomal studies in Callicebus donacophilus pallescens, with classic and molecular cytogenetic approaches: Multicolour FISH using human and Saguinus oedipus painting probes. Chromosom. Res. 2003, 11, 327–334. [Google Scholar] [CrossRef]
- Dumas, F.; Bigoni, F.; Stone, G.; Sineo, L.; Stanyon, R. Mapping genomic rearrangements in titi monkeys by chromosome flow sorting and multidirectional in-situ hybridization. Chromosom. Res. 2005, 13, 85–96. [Google Scholar] [CrossRef]
- Stanyon, R.; Bonvicino, C.R.; Svartman, M. Chromosome painting in Callicebus lugens, the species with the lowest diploid number (2n = 16) known in primates. Chromosom. Res. 2003, 112, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Pieczarka, J.C.; Pissinati, A.; De Oliveira, E.H.C.; Rissino, J.D.D.; Nagamachi, C.Y. Genomic mapping of human chromosome paints on the threatened masked Titi monkey (Callicebus personatus). Cytogenet. Genome Res. 2011, 133, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Araújo, N.; Santo, A.A.D.E.; Pereira, V.D.S.; Stanyon, R.; Svartman, M. Chromosome painting in Callicebus nigrifrons provides insights into the genome evolution of Titi Monkeys and the ancestral callicebinae karyotype. Cytogenet. Genome Res. 2017, 151, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Cardone, M.F.; Ventura, M.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Analysis of chromosome conservation in Lemur catta studied by chromosome paints and BAC/PAC probes. Chromosom. Res. 2002, 111, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Warter, S.; Hauwy, M.; Dutrillaux, B.; Rumpler, Y. Application of molecular cytogenetics for chromosomal evolution of the Lemuriformes (Prosimians). Cytogenet. Genome Res. 2004, 108, 197–203. [Google Scholar] [CrossRef]
- Rumpler, Y.; Warter, S.; Hauwy, M.; Fausser, J.-L.; Roos, C.; Zinner, D. Comparing chromosomal and mitochondrial phylogenies of sportive lemurs (Genus Lepilemur, Primates). Chromosom. Res. 2008, 16, 1143–1158. [Google Scholar] [CrossRef]
- Stanyon, R.; Dumas, F.; Stone, G.; Bigoni, F. Multidirectional chromosome painting reveals a remarkable syntenic homology between the greater galagos and the slow loris. Am. J. Primatol. 2006, 68, 349–359. [Google Scholar] [CrossRef]
- Nie, W.; O’Brien, P.C.; Fu, B.; Wang, J.; Su, W.; Robinson, T.; Yang, F.; Ferguson-Smith, M.A. Chromosome painting between human and lorisiform prosimians: Evidence for the HSA 7/16 synteny in the primate ancestral karyotype. Am. J. Phys. Anthr. 2006, 129, 250–259. [Google Scholar] [CrossRef]
- Stanyon, R.; Koehler, U.; Consigliere, S. Chromosome painting reveals that galagos have highly derived karyotypes. Am. J. Phys. Anthr. 2002, 117, 319–326. [Google Scholar] [CrossRef]
- Nie, W.; Fu, B.; O’Brien, P.C.; Wang, J.; Su, W.; Tanomtong, A.; Volobouev, V.; Ferguson-Smith, M.; Yang, F. Flying lemurs—The ’flying tree shrews’? Molecular cytogenetic evidence for a Scandentia-Dermoptera sister clade. BMC Boil. 2008, 6, 18. [Google Scholar] [CrossRef]
- Muller, S.; Stanyon, R.; Ferguson-Smith, M.A.; Plesker, R.; Wienberg, J.; O’Brien, P.C.M. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosom. Res. 1999, 108, 393–400. [Google Scholar] [CrossRef]
- Dumas, F.; Houck, M.; Bigoni, F.; Perelman, P.; Romanenko, S.; Stanyon, R. Chromosome painting of the pygmy tree shrew shows that no derived cytogenetic traits link primates and scandentia. Cytogenet. Genome Res. 2012, 136, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Korstanje, R.; O’Brien, P.; Yang, F.; Rens, W.; Bosma, A.; Van Lith, H.; Van Zutphen, L.; Ferguson-Smith, M. Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet. Cell Genet. 1999, 86, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.; Ferguson-Smith, M.; Stanyon, R. A Short introduction to cytogenetic studies in mammals with reference to the present volume. Cytogenet. Genome Res. 2012, 137, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.; Yang, F.; Dobigny, G.; Romanenko, S.A.; Biltueva, L.S.; Perelman, P.L.; Beklemisheva, V.R.; Alkalaeva, E.; Serdukova, N.A.; Ferguson-Smith, M.A.; et al. Tracking genome organization in rodents by Zoo-FISH. Chromosom. Res. 2008, 16, 261–274. [Google Scholar] [CrossRef]
- Stanyon, R.; Stone, G.; Garcia, M.; Froenicke, L.; García, M. Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics 2003, 82, 245–249. [Google Scholar] [CrossRef]
- Li, T.; O’Brien, P.; Biltueva, L.; Fu, B.; Wang, J.; Nie, W.; Ferguson-Smith, M.; Graphodatsky, A.; Yang, F. Evolution of genome organizations of Squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosom. Res. 2004, 12, 317–335. [Google Scholar] [CrossRef]
- Yang, F.; Graphodatsky, A.; Li, T.; Fu, B.; Dobigny, G.; Wang, J.; Perelman, P.L.; Serdukova, N.A.; Su, W.; O’Brien, P.C.; et al. Comparative genome maps of the pangolin, hedgehog, sloth, anteater and human revealed by cross-species chromosome painting: Further insight into the ancestral karyotype and genome evolution of eutherian mammals. Chromosom. Res. 2006, 14, 283–296. [Google Scholar] [CrossRef]
- Nie, W.; Wang, J.; Su, W.; Wang, Y.; Yang, F. Chromosomal rearrangements underlying karyotype differences between Chinese pangolin (Manis pentadactyla) and Malayan pangolin (Manis javanica) revealed by chromosome painting. Chromosom. Res. 2009, 17, 321–329. [Google Scholar] [CrossRef]
- Breen, M.; Thomas, R.; Binns, M.M.; Carter, N.P.; Langford, C.F. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics 1999, 61, 145–155. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.; Milne, B.; Graphodatsky, A.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.R.; Ferguson-Smith, M. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 1999, 62, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Graphodatsky, A.; O’Brien, P.C.M.; Colabella, A.; Solanky, N.; Squire, M.; Sargan, D.R.; Ferguson-Smith, M.A. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosom. Res. 2000, 8, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Cavagna, P.; Menotti, A.; Stanyon, R. Genomic homology of the domestic ferret with cats and humans. Mamm. Genome 2000, 11, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Perelman, P.L.; Graphodatsky, A.; Dragoo, J.W.; Serdyukova, N.A.; Stone, G.; Cavagna, P.; Menotti, A.; Nie, W.; O’Brien, P.C.M.; Wang, J.; et al. Chromosome painting shows that skunks (Mephitidae, Carnivora) have highly rearranged karyotypes. Chromosom. Res. 2008, 16, 1215–1231. [Google Scholar] [CrossRef]
- Yang, F.; Fu, B.; O’Brien, P.C.M.; Nie, W.; Ryder, O.A.; Ferguson-Smith, M.A. Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: Insight into the occasional fertility of mules. Chromosom. Res. 2004, 12, 65–76. [Google Scholar] [CrossRef]
- Balmus, G.; Trifonov, V.; Biltueva, L.S.; O’Brien, P.C.; Alkalaeva, E.; Fu, B.; Skidmore, J.A.; Allen, T.; Graphodatsky, A.; Yang, F.; et al. Cross-species chromosome painting among camel, cattle, pig and human: Further insights into the putative Cetartiodactyla ancestral karyotype. Chromosom. Res. 2007, 15, 499–514. [Google Scholar] [CrossRef]
- Kulemzina, A.I.; Trifonov, V.; Perelman, P.L.; Rubtsova, N.V.; Volobuev, V.; Ferguson-Smith, M.A.; Stanyon, R.; Yang, F.; Graphodatsky, A. Cross-species chromosome painting in Cetartiodactyla: Reconstructing the karyotype evolution in key phylogenetic lineages. Chromosom. Res. 2009, 17, 419–436. [Google Scholar] [CrossRef]
- Volleth, M.; Heller, K.-G.; Pfeiffer, R.; Hameister, H. A comparative ZOO-FISH analysis in bats elucidates the phylogenetic relationships between Megachiroptera and five microchiropteran families. Chromosom. Res. 2002, 10, 477–497. [Google Scholar] [CrossRef]
- Mao, X.; Nie, W.; Wang, J.; Su, W.; Feng, Q.; Wang, Y.; Dobigny, G.; Yang, F. Comparative cytogenetics of bats (Chiroptera): The prevalence of Robertsonian translocations limits the power of chromosomal characters in resolving interfamily phylogenetic relationships. Chromosom. Res. 2008, 16, 155–170. [Google Scholar] [CrossRef]
- Mao, X.; Nie, W.; Wang, J.; Su, W.; Ao, L.; Feng, Q.; Wang, Y.; Volleth, M.; Yang, F. Karyotype evolution in Rhinolophus bats (Rhinolophidae, Chiroptera) illuminated by cross-species chromosome painting and G-banding comparison. Chromosom. Res. 2007, 15, 835–848. [Google Scholar] [CrossRef]
- Ye, J.; Biltueva, L.; Huang, L.; Nie, W.; Wang, J.; Jing, M.; Su, W.; Vorobieva, N.V.; Jiang, X.; Graphodatsky, A.; et al. Cross-species chromosome painting unveils cytogenetic signatures for the Eulipotyphla and evidence for the polyphyly of Insectivora. Chromosom. Res. 2006, 14, 151–159. [Google Scholar] [CrossRef]
- Volleth, M.; Müller, S. Zoo-FISH in the European mole (Talpa europaea) detects all ancestral Boreo-Eutherian human homologous chromosome associations. Cytogenet. Genome Res. 2006, 115, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Alkalaeva, E.Z.; Perelman, P.L.; Pardini, A.T.; Harrison, W.R.; O’Brien, P.C.M. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ances-tral karyotype. PNAS 2003, 100, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, M.E.; Burkett, S.; Dennis, T.R.; Stone, G.; Gray, B.A.; McGuire, P.M.; Zori, R.; Stanyon, R. Chromosome painting in the manatee supports Afrotheria and Paenungulata. BMC Evol. Boil. 2007, 7, 6. [Google Scholar]
- Ruiz-Herrera, A.; Robinson, T. Chromosomal instability in Afrotheria: Fragile sites, evolutionary breakpoints and phylogenetic inference from genome sequence assemblies. BMC Evol. Boil. 2007, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.; Fu, B.; Ferguson-Smith, M.A.; Yang, F. Cross-species chromosome painting in the golden mole and elephant-shrew: Support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. R. Soc. 2004, 271, 1477–1484. [Google Scholar] [CrossRef] [PubMed]
- Svartman, M.; Stone, G.; Page, J.E.; Stanyon, R. A chromosome painting test of the basal eutherian karyotype. Chromosom. Res. 2004, 12, 45–53. [Google Scholar] [CrossRef]
- Svartman, M.; Stone, G.; Stanyon, R. The Ancestral eutherian karyotype is present in Xenarthra. PLoS Genet. 2006, 2, e109. [Google Scholar] [CrossRef]
- Azevedo, N.F.; Svartman, M.; Manchester, A.; Moraes-Barros, N.; Stanyon, R.; Vianna-Morgante, A.M. Chromosome painting in three-toed sloths: A cytogenetic signature and ancestral karyotype for Xenarthra. BMC Evol. Boil. 2012, 12, 36. [Google Scholar] [CrossRef]
- Graphodatsky, A.; Perelman, P.L.; Sokolovskaya, N.V.; Beklemisheva, V.R.; Serdukova, N.A.; Dobigny, G.; O’Brien, S.; Ferguson-Smith, M.A.; Yang, F. Phylogenomics of the dog and fox family (Canidae, Carnivora) revealed by chromosome painting. Chromosom. Res. 2008, 16, 129–143. [Google Scholar] [CrossRef]
- Robinson, T.; Ruiz-Herrera, A. Defining the ancestral eutherian karyotype: A cladistic interpretation of chromosome painting and genome sequence assembly data. Chromosom. Res. 2008, 16, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Byrne, H.; Rylands, A.B.; Carneiro, J.C.; Alfaro, J.W.; Bertuol, F.; da Silva, M.N.; Messias, M.; Groves, C.P.; Mittermeier, R.A.; Farias, I.; et al. Phylogenetic relationship of the New World Titi monkeys (Callicebus): First appraisal of taxonomy based on molecular evidence. Front. Zool. 2016, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Perelman, P.; Johnson, W.; Roos, C.; Seuanez, H.N.; Horvath, J.E.; Moreira, M.A.M.; Kessing, B.; Pontius, J.; Roelke, M.; Rumpler, Y.; et al. A molecular phylogeny of living primates. PLoS Genet. 2011, 7, e1001342. [Google Scholar] [CrossRef] [PubMed]
- Clemente, I.C.; Egozcue, J.; García, M.; García, M. Evolution of the Simiiformes and the phylogeny of human chromosomes. Qual. Life Res. 1990, 84, 493–506. [Google Scholar] [CrossRef]
| Family | Latin Name | Code | Cell Type | Sample/Cell Line Acknowledgement |
|---|---|---|---|---|
| Cebidae | Saguinus oedipus | SOE | fibroblast cell line | Melody Roelke (Frederick National Laboratory of Cancer Research, Leidos Biomedical Research, Frederick, MD, USA), June Bellizzi and Director Richard Hann (Catoctin Wildlife Park and Zoo, Thumont, MD, USA) |
| Cebidae | Callithrix argentata | CAR | fibroblast cell line | Stephen O’Brien (Laboratory of Genomic Diversity, National Cancer Institute, Frederick, MD, USA) and Hector Seuánez (Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Brazil) |
| Atelidae | Alouatta belzebul | ABE | fibroblast cell line |
| Species | Chromosome Morphology | Chr. Num. | Human Chromosome Association | Reference | Method |
|---|---|---|---|---|---|
| MAMMALIA | |||||
| EUTHERIA | |||||
| BOREOEUTHERIA | |||||
| EUARCHONTOGLIRES | |||||
| PRIMATES | |||||
| Catarrhini | |||||
| Homo sapiens | A | 13 | |||
| Pan troglodytes | A | 14 | [7] | CP | |
| Gorilla gorilla | A | 14 | [7] | CP | |
| Pongo pygmaeus | A | 14 | [7] | CP | |
| Hylobates concolor | M | 5, 9 | 1/13; 1/4/10/13 | [33] | CP |
| Hylobates klossii | 4q | 3/13 | [34] | CP | |
| Hylobates moloch | 4q | 3/13 | [34] | CP | |
| Hylobates lar | 4q | 3/13 | [7] | CP | |
| Symphalangus syndactylus | M | 15 | [7] | CP | |
| Pygathrix nemaeus | SM | 17 | [35] | CP | |
| Nasalis larvatus | M | 15 | [36] | CP | |
| Semnopithecus francoisi | M | 9 | [37] | CP | |
| Semnopithecus phayrei | M | 9 | [34] | CP | |
| Presbytis cristata | M | 19 | [38] | CP | |
| Colobus guereza | M | 19 | [39] | CP | |
| Erythrocebus patas | SM | 15 | [40] | RP | |
| Chlorocebus aethiops | M | 3 | [41] | CP | |
| Cercopithecus erythrogaster | SM | 12 | [42] | Z-F | |
| Cercopithecus stampflii | SM | 13 | [42] | Z-F | |
| Cercopithecus neglectus | M | 19 | [40] | RP | |
| Macaca fuscata | SM | 16 | [43] | CP | |
| Platyrrhini | |||||
| Cebuella pygmaea | SMs | 1, 4 | 13/9/22, 20/17/13 | [44] [8] | CP RP |
| Callithrix argentata | SMs | 2, 1 | 13/9/22, 20/17/13 | ||
| Callithrix jacchus | SMs | 1, 5 | 13/9/22, 20/17/13 | ||
| Callimico goeldii | As | 19, 17 | 13/9/22, 13/17 | ||
| Saguinus oedipus | SMs | 1, 2 | 9/13/22, 20/17/13 | ||
| Leontopithecus chrysomelas | SMs | 1, 2 | 9/13/22,13/17/20 | [45] | CP |
| Aotus nancymaae | A | 19 | [46] | CP | |
| Aotus infulatus | A | 14 | [47] | CP | |
| Aotus lemurinus griseimembra | A | 17 | [46,48,49] | CP | |
| Saimiri sciureus | A | 16 | [8,50] | CP | |
| Cebus (Sapajus) apella | A | 17 | [51,52] | CP, Z-F | |
| Sapajus a. paraguayanus | A | 17 | [52] | Z-F, CP | |
| Sapajus a. robustus | A | 17 | |||
| Cebus capucinus | A | 11 | |||
| Cebus nigrivitatus | A | 17 | [53] | CP | |
| Lagothrix lagotricha | SM | 8 | [54] | CP | |
| Brachyteles arachnoides | A | 20 | [55] | CP | |
| Ateles paniscus paniscus | M | 4 | 13a/13b/3c/7b/1a2 | ||
| Ateles belzebuth marginatus | SM | 12 | |||
| Ateles geoffroyi | SM | 12 | [56] | CP | |
| Ateles belzebuth hybridus | A | 12 | [53] | CP | |
| Alouatta belzebul | A | 14 | [57] | CP | |
| Alouatta seniculus sara | 12 | [57] | CP | ||
| Alouatta seniculus arctoidea | 16 | [57] | CP | ||
| Alouatta caraya | A | 15 (20 *) | (* [48]) [49,58] | CP | |
| Alouatta seniculus macconnelli | SM | 4q | 13/19 | [58] | CP |
| Alouatta guariba guariba | A | 14 | [48,49] | CP | |
| Cacajao calvus rubicundus | A | 13 | [59] | CP | |
| Chiropotes israelita | A | 15 | [46] | CP | |
| Chiropotes utahicki | A | 15 | [46] | CP | |
| Pithecia irrorata | SM | 8 | 22/13 | [59] | CP |
| Plecturocebus (Callicebus) donacophilus pallescens | A | 15 | [60] | CP | |
| Plecturocebus (Callicebus) cupreus | SMA | 7, 17 | 3/21/13, 13/17 | [61] | CP |
| Plecturocebus (Callicebus) pallescens | A | 18, 21 | 13/17, 13 | [61] | CP, |
| Cheracebus (Callicebus) lugens | SM | 1 | 1/13 - 12/13 | [62] | CP |
| Callicebus moloch | A | 21 | [50] | CP | |
| Callicebus personatus | M | 1 | 13/20 | [63] | CP |
| Callicebus nigrifrons | A | 4, 17 | 13/20, 13/17 | [64] | CP |
| Strepsirrhini | |||||
| Lemur catta | A | 13 | [65] [66] | BAC CP | |
| Hapalemur griseus griseus | 15 | [66] | CP | ||
| Eulemur fulvus | A | 12 | [66] | CP | |
| Microcebus murinus | SM | 13 | [67] [66] | CP | |
| Lepilemur edwardsi | 6p | [67] | CP | ||
| Lepilemur ankaranensis | 14 | [67] | CP | ||
| Lepilemur jamesi | 5q ter | [67] | CP | ||
| Lepilemur leucopus | 1q ter | [67] | CP | ||
| Lepilemur microdon | 5p | [67] | CP | ||
| Lepilemur mittermeieri | 7p | [67] | CP | ||
| Lepilemur dorsalis | 6p | [66,67] | CP | ||
| Lepilemur mustelinus | 8 ter | [66,67] | CP | ||
| Lepilemur ruficaudatus | 5q prox | [66,67] | CP | ||
| Lepilemur septentrionalis | 14 | [67] [66] | CP | ||
| Lepilemur dorsalis | 6p | [67] [66] | CP | ||
| Lepilemur mustelinus | 8 ter | [67] [66] | CP | ||
| Lepilemur ruficaudatus | 5q prox | [67] [66] | CP | ||
| Lepilemur septentrionalis | 14 | [67] [66] | CP | ||
| Indri indri | SM | 3p | 13/17 | [66] | CP |
| Propithecus verreauxi | 6q | 5/13 | [66] | CP | |
| Avahi laniger | 12 | [66] | CP | ||
| Daubentonia madagascariensis | 8p | 10/13 | [66] | CP | |
| Nycticebus coucang | SM | 18 17 | [68] [69] | RP CP | |
| Galago moholi | M | 5 | 13/16/12 | [70] | CP |
| Otolemur garnettii | SM | 14 | [68] | RP | |
| Otolemur crassicaudatus | A | 14 | [70] | CP | |
| DERMOPTERA | |||||
| Galeopterus variegatus | A | 13 | [71] | RP | |
| SCANDENTIA | |||||
| Tupaia belangeri | A | 17 | [72] | CP | |
| Tupaia minor | A | 16 | [73] | CP | |
| LAGOMORPHA | |||||
| Oryctolagus cuniculus | SM | 8 | 13/12 | [74] [75] | RP SA |
| RODENTIA | |||||
| Mus musculus | 3, 5, 8, 14, 14 | [20,21] [23,75] | SA | ||
| Rattus norvegicus | 2, 12, 15, 15, 16 | [20] [23] | SA | ||
| Pedetes capensis | SM | 6 | 13/12/22 | [76] | CP |
| Sicista betulina | M, SM | 1, 9 | 13/4/10/11/9/10, 3/6/313/19 | [76] | CP |
| Castor fiber | SM | 4 | 8/13 | [76] | CP |
| Sciurus carolinensis | SM | 6 | 10/13 | [77,78] | RP |
| Petaurista albiventer | M | 11 | 10/13 | [78] | CP |
| Tamias sibiricus | M | 10 | 10/13 | [78] | CP |
| LAURASIATHERIA | |||||
| PHOLIDOTA | |||||
| Manis javanica | SM M | 1, 9q | 13/5/2p, 18/13 | [79] [80] | CP CP |
| Manis pentadactyla | SM A | 1q, 17 | 13/5/2, 13 | [80] | CP |
| CARNIVORA | |||||
| Canis familiaris | As | (25 *) 22, 28 | (*[81]), [82] [75,83] [23] | RP CP Z-F SA | |
| Vulpes vulpes | SMs | 6, 9 | 13/14, 2/8/13/3/19 | [82] | RP |
| Mustela putorius | SM | 8 | [84] | CP | |
| Procyon lotor | M | 3 | 13/2 | [85] | CP |
| Mephitis mephitis | SM | 19 | [85] | CP | |
| Felis catus | A1p | 13/5 | [82] [19,20], [75] | CP SA | |
| PERISSODACTYLA | |||||
| Tapirus indicus | A | 18 | [9] | Z-F | |
| Diceros bicornis | A | 10 | [9] | Z-F | |
| Ceratotherium simum | 10 | [9] | Z-F | ||
| Equus caballus | A | 17 | [86] [20,23] [9] [27] | RP SA Z-F BAC | |
| Equus burchelli | SM | 6q | 13/9 | [9] | RP Z-F |
| Equus asinus | 11 | [9] | Z-F | ||
| Equus grevyi | SM | 6q | 13/9 | [9] | Z-F |
| Equus zebra hartmannae | 15 | [9] | Z-F | ||
| Equus hemionus onager | 5q | 12/13/22 | [9] | Z-F | |
| Equus przewalskii | 16 | [9] | Z-F | ||
| CETARTIODACTYLA | |||||
| Bos taurus | A | 12 | [19] [23] [87] | SA RP | |
| Moschus moschiferus | A | 17 | [88] | Z-F | |
| Okapia johnstoni | A | 11 | [88] | Z-F | |
| Giraffa camelopardalis | M | 12 | 14/15/13 | [88] | Z-F |
| Globicephala melas | M | 15 | [88] | Z-F | |
| Hippopotamus amphibious | M | 15 | [88] | Z-F | |
| Sus scrofa | M | 11 | [19] [23] [87] | SA RP | |
| Camelus dromedarius | M | 14 | [87] | RP | |
| CHIROPTERA | |||||
| Mormopteurus planiceps | M | M7 | 13/18 | [89] | CP |
| Myotis myotis | M | V5/6 | 4/8/13/12/22 | [89] | CP |
| Taphozous melanopogon | SM | 1 | 4c/8b/13/16b/7c/5a | [90] | CP |
| Megaderma spasma | M | 12 | 20/13/8b/4c | [90] | CP |
| Rhinolophus mehelyi | A | R6 | 13/4/8/13 | [89] | CP |
| Aselliscus stoliczkanus | M | 1 | 22/12/13/4/8/13 | [91] | CP |
| Hipposideros larvatus | M | H1 | 13/3/21 | [89] [91] | CP |
| Eonycteris spelaea | SM | E11 | 13/4/8/13 | [89] | CP |
| EULIPOTYPHLA | |||||
| Hemiechinus auritus | SMs | 5q, 6 | 5/13, 2/22/12/13/12 | [79] | CP |
| Neotetracus sinensis | SM A | 3, 10 | 13/4/20/10,1/13/10/12/22 | [92] | CP |
| Sorex araneus | M | bc | 9/5/2/13/8/7 | [23,92] | CP, SA |
| Blarinella griselda | SM | 3 | 13/10/13/4/5 | [92] | CP |
| Talpa europaea | M | 6 | 2/13 | [93] | CP |
| ATLANTOGENATA | |||||
| AFROTHERIA | |||||
| PROBOSCIDEA | |||||
| Loxodonta africana | A, SM | 16, 26 | 13, 6/13/3 | [19,94] [23] | CP SA |
| Elephas maximus | A, SM | 16, 26 | 13, 6/13/3 | [94] | CP |
| SIRENIA | |||||
| Trichechus manatus | M | 19 | 13/3 | [95] | CP |
| TUBULIDENTATA | |||||
| Orycteropus afer | SM | 1 | 19/16/13/2/8/4 | [94] [96] | CP SA |
| MACROSCELIDEA | |||||
| Elephantulus rupestris Elephantulus edwardii | SM | 2 | 13/3/21/5 | [97] [96] | CP SA |
| Macroscelides proboscideus | SM | 2 | 13/3/21/5 | [98] | CP |
| AFROSORICIDA | |||||
| Chrysochloris asiatica | M | 8 | 13/18 | [97] [96] | RP SA |
| XENARTHRA | |||||
| CINGULATA | |||||
| Dasypus novemcinctus | SM | 19 | [99] | CP | |
| PILOSA | |||||
| Tamandua tetradactyla | M | 4, (2 *) | 13/1 | [79], (* [99]) | CP |
| Choloepus didactylus | A | 17 | [79] | CP | |
| Choloepus hoffmanni | A | 12 | [99] | CP | |
| Bradypus torquatus | A | 12 | [100] | CP | |
| Bradypus variegatus | A | 17 | |||
| METATHERIA | |||||
| MARSUPIALIA | |||||
| DIDELPHIMORPHIA | |||||
| Monodelphis domestica | SMs | 4, 7 | [101,102] | SA | |
| PROTOTHERIA | |||||
| MONOTREMATA | |||||
| Ornithorhynchus anatinus | SM Ms | 2, 10, 20 | [101] | SA | |
| AVES | |||||
| GALLIFORMES | |||||
| Gallus gallus | 1 | [101,102] | SA | ||
| BAC Clone | Start Coordinates | Mapping Position | |||
|---|---|---|---|---|---|
| HSA | SOE | ABE | CAR | ||
| A6 CHORI RP11-35m5 | 27534229 | 13 tel | 1p tel/ three bands | 14 tel | 1p tel |
| A7 CHORI RP11-85p8 | 27475788 | ||||
| A11 CHORI RP11-14a4 | 39027477 | 13 tel | 1 p tel | 14 tel | 1p tel |
| B8 CHORI RP11-30n18 | 48823331 | 13 cen | 2q tel | 4 interstitial below dark band | 2q tel |
| B11 CHORI RP11-54g17 | 44481779 | 13 cen | 2q tel | 14 interstitial below dark band | 2q tel |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scardino, R.; Milioto, V.; Proskuryakova, A.A.; Serdyukova, N.A.; Perelman, P.L.; Dumas, F. Evolution of the Human Chromosome 13 Synteny: Evolutionary Rearrangements, Plasticity, Human Disease Genes and Cancer Breakpoints. Genes 2020, 11, 383. https://doi.org/10.3390/genes11040383
Scardino R, Milioto V, Proskuryakova AA, Serdyukova NA, Perelman PL, Dumas F. Evolution of the Human Chromosome 13 Synteny: Evolutionary Rearrangements, Plasticity, Human Disease Genes and Cancer Breakpoints. Genes. 2020; 11(4):383. https://doi.org/10.3390/genes11040383
Chicago/Turabian StyleScardino, Rita, Vanessa Milioto, Anastasia A. Proskuryakova, Natalia A. Serdyukova, Polina L. Perelman, and Francesca Dumas. 2020. "Evolution of the Human Chromosome 13 Synteny: Evolutionary Rearrangements, Plasticity, Human Disease Genes and Cancer Breakpoints" Genes 11, no. 4: 383. https://doi.org/10.3390/genes11040383
APA StyleScardino, R., Milioto, V., Proskuryakova, A. A., Serdyukova, N. A., Perelman, P. L., & Dumas, F. (2020). Evolution of the Human Chromosome 13 Synteny: Evolutionary Rearrangements, Plasticity, Human Disease Genes and Cancer Breakpoints. Genes, 11(4), 383. https://doi.org/10.3390/genes11040383





