Arabidopsis thaliana MLK3, a Plant-Specific Casein Kinase 1, Negatively Regulates Flowering and Phosphorylates Histone H3 In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plasmid Constructs and Plant Transformation
2.3. In Vitro Protein Kinase Activity Assay
2.4. mRNA Sequencing and Data Analysis
2.5. Immunoblot Analysis
2.6. Subcellular Localization and Nuclear Staining
3. Results
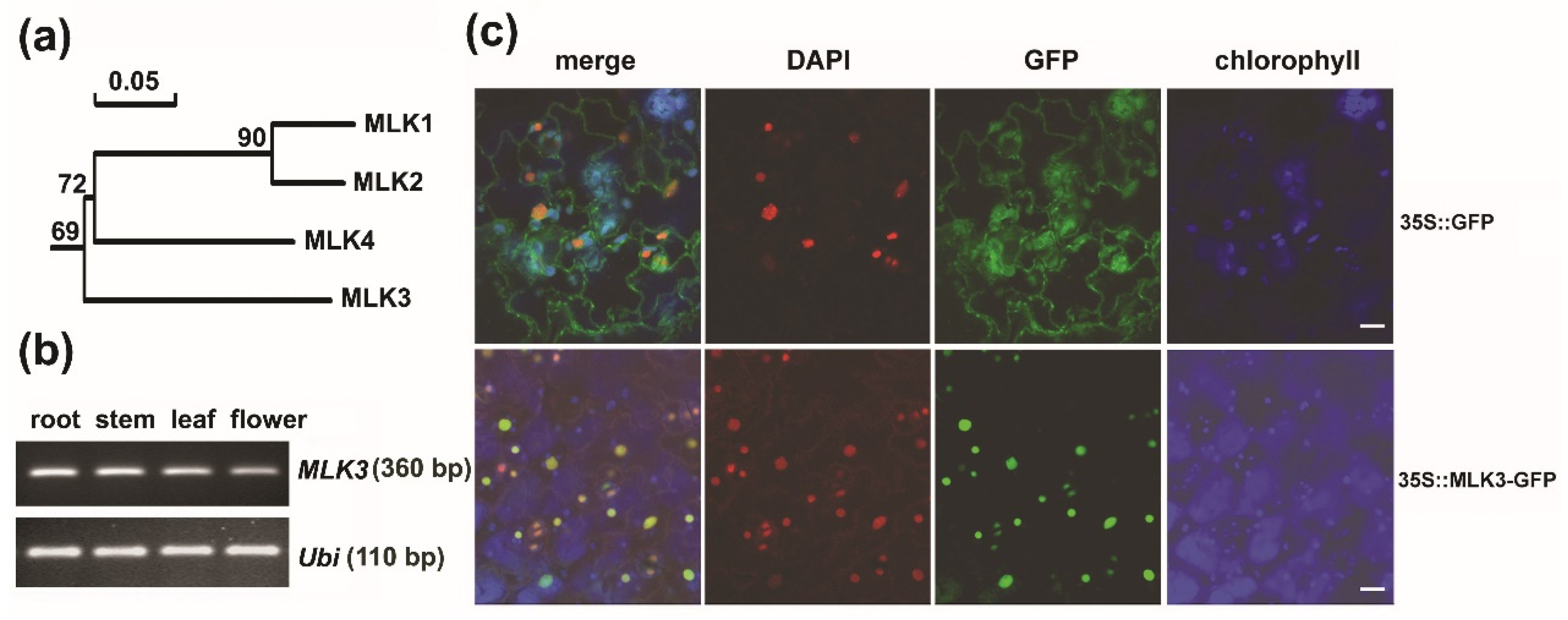
3.1. MLK3 Belonged to a Subgroup Divergent from Its Paralogs
3.2. MLK3 Was a Nuclear Protein and MLK3 Was Expressed Ubiquitously
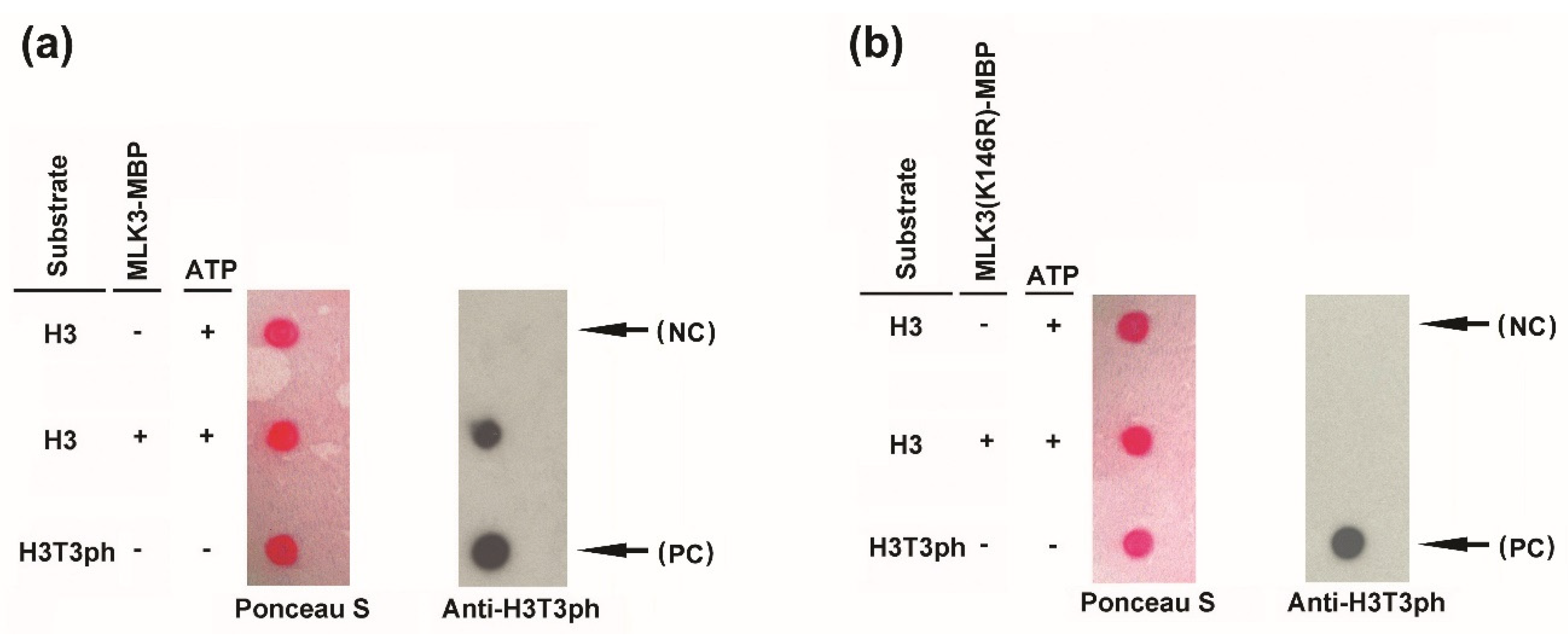
3.3. MLK3 Phosphorylated Histone H3 and an Intact Lysine (K146) Was Required for In Vitro Activity
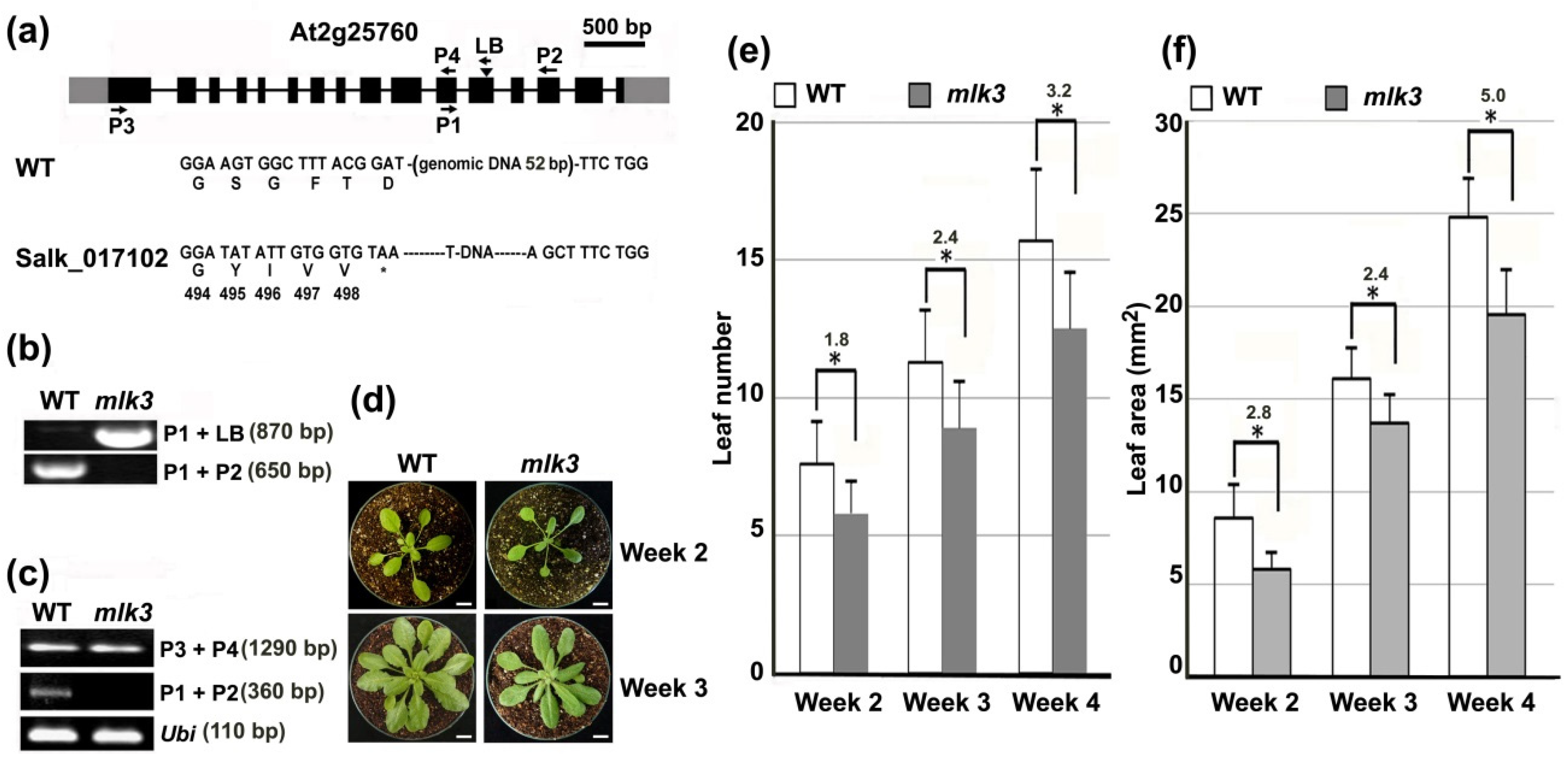
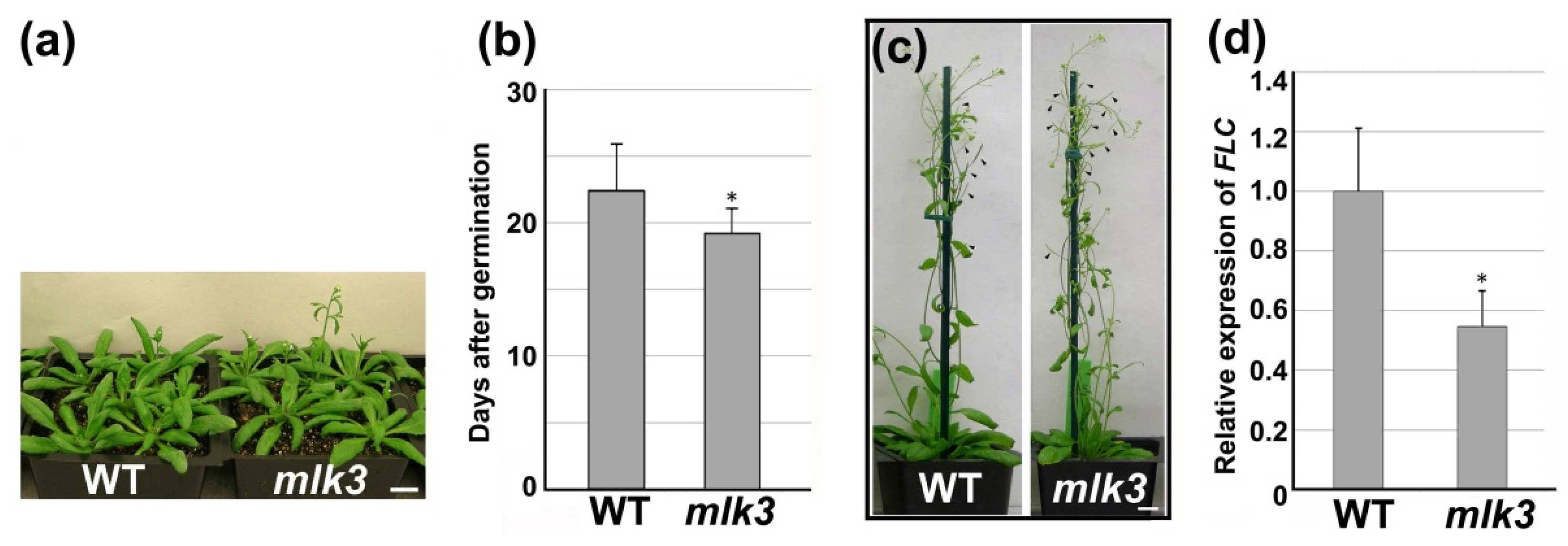
3.4. MLK3 Affected Leaf Growth and Flowering Time
3.5. Disruption of MLK3 Repressed the Expression of FLC
3.6. The Negative Role of MLK3 in Flowering Required the Intact Lysine (K) 146
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Lohler, J.; Stoter, M. The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005, 17, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, H.; Casas-Mollano, J.A. Histone H3 phosphorylation: Universal code or lineage specific dialects? Epigenetics 2009, 4, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef]
- Kang, J.M.; Wang, Z. Mut9p-LIKE KINASE family members: New roles of the plant-specific casein kinase I in plant growth and development. Int. J. Mol. Sci. 2020, 21, 1562. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.N.; Mizutani, Y.; Kuwata, K.; Hirota, T.; Sato, A.; Mizoi, J.; Takao, S.; Matsuo, H.; Suzuki, T.; Ito, S.; et al. Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 11528–11536. [Google Scholar] [CrossRef]
- Wang, Z.; Casas-Mollano, J.A.; Xu, J.P.; Riethoven, J.J.; Zhang, C.; Cerutti, H. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8487–8492. [Google Scholar] [CrossRef] [PubMed]
- Casas-Mollano, J.A.; Jeong, B.R.; Xu, J.; Moriyama, H.; Cerutti, H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 6486–6491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Q.; Wang, X.; Zuo, Z.; Oka, Y.; Lin, C. New insights into the mechanisms of phytochrome-cryptochrome coaction. New Phytol. 2018, 217, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Q.; Deng, W.; Wang, X.; Piao, M.; Cai, D.; Li, Y.; Barshop, W.D.; Yu, X.; Zhou, T.; et al. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 2017, 8, 15234. [Google Scholar] [CrossRef]
- Huang, H.; Alvarez, S.; Bindbeutel, R.; Shen, Z.; Naldrett, M.J.; Evans, B.S.; Briggs, S.P.; Hicks, L.M.; Kay, S.A.; Nusinow, D.A. Identification of Evening Complex Associated Proteins in Arabidopsis by Affinity Purification and Mass Spectrometry. Mol. Cell. Proteom. 2016, 15, 201–217. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; Gonzalez-Grandio, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Qu, L.; Xu, Z.H.; Zhu, J.K.; Xue, H.W. EL1-like Casein Kinases Suppress ABA Signaling and Responses by Phosphorylating and Destabilizing the ABA Receptors PYR/PYLs in Arabidopsis. Mol. Plant 2018, 11, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Wirthmueller, L.; Asai, S.; Rallapalli, G.; Sklenar, J.; Fabro, G.; Kim, D.S.; Lintermann, R.; Jaspers, P.; Wrzaczek, M.; Kangasjarvi, J.; et al. Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to radical-induced cell death1. New Phytol. 2018, 220, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Xue, H.W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.T.; Koo, B.H.; Kim, D.; Yoo, S.C.; Paek, N.C. Casein kinases I and 2alpha phosphorylate oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol. Cells 2015, 38, 81–88. [Google Scholar]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013, 76, 36–46. [Google Scholar] [CrossRef]
- Su, Y.; Wang, S.; Zhang, F.; Zheng, H.; Liu, Y.; Huang, T.; Ding, Y. Phosphorylation of Histone H2A at Serine 95: A Plant-Specific Mark Involved in Flowering Time Regulation and H2A.Z Deposition. Plant Cell 2017, 29, 2197–2213. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–570. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Van Dijk, K.; Marley, K.E.; Jeong, B.R.; Xu, J.; Hesson, J.; Cerny, R.L.; Waterborg, J.H.; Cerutti, H. Monomethyl histone H3 lysine 4 as an epigenetic mark for silenced euchromatin in Chlamydomonas. Plant Cell 2005, 17, 2439–2453. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, F.; Wang, S.; Su, Y.; Jiang, P.; Cheng, R.; Ji, X.; Hou, S.; Ding, Y. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 2017, 30, 67–82. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; McKay, R.M.; McKay, J.P.; Graff, J.M. Casein kinase I transduces Wnt signals. Nature 1999, 401, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Fujiwara, S.; Kamada, H.; Coupland, G.; Mizoguchi, T. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 2004, 557, 259–264. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.; Cui, H.; Jia, S.; Liu, W.; Yu, R.; Wu, Z.; Wang, Z. Arabidopsis thaliana MLK3, a Plant-Specific Casein Kinase 1, Negatively Regulates Flowering and Phosphorylates Histone H3 In Vitro. Genes 2020, 11, 345. https://doi.org/10.3390/genes11030345
Kang J, Cui H, Jia S, Liu W, Yu R, Wu Z, Wang Z. Arabidopsis thaliana MLK3, a Plant-Specific Casein Kinase 1, Negatively Regulates Flowering and Phosphorylates Histone H3 In Vitro. Genes. 2020; 11(3):345. https://doi.org/10.3390/genes11030345
Chicago/Turabian StyleKang, Junmei, Huiting Cui, Shangang Jia, Wenwen Liu, Renjie Yu, Zhihai Wu, and Zhen Wang. 2020. "Arabidopsis thaliana MLK3, a Plant-Specific Casein Kinase 1, Negatively Regulates Flowering and Phosphorylates Histone H3 In Vitro" Genes 11, no. 3: 345. https://doi.org/10.3390/genes11030345
APA StyleKang, J., Cui, H., Jia, S., Liu, W., Yu, R., Wu, Z., & Wang, Z. (2020). Arabidopsis thaliana MLK3, a Plant-Specific Casein Kinase 1, Negatively Regulates Flowering and Phosphorylates Histone H3 In Vitro. Genes, 11(3), 345. https://doi.org/10.3390/genes11030345




