Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism
Abstract
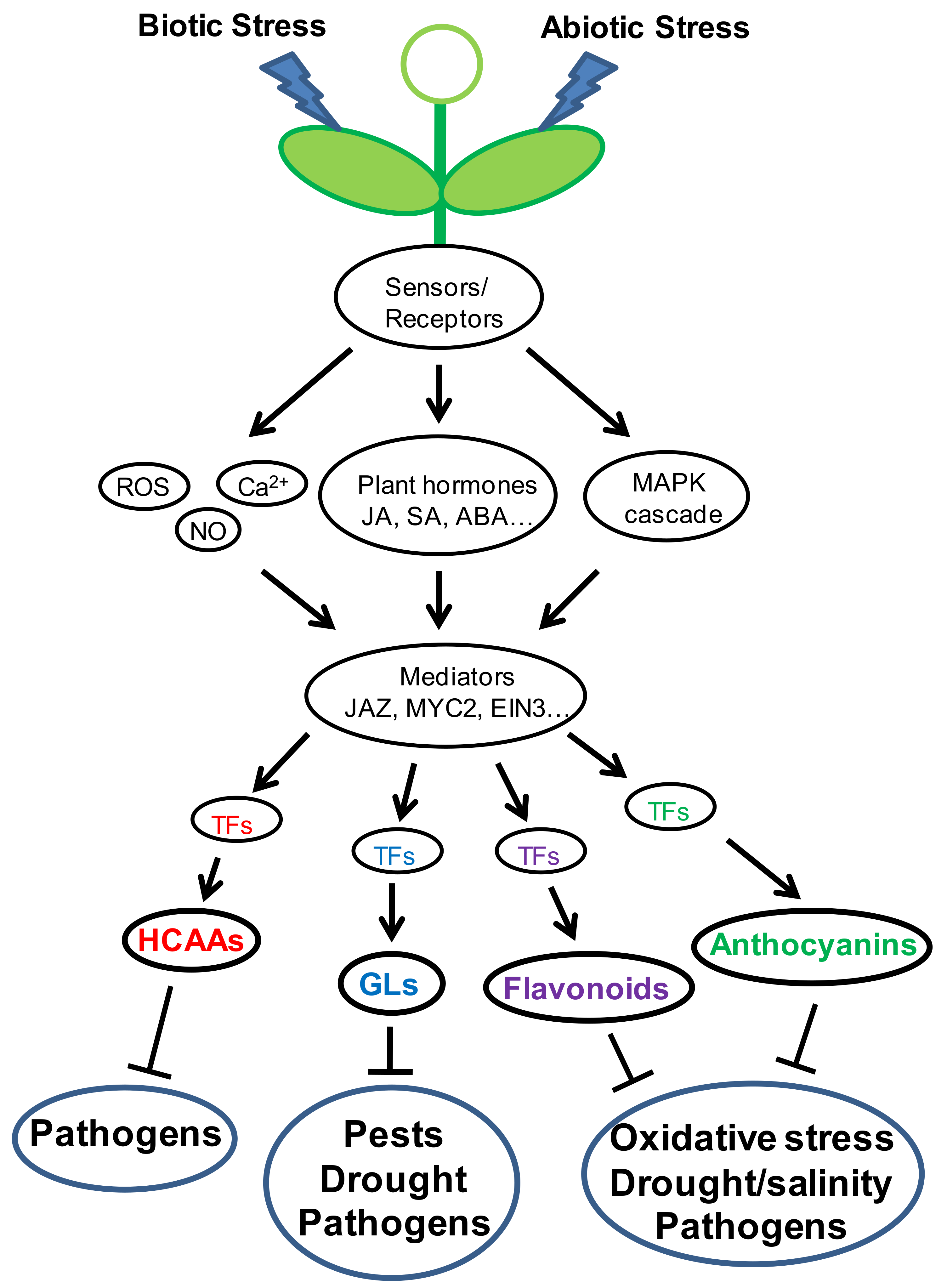
1. Introduction
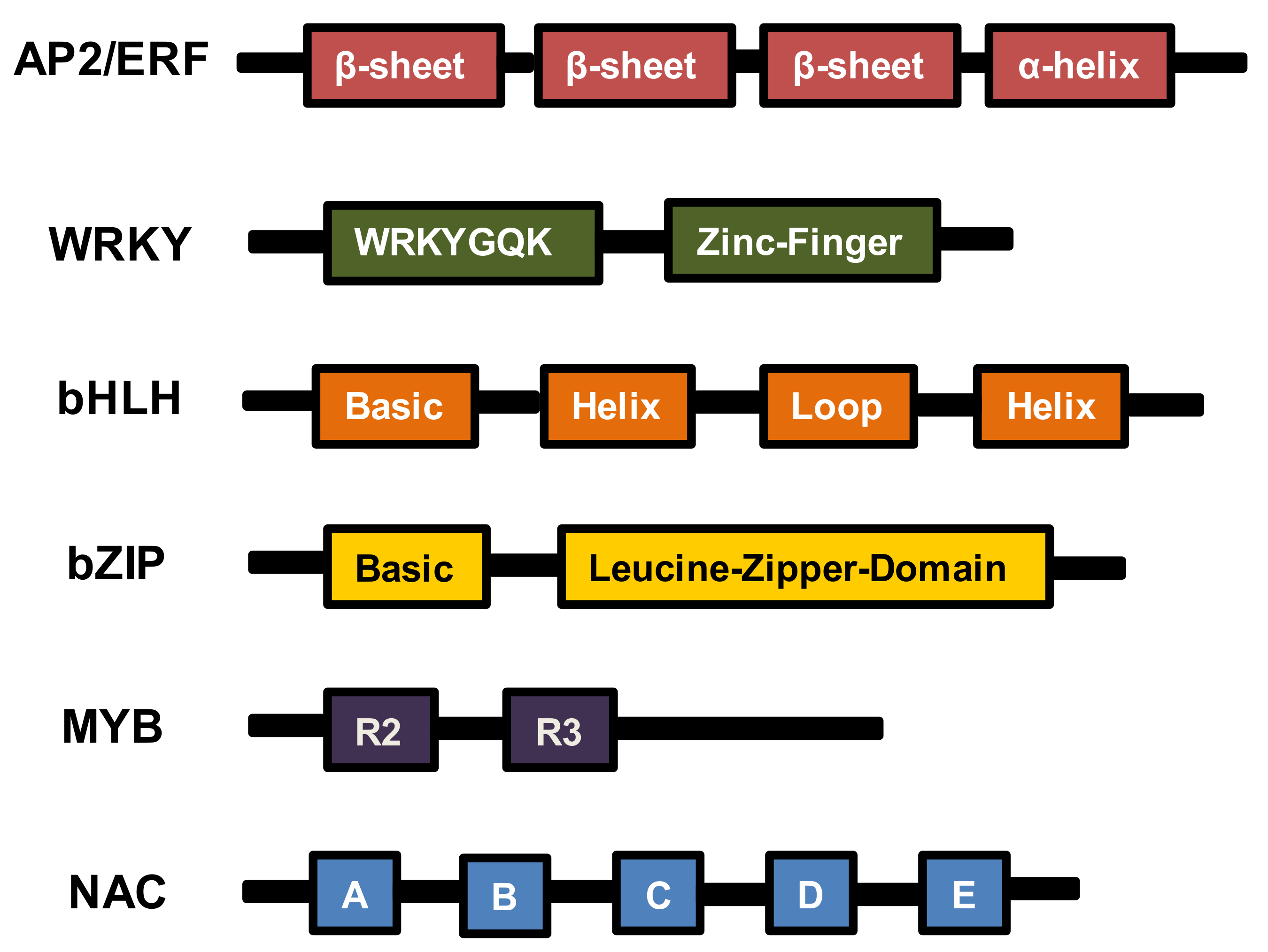
2. AP2/ERF TFs
3. WRKY TFs
4. bHLH TFs
5. bZIP TFs
6. MYB TFs
7. NAC TFs
8. Cross Interactions of Diverse Transcription Proteins Show the Complexity of Phytoprotective Regulation
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Knight, M.R. Abiotic stress signalling pathways: Specificity and cross-talk. Trends Plant Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef]
- Camejo, D.; Guzmán-Cedeño, Á.; Moreno, A. Reactive oxygen species, essential molecules, during plant–pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qi, M.; Mei, C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004, 40, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Menke, F.L.; Champion, A.; Kijne, J.W.; Memelink, J. A novel jasmonate-and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate-and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999, 18, 4455–4463. [Google Scholar] [CrossRef]
- Treisman, R. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 1996, 8, 205–215. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Roze, L.V.; Linz, J.E. Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins 2013, 5, 683–702. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, W.; Kinkema, M.; Li, X.; Dong, X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 1999, 96, 6523–6528. [Google Scholar] [CrossRef]
- Loake, G.; Grant, M. Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef]
- Fan, W.; Dong, X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 2002, 14, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Jiang, H.; Li, C.-B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H. Glucosinolate regulation in a complex relationship–MYC and MYB–no one can act without each other. In Advances in Botanical Research; Elsevier: London, UK, 2016; Volume 80, pp. 57–97. [Google Scholar]
- Maróti, G.; Kereszt, A.; Kondorosi, E.; Mergaert, P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 2011, 162, 363–374. [Google Scholar] [CrossRef]
- Bednarek, P. Chemical warfare or modulators of defence responses–the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 407–414. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef]
- Voigt, C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014, 5, 168. [Google Scholar] [CrossRef]
- Hartmann, T. The lost origin of chemical ecology in the late 19th century. Proc. Natl. Acad. Sci. USA 2008, 105, 4541–4546. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. N. Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef]
- Osbourn, A.E. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 1996, 8, 1821. [Google Scholar] [CrossRef]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2016, 68, 1303–1321. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Paul, P.; Singh, S.K.; Patra, B.; Liu, X.; Pattanaik, S.; Yuan, L. Mutually Regulated AP2/ERF Gene Clusters Modulate Biosynthesis of Specialized Metabolites in Plants. Plant Physiol. 2020, 182, 840–856. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Singh, S.K.; Patra, B.; Sui, X.Y.; Pattanaik, S.; Yuan, L. A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. N. Phytol. 2017, 213, 1107–1123. [Google Scholar] [CrossRef] [PubMed]
- Roepke, J.; Salim, V.; Wu, M.; Thamm, A.M.; Murata, J.; Ploss, K.; Boland, W.; De Luca, V. Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc. Natl. Acad. Sci. USA 2010, 107, 15287–15292. [Google Scholar] [CrossRef]
- Nakayasu, M.; Umemoto, N.; Ohyama, K.; Fujimoto, Y.; Lee, H.J.; Watanabe, B.; Muranaka, T.; Saito, K.; Sugimoto, Y.; Mizutani, M. A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol. 2017, 175, 120–133. [Google Scholar] [CrossRef]
- Friedman, M. Tomato glycoalkaloids: Role in the plant and in the diet. J. Agric. Food Chem. 2002, 50, 5751–5780. [Google Scholar] [CrossRef]
- Friedman, M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J. Agric. Food Chem. 2006, 54, 8655–8681. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.; Shinde, B.; Cardenas, P.; Bocobza, S.; Unger, T.; Malitsky, S.; Finkers, R. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Cárdenas, P.D.; Sonawane, P.D.; Pollier, J.; Bossche, R.V.; Dewangan, V.; Weithorn, E.; Tal, L.; Meir, S.; Rogachev, I.; Malitsky, S. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 2016, 7, 10654. [Google Scholar] [CrossRef]
- Thagun, C.; Imanishi, S.; Kudo, T.; Nakabayashi, R.; Ohyama, K.; Mori, T.; Kawamoto, K.; Nakamura, Y.; Katayama, M.; Nonaka, S. Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 2016, 57, 961–975. [Google Scholar] [CrossRef]
- Steppuhn, A.; Gase, K.; Krock, B.; Halitschke, R.; Baldwin, I.T. Nicotine’s defensive function in nature. PLoS Biol. 2004, 2, e217. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- De Boer, K.; Tilleman, S.; Pauwels, L.; Vanden Bossche, R.; De Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Sears, M.T.; Zhang, H.; Rushton, P.J.; Wu, M.; Han, S.; Spano, A.J.; Timko, M.P. NtERF32: A non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 2014, 84, 49–66. [Google Scholar] [CrossRef]
- Papadopoulou, K.; Melton, R.; Leggett, M.; Daniels, M.; Osbourn, A. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 1999, 96, 12923–12928. [Google Scholar] [CrossRef]
- Avato, P.; Bucci, R.; Tava, A.; Vitali, C.; Rosato, A.; Bialy, Z.; Jurzysta, M. Antimicrobial activity of saponins from Medicago sp.: Structure-activity relationship. Phytother. Res. 2006, 20, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Huang, Z.; Ge, F.; Liu, D.; Lu, R.; Chen, C. An AP2/ERF family transcription factor PnERF1 raised the biosynthesis of saponins in panax notoginseng. J. Plant Growth Regul. 2017, 36, 691–701. [Google Scholar] [CrossRef]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef]
- Young, D.; Michelotti, E.; Swindell, C.; Krauss, N. Antifungal properties of taxol and various analogues. Experientia 1992, 48, 882–885. [Google Scholar] [CrossRef]
- Zhang, M.; Li, S.; Nie, L.; Chen, Q.; Xu, X.; Yu, L.; Fu, C. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 2015, 89, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, K.; Meng, Y.; Hu, J.; Ding, M.; Bian, J.; Yan, M.; Han, J.; Zhou, M. Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA 59 transcription factor. Plant J. 2018, 95, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Montero, C.; Cristescu, S.; Jiménez, J.; Orea, J.; te Lintel Hekkert, S.; Harren, F.; Urena, A.G. Trans-resveratrol and grape disease resistance. A dynamical study by high-resolution laser-based techniques. Plant Physiol. 2003, 131, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y. Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep. 2019, 38, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, H.; Li, F.; Chen, Z.; Li, X.; Zhu, L.; Wang, G.; Yu, J.; Huang, D.; Lang, Z. The maize transcription factor EREB 58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 2015, 84, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Yogendra, K.N.; Kumar, A.; Sarkar, K.; Li, Y.; Pushpa, D.; Mosa, K.A.; Duggavathi, R.; Kushalappa, A.C. Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 2015, 66, 7377–7389. [Google Scholar] [CrossRef]
- Yogendra, K.N.; Dhokane, D.; Kushalappa, A.C.; Sarmiento, F.; Rodriguez, E.; Mosquera, T. StWRKY8 transcription factor regulates benzylisoquinoline alkaloid pathway in potato conferring resistance to late blight. Plant Sci. 2017, 256, 208–216. [Google Scholar] [CrossRef]
- Fu, J.; Liu, Q.; Wang, C.; Liang, J.; Liu, L.; Wang, Q. ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J. Exp. Bot. 2017, 69, 497–510. [Google Scholar] [CrossRef]
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Yu, L. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. 2013, 15, 19–26. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. N. Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef]
- Kage, U.; Yogendra, K.N.; Kushalappa, A.C. TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 2017, 7, 42596. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; De Tommasi, N.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef] [PubMed]
- Vannozzi, A.; Wong, D.C.J.; Höll, J.; Hmmam, I.; Matus, J.T.; Bogs, J.; Ziegler, T.; Dry, I.; Barcaccia, G.; Lucchin, M. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 2018, 59, 1043–1059. [Google Scholar] [CrossRef]
- Jiang, J.; Xi, H.; Dai, Z.; Lecourieux, F.; Yuan, L.; Liu, X.; Patra, B.; Wei, Y.; Li, S.; Wang, L. VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 2018, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Karre, S.; Kumar, A.; Yogendra, K.; Kage, U.; Kushalappa, A.; Charron, J.-B. HvWRKY23 regulates flavonoid glycoside and hydroxycinnamic acid amide biosynthetic genes in barley to combat Fusarium head blight. Plant Mol. Biol. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Samira, R.; Li, B.; Kliebenstein, D.; Li, C.; Davis, E.; Gillikin, J.W.; Long, T.A. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. 2018, 97, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Berger, B.; Gigolashvili, T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014, 166, 349–369. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 125, 387–398. [Google Scholar] [CrossRef]
- An, J.-P.; Li, H.-H.; Song, L.-Q.; Su, L.; Liu, X.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 2016, 108, 24–31. [Google Scholar] [CrossRef]
- Yamamura, C.; Mizutani, E.; Okada, K.; Nakagawa, H.; Fukushima, S.; Tanaka, A.; Maeda, S.; Kamakura, T.; Yamane, H.; Takatsuji, H. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015, 84, 1100–1113. [Google Scholar] [CrossRef]
- Mertens, J.; Pollier, J.; Bossche, R.V.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Goossens, A. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016, 170, 194–210. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-N.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticult. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Chi, C.; Jin, L.J.; Zhu, J.; Yu, J.Q.; Zhou, Y.H. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Sun, T.-H.; Zhao, L.; Pan, X.-W.; Lu, S. The bZIP transcription factor HY5 interacts with the promoter of the monoterpene synthase gene QH6 in modulating its rhythmic expression. Front. Plant Sci. 2015, 6, 304. [Google Scholar] [CrossRef]
- Miyamoto, K.; Matsumoto, T.; Okada, A.; Komiyama, K.; Chujo, T.; Yoshikawa, H.; Nojiri, H.; Yamane, H.; Okada, K. Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor-induced hyperaccumulation of diterpenoid phytoalexins in rice cells. PLoS ONE 2014, 9, e105823. [Google Scholar] [CrossRef]
- Okada, A.; Okada, K.; Miyamoto, K.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. OsTGAP1, a bZIP Transcription Factor, Coordinately Regulates the Inductive Production of Diterpenoid Phytoalexins in Rice. J. Biol. Chem. 2009, 284, 26510–26518. [Google Scholar] [CrossRef]
- Yoshida, Y.; Miyamoto, K.; Yamane, H.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Okada, K. OsTGAP1 is responsible for JA-inducible diterpenoid phytoalexin biosynthesis in rice roots with biological impacts on allelopathic interaction. Physiol. Plant. 2017, 161, 532–544. [Google Scholar] [CrossRef]
- Miyamoto, K.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Yamane, H.; Okada, K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2015, 173, 19–27. [Google Scholar] [CrossRef]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef]
- Frerigmann, H.; Piślewska-Bednarek, M.; Sánchez-Vallet, A.; Molina, A.; Glawischnig, E.; Gigolashvili, T.; Bednarek, P. Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 2016, 9, 682–695. [Google Scholar] [CrossRef]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3, 7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 2014, 65, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ran, L.; Hou, Y.; Tian, Q.; Li, C.; Liu, R.; Fan, D.; Luo, K. The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. N. Phytol. 2017, 215, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Vialet, S.; Guiraud, J.L.; Torregrosa, L.; Bertrand, Y.; Cheynier, V.; This, P.; Terrier, N. A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. N. Phytol. 2014, 201, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Sun, T.; Pan, Q.; Anupol, N.; Chen, H.; Shi, J.; Liu, F.; Deqiang, D.; Wang, C.; Zhao, J. RrMYB5-and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 2019, 17, 2078–2095. [Google Scholar] [CrossRef]
- Liu, C.; Long, J.; Zhu, K.; Liu, L.; Yang, W.; Zhang, H.; Li, L.; Xu, Q.; Deng, X. Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 2016, 6, 25352. [Google Scholar] [CrossRef]
- Misra, P.; Pandey, A.; Tiwari, M.; Chandrashekar, K.; Sidhu, O.P.; Asif, M.H.; Chakrabarty, D.; Singh, P.K.; Trivedi, P.K.; Nath, P.; et al. Modulation of Transcriptome and Metabolome of Tobacco by Arabidopsis Transcription Factor, AtMYB12, Leads to Insect Resistance. Plant Physiol. 2010, 152, 2258–2268. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Khan, M.P.; Swarnkar, G.; Tewari, M.C.; Bhambhani, S.; Trivedi, R.; Chattopadhyay, N.; Trivedi, P.K. Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol. J. 2014, 12, 69–80. [Google Scholar] [CrossRef]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Wang, S.; Ning, J.; Ding, X.; Chu, Z. AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco. Plant Cell Tissue Organ Culture 2015, 122, 309–319. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Bhambhani, S.; Bhatia, C.; Trivedi, P.K. Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014, 4, 5018. [Google Scholar] [CrossRef]
- Yuan, Y.; Qi, L.; Yang, J.; Wu, C.; Liu, Y.; Huang, L. A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 120, 961–972. [Google Scholar] [CrossRef]
- Wang, W.-L.; Wang, Y.-X.; Li, H.; Liu, Z.-W.; Cui, X.; Zhuang, J. Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2018, 18, 288. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Seo, S.; Takahashi, A.; Hirochika, H. The MAMP-Responsive MYB Transcription Factors MYB30, MYB55 and MYB110 Activate the HCAA Synthesis Pathway and Enhance Immunity in Rice. Plant Cell Physiol. 2018, 59, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Fu, B.; Sun, P.; Xiao, C.; Liu, J.-H. A NAC transcription factor represses putrescine biosynthesis and affects drought tolerance. Plant Physiol. 2016, 172, 1532–1547. [Google Scholar] [CrossRef]
- Dalman, K.; Wind, J.J.; Nemesio-Gorriz, M.; Hammerbacher, A.; Lundén, K.; Ezcurra, I.; Elfstrand, M. Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Cao, Y.; Zhai, J.; Wang, Q.; Yuan, H.; Huang, X. Function of Hevea brasiliensis NAC1 in dehydration-induced laticifer differentiation and latex biosynthesis. Planta 2017, 245, 31–44. [Google Scholar] [CrossRef]
- Duan, M.; Zhang, R.; Zhu, F.; Zhang, Z.; Gou, L.; Wen, J.; Dong, J.; Wang, T. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 2017, 29, 1748–1772. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Outchkourov, N.S.; Carollo, C.A.; Gomez-Roldan, V.; de Vos, R.C.; Bosch, D.; Hall, R.D.; Beekwilder, J. Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves. Front. Plant Sci. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- Nemesio-Gorriz, M.; Blair, P.B.; Dalman, K.; Hammerbacher, A.; Arnerup, J.; Stenlid, J.; Mukhtar, S.M.; Elfstrand, M. Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway. Front. Plant Sci. 2017, 8, 305. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Lv, Z.; Lu, X.; Shen, Q.; Zhang, L.; Zhu, M.; Wang, G.; Sun, X.; Liao, Z. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 163–175. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Ji, A.; Luo, H.; Song, J. Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm. Sin. B 2018, 8, 295–305. [Google Scholar] [CrossRef]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef]
- Bednarek, P.; Osbourn, A. Plant-Microbe Interactions: Chemical Diversity in Plant Defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef]
- Nisha, S.N.; Prabu, G.; Mandal, A.K.A. Biochemical and molecular studies on the resistance mechanisms in tea [Camellia sinensis (L.) O. Kuntze] against blister blight disease. Physiol. Mol. Biol. Plants 2018, 24, 867–880. [Google Scholar] [CrossRef]
- Li, B.; Fan, R.; Guo, S.; Wang, P.; Zhu, X.; Fan, Y.; Chen, Y.; He, K.; Kumar, A.; Shi, J. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 2019, 166, 103807. [Google Scholar] [CrossRef]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- McGrann, G.R.; Steed, A.; Burt, C.; Goddard, R.; Lachaux, C.; Bansal, A.; Corbitt, M.; Gorniak, K.; Nicholson, P.; Brown, J.K. Contribution of the drought tolerance-related Stress-responsive NAC 1 transcription factor to resistance of barley to R amularia leaf spot. Mol. Plant Pathol. 2015, 16, 201–209. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Danielsson, M.; Lundén, K.; Elfstrand, M.; Hu, J.; Zhao, T.; Arnerup, J.; Ihrmark, K.; Swedjemark, G.; Borg-Karlson, A.-K.; Stenlid, J. Chemical and transcriptional responses of Norway spruce genotypes with different susceptibility to Heterobasidion spp. infection. BMC Plant Biol. 2011, 11, 154. [Google Scholar] [CrossRef]
- Tian, W.-M.; Yang, S.-G.; Shi, M.-J.; Zhang, S.-X.; Wu, J.-L. Mechanical wounding-induced laticifer differentiation in rubber tree: An indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol. 2015, 182, 95–103. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.H.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Kaplan, F.; Huffaker, A.; Dafoe, N.J.; Vaughan, M.M.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 5455–5460. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.M.; Christensen, S.; Schmelz, E.A.; Huffaker, A.; McAuslane, H.J.; Alborn, H.T.; Romero, M.; Allen, L.H.; Teal, P.E. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 2015, 38, 2195–2207. [Google Scholar] [CrossRef] [PubMed]

| Families | Transcriptional Factors | Plant Species | Metabolites | Resistance | References |
|---|---|---|---|---|---|
| AP2/ERF | JRE4 (GAME9) | S. lycopersicum, S. tuberosum | SGAs | S. litura | [47,51] |
| NtERF32 | N. tabaccum | Nicotine | Toxic against herbivory | [56] | |
| GbERF1 | G. barbadense | Lignin | V. dahliae | [60] | |
| ORA59 | A. thaliana | HCAAs | A. brassicicola, B. cineria | [63] | |
| TcERF12/ TcERF15 | T. chinensis | Taxol | P. capsici | [62] | |
| VqERF114 | V. quinquangularis | Resveratrol | B. cineria | [65] | |
| EREB58 | Z. mays | Sesquiterpenes | Defence against herbivory | [66] | |
| PnERF1 | P. notoginseng | Saponins | Anti-microbial | [59] | |
| WRKY | StWRKY1 | S. tuberosum | HCAAs | Anti-microbial | [67] |
| StWRKY8 | S. tuberosum | BIAs | P. infestance | [68] | |
| ZmWRKY79 | Z. mays | Terpenoid phytoalexins | Anti-microbial | [69] | |
| TcWRKY1 | T. chinenesis | Taxol | Anti-microbial | [70] | |
| WsWRKY1 | W. somnifera | Phytosterol | Bacteria, Fungi and Insect | [71] | |
| TaWRKY70 | T. aestivum | HCAAs | Fungi | [72] | |
| SsWRKY18/ 40 | S. sclarea | Diterpenoids | Bacteria and Fungi | [73] | |
| VviWRKY24/03/VvWRKY8 | V. vinifera | Resveratrol | B. cineria | [74,75] | |
| HvWRKY23 | H. vulgare | HCAAs | Fusarium head blight | [76] | |
| bHLH | ILR3/bHLH104, bHLH04/05/06 | A. thaliana | GLs | H. schachtii | [77,78] |
| VvbHLH1 | A. thaliana | Flavoniods | Drought and Salt | [79] | |
| MdMYC2 | M. domestica | Anthocyanin | Anti-pathogenic, Drought and Salinity | [80] | |
| DPF | O. sativa | Diterpenoid phytoalexins | Anti-Pathogenic | [81] | |
| TSAR1/TSAR2 | M. falcata | Saponins | Anti-microbial | [82] | |
| bZIP | MdHY5 | M. domestica | Anthocyanin | Drought, Pathogen and Salinity | [83] |
| SlHY5 | S. lycopersicum | AnthocyaninMonoterpenoids | Drought, Pathogen and SalinityAnti-pathogenic | [84,85] | |
| OsTGAP1 | O. sativa | Diterpenoid phytoalexins | Anti-pathogenic | [86,87,88] | |
| OsbZIP79 | O. sativa | Diterpenoid phytoalexins | Anti-pathogenic | [89] | |
| MYB | AtMYB34/51/112 | A. thaliana | IGS | Plectospharella cucumerina | [90,91] |
| AtMYB75 | A. thaliana | Anthocyanin | Pieris brassicae | [92] | |
| PtMYB115 | P. tomentosa | Proanthocyanin | Dothiorella gregaria | [93] | |
| VvMYBC2-L1 | V. vinifera | Proanthocyanin | Wounding and Oxidative stress | [94] | |
| VvMYB14/VviMYB14 | V. vinifera | Resveratrol | B. cineria | [74,75] | |
| RrMYB5/ RrMYB10 | R. rugosa | Proanthocyanin | Wounding and Oxidative stress | [95] | |
| CsMYBF1 | Citrus sinensis | Flavonoids and HCAAs | Antimicrobial | [96] | |
| AtMYB11/12/111 | A. thaliana | Flavonoids | Antimicrobial, Salinity | [97,98,99,100,101,102] | |
| SbMYB8 | S. baicalensis | Flavonoids | Drought | [103] | |
| CsMYB2/26 | C. sinensis | Flavonoids | Blister Blight | [104] | |
| OsMYB30/55/110 | O. sativa | HCAAs | Fungi and Bacteria | [105] | |
| NAC | PtrNAC72 | P. trifoliata | Putriscene | Drought | [106] |
| PaNAC03 | P. abies | Flavonoid | H. annosum | [107] | |
| ANACO32 | A. thaliana | Anthocyanin | Drought and Salinity | [108] | |
| HbNAC1 | H. brasiliensis | Latex | Drought | [109] | |
| MfNACsa | M. falcata | Glutathione | Drought | [110] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes 2020, 11, 346. https://doi.org/10.3390/genes11040346
Meraj TA, Fu J, Raza MA, Zhu C, Shen Q, Xu D, Wang Q. Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes. 2020; 11(4):346. https://doi.org/10.3390/genes11040346
Chicago/Turabian StyleMeraj, Tehseen Ahmad, Jingye Fu, Muhammad Ali Raza, Chenying Zhu, Qinqin Shen, Dongbei Xu, and Qiang Wang. 2020. "Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism" Genes 11, no. 4: 346. https://doi.org/10.3390/genes11040346
APA StyleMeraj, T. A., Fu, J., Raza, M. A., Zhu, C., Shen, Q., Xu, D., & Wang, Q. (2020). Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes, 11(4), 346. https://doi.org/10.3390/genes11040346





