Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family
Abstract
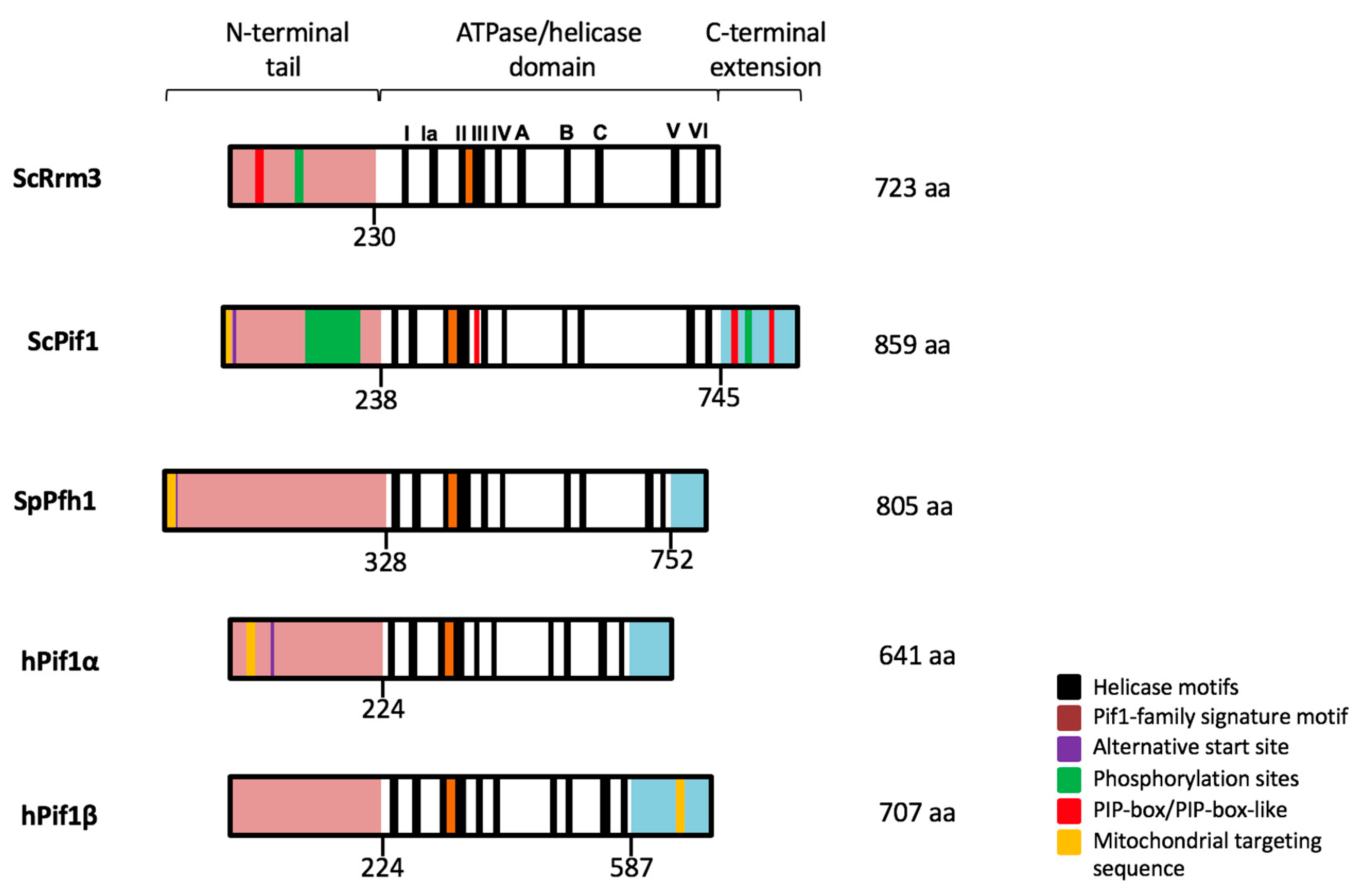
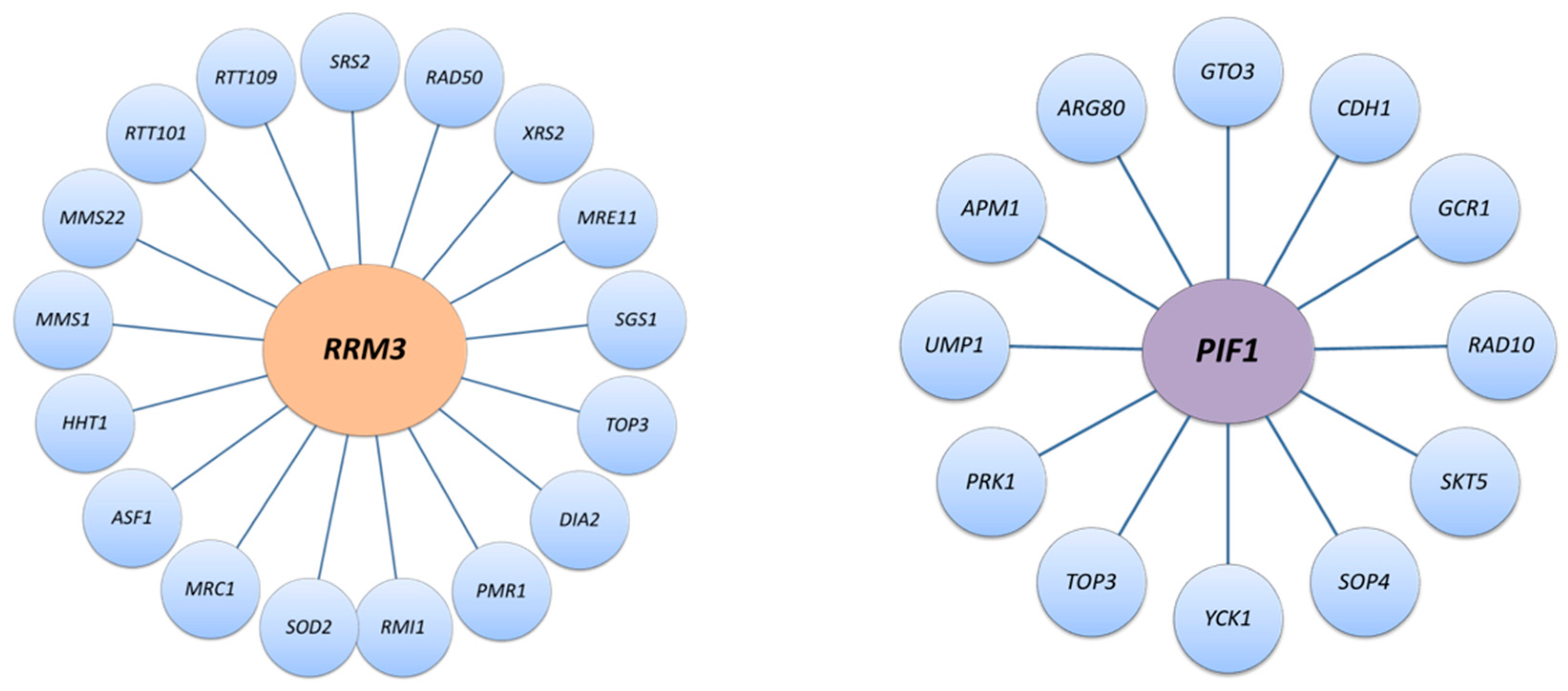
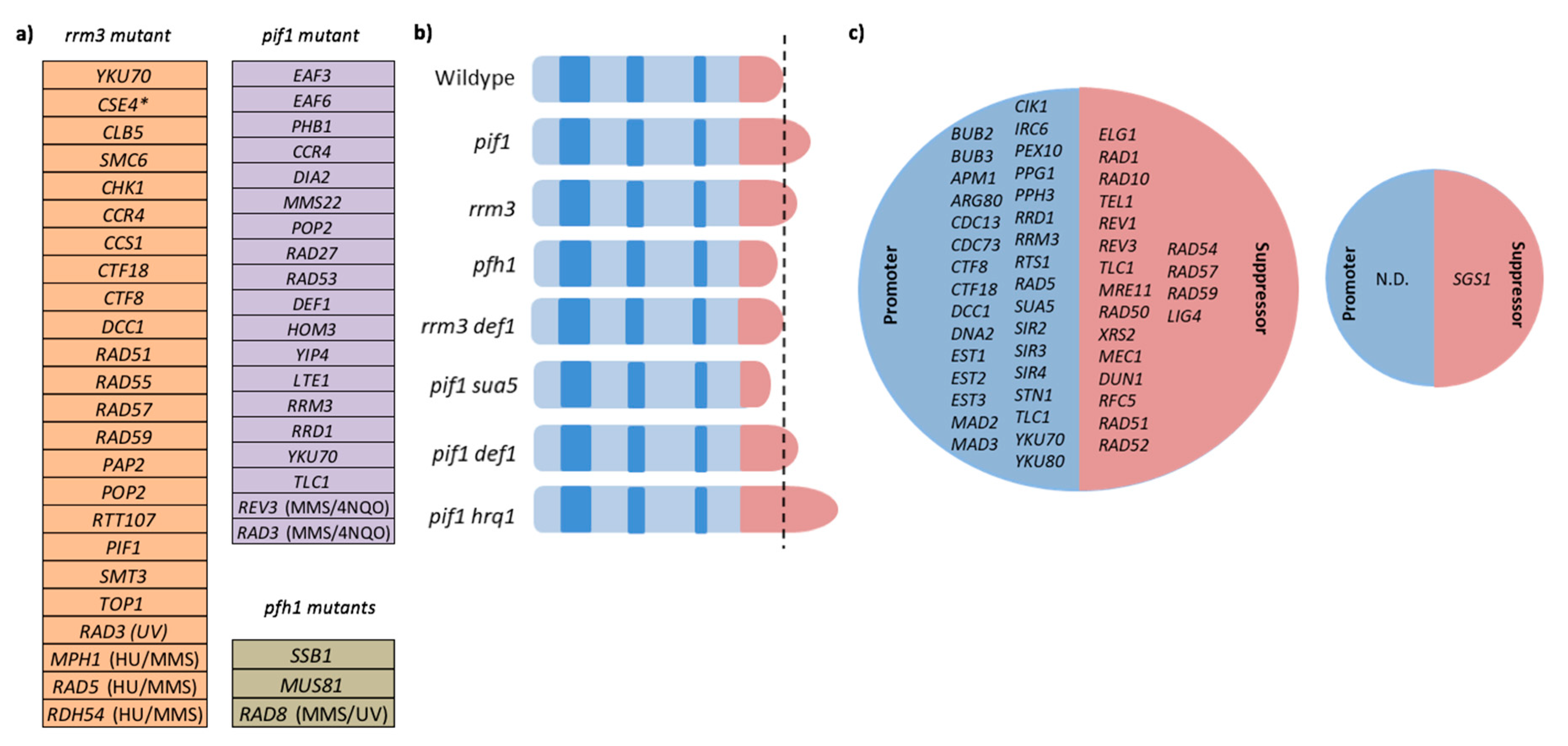
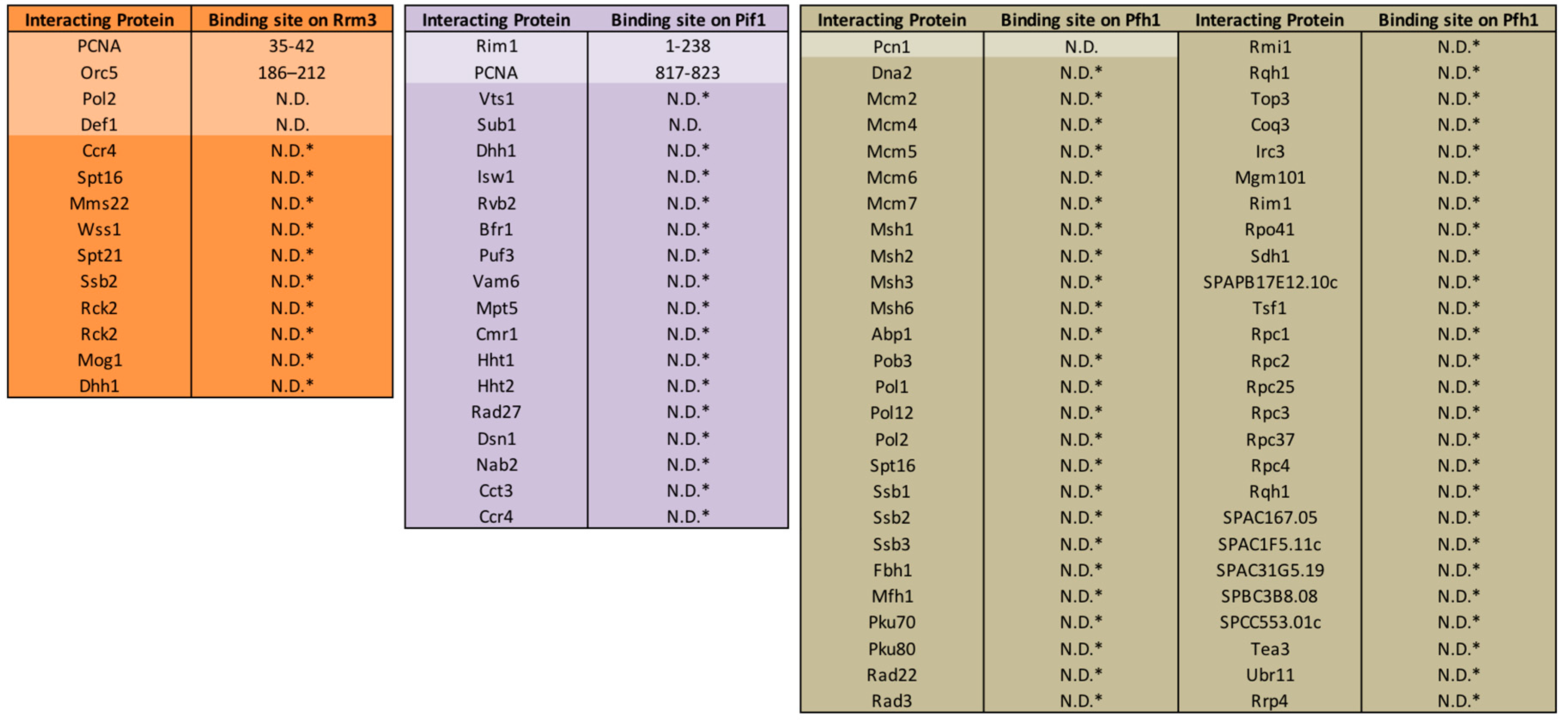
1. History of the PIF1 DNA Helicase Family
2. Replication Through the rDNA Replication Fork Barrier
3. Termination of DNA Replication
4. Replication Through transfer RNA (tRNA) Genes
5. Telomere Length Maintenance
6. Okazaki Fragment Maturation
7. Resolution of G4 Structures
8. Cellular Response to Replication Fork Stalling in the Absence of Rrm3
9. A Helicase-Independent Function of Rrm3 During Replication Stress
10. Protection of Mitochondrial DNA
11. Localization to Centromeres
12. DNA Break Repair
13. Regulation of Ty1 Transposition
14. Fragile Site Expression
15. Yeast as a Model System for the Functional Evaluation of hPif1 Mutations
16. Concluding Remarks
Funding
Conflicts of Interest
References
- Boule, J.B.; Zakian, V.A. Roles of Pif1-Like Helicases in the Maintenance of Genomic Stability. Nucleic Acids Res. 2006, 34, 4147–4153. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Judge, C.P.; Zakian, V.A. The Pif1 Family in Prokaryotes: What Are Our Helicases Doing in Your Bacteria? Mol. Biol. Cell 2011, 22, 1955–1959. [Google Scholar] [CrossRef] [PubMed]
- Foury, F.; Kolodynski, J. Pif Mutation Blocks Recombination between Mitochondrial Rho+ and Rho- Genomes Having Tandemly Arrayed Repeat Units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1983, 80, 5345–5349. [Google Scholar] [CrossRef] [PubMed]
- Bessler, J.B.; Zakian, V.A. The Pif1p Subfamily of Helicases: Region-Specific DNA Helicases? Trends Cell Biol. 2001, 11, 60–65. [Google Scholar] [CrossRef]
- Keil, R.L.; McWilliams, A.D. A Gene with Specific and Global Effects on Recombination of Sequences from Tandemly Repeated Genes in Saccharomyces cerevisiae. Genetics 1993, 135, 711–718. [Google Scholar]
- Enemark, E.J.; Joshua-Tor, L. On Helicases and Other Motor Proteins. Curr. Opin. Struct. Biol. 2008, 18, 243–257. [Google Scholar] [CrossRef]
- Lohman, T.M.; Tomko, E.J.; Wu, C.G. Non-Hexameric DNA Helicases and Translocases: Mechanisms and Regulation. Nat. Rev. Mol. Cell Biol. 2008, 9, 391–401. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Derry, K.L.; Kolodner, R.D. Saccharomyces cerevisiae Rrm3, a 5′ to 3′ DNA Helicase, Physically Interacts with Proliferating Cell Nuclear Antigen. J. Biol. Chem. 2002, 277, 45331–45337. [Google Scholar] [CrossRef]
- Rossi, S.E.; Ajazi, A.; Carotenuto, W.; Foiani, M.; Giannattasio, M. Rad53-Mediated Regulation of Rrm3 and Pif1 DNA Helicases Contributes to Prevention of Aberrant Fork Transitions under Replication Stress. Cell Rep. 2015, 13, 80–92. [Google Scholar] [CrossRef]
- Buzovetsky, O.; Kwon, Y.; Pham, T.N.; Kim, C.; Ira, G.; Sung, P.; Xiong, Y. Role of the Pif1-Pcna Complex in Pol δ-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication. Cell Rep. 2017, 21, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Dahan, D.; Tsirkas, I.; Dovrat, D.; Sparks, M.A.; Singh, S.P.; Galletto, R.; Aharoni, A. Pif1 Is Essential for Efficient Replisome Progression through Lagging Strand G-Quadruplex DNA Secondary Structures. Nucleic Acids Res. 2018, 46, 11847–11857. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Monson, E.K.; Teng, S.C.; Schulz, V.P.; Zakian, A.V. Pif1p Helicase, a Catalytic Inhibitor of Telomerase in Yeast. Science 2000, 289, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Reyes, A.; Duncan, A.L.; Rorbach, J.; Wood, S.R.; Brea-Calvo, G.; Gammage, P.A.; Robinson, A.J.; Minczuk, M.; Holt, I.J. Alternative Translation Initiation Augments the Human Mitochondrial Proteome. Nucleic Acids Res. 2013, 41, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Horiuchi, T. A Yeast Gene Product, Fob1 Protein, Required for Both Replication Fork Blocking and Recombinational Hotspot Activities. Genes Cells 1996, 1, 465–474. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Zhou, J.Q.; Zakian, A.V. The Saccharomyces Pif1p DNA Helicase and the Highly Related Rrm3p Have Opposite Effects on Replication Fork Progression in Ribosomal DNA. Cell 2000, 100, 479–489. [Google Scholar] [CrossRef]
- Sabouri, N.; McDonald, K.R.; Webb, C.J.; Cristea, I.M.; Zakian, A.V. DNA Replication through Hard-to-Replicate Sites, Including Both Highly Transcribed Rna Pol Ii and Pol Iii Genes, Requires the S. Pombe Pfh1 Helicase. Genes Dev. 2012, 26, 581–593. [Google Scholar] [CrossRef]
- Torres, J.Z.; Bessler, J.B.; Zakian, V.A. Local Chromatin Structure at the Ribosomal DNA Causes Replication Fork Pausing and Genome Instability in the Absence of the S. cerevisiae DNA Helicase Rrm3p. Genes Dev. 2004, 18, 498–503. [Google Scholar] [CrossRef]
- Syed, S.; Desler, C.; Rasmussen, J.L.; Schmidt, H.K. A Novel Rrm3 Function in Restricting DNA Replication Via an Orc5-Binding Domain Is Genetically Separable from Rrm3 Function as an ATPase/Helicase in Facilitating Fork Progression. PLoS Genet. 2016, 12, e1006451. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Bairwa, N.K.; Bastia, D. The Tof1p-Csm3p Protein Complex Counteracts the Rrm3p Helicase to Control Replication Termination of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2006, 103, 897–902. [Google Scholar] [CrossRef]
- Crickard, J.B.; Xue, C.; Wang, W.; Kwon, Y.; Sung, P.; Greene, E.C. The Recq Helicase Sgs1 Drives ATP-Dependent Disruption of Rad51 Filaments. Nucleic Acids Res. 2019, 47, 4694–4706. [Google Scholar] [CrossRef] [PubMed]
- Krejci, L.; Van Komen, S.; Li, Y.; Villemain, J.; Reddy, M.S.; Klein, H.; Ellenberger, T.; Sung, P. DNA Helicase Srs2 Disrupts the Rad51 Presynaptic Filament. Nature 2003, 423, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Calzada, A.; Hodgson, B.; Kanemaki, M.; Bueno, A.; Labib, K. Molecular Anatomy and Regulation of a Stable Replisome Eukaryotic DNA at a Paused Replication Fork. Genes Dev. 2005, 19, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.J.; Elledge, S.J. Mrc1 Is a Replication Fork Component Whose Phosphorylation in Response to DNA Replication Stress Activates Rad53. Genes Dev. 2003, 17, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.H.; Kolodner, R.D. Suppression of Spontaneous Genome Rearrangements in Yeast DNA Helicase Mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 18196–18201. [Google Scholar] [CrossRef]
- Szyjka, S.J.; Viggiani, C.J.; Aparicio, O.M. Mrc1 Is Required for Normal Progression of Replication Forks Throughout Chromatin in S. cerevisiae. Mol. Cell 2005, 19, 691–697. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Zhou, J.Q.; Schulz, V.P.; Monson, E.K.; Zakian, A.V. Saccharomyces Rrm3p, a 5′ to 3′ DNA Helicase That Promotes Replication Fork Progression through Telomeric and Subtelomeric DNA. Genes Dev. 2002, 16, 1383–1396. [Google Scholar] [CrossRef]
- Pinter, S.F.; Aubert, S.D.; Zakian, A.V. The Schizosaccharomyces Pombe Pfh1p DNA Helicase Is Essential for the Maintenance of Nuclear and Mitochondrial DNA. Mol. Cell. Biol. 2008, 28, 6594–6608. [Google Scholar] [CrossRef]
- Prokisch, H.; Scharfe, C.; Camp, D.G. Integrative Analysis of the Mitochondrial Proteome in Yeast. PLoS Biol. 2004, 2, e160. [Google Scholar] [CrossRef]
- Schulz, V.P.; Zakian, V.A. The Saccharomyces Pif1 DNA Helicase Inhibits Telomere Elongation and De Novo Telomere Formation. Cell 1994, 76, 145–155. [Google Scholar] [CrossRef]
- Boule, J.B.; Vega, L.R.; Zakian, A.V. The Yeast Pif1p Helicase Removes Telomerase from Telomeric DNA. Nature 2005, 438, 57–61. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.R.; Sabouri, N.; Webb, J.C.; Zakian, A.V. The Pif1 Family Helicase Pfh1 Facilitates Telomere Replication and Has an Rpa-Dependent Role During Telomere Lengthening. DNA Repair 2014, 24, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Dunaway, S.; Ivessa, S.A. The Role of Pif1p, a DNA Helicase in Saccharomyces cerevisiae, in Maintaining Mitochondrial DNA. Mitochondrion 2007, 7, 211–222. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, A.V. The Saccharomyces cerevisiae Helicase Rrm3p Facilitates Replication Past Nonhistone Protein-DNA Complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Pohl, T.J.; Chen, C.F.; Chan, A.; Pott, S.; Zakian, A.V. Pif1 Family DNA Helicases Suppress R-Loop Mediated Genome Instability at Trna Genes. Nat. Commun. 2017, 8, 15025. [Google Scholar] [CrossRef] [PubMed]
- Osmundson, J.S.; Kumar, J.; Yeung, R.; Smith, J.D. Pif1-Family Helicases Cooperatively Suppress Widespread Replication-Fork Arrest at tRNA Genes. Nat. Struct. Mol. Biol. 2017, 24, 162–170. [Google Scholar] [CrossRef]
- Chen, C.F.; Pohl, T.J.; Pott, S.; Zakian, A.V. Two Pif1 Family DNA Helicases Cooperate in Centromere Replication and Segregation in Saccharomyces cerevisiae. Genetics 2019, 211, 105–119. [Google Scholar] [CrossRef]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, A.V. Highly Transcribed Rna Polymerase Ii Genes Are Impediments to Replication Fork Progression in Saccharomyces cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef]
- Paeschke, K.; Capra, J.A.; Zakian, A.V. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef]
- McDonald, K.R.; Guise, A.J.; Pourbozorgi-Langroudi, P.; Cristea, I.M.; Zakian, V.A.; Capra, J.A.; Sabouri, N. Pfh1 Is an Accessory Replicative Helicase That Interacts with the Replisome to Facilitate Fork Progression and Preserve Genome Integrity. PLoS Genet. 2016, 12, e1006238. [Google Scholar] [CrossRef]
- Wallgren, M.; Mohammad, J.B.; Yan, K.P.; Pourbozorgi-Langroudi, P.; Ebrahimi, M.; Sabouri, N. G-Rich Telomeric and Ribosomal DNA Sequences from the Fission Yeast Genome Form Stable G-Quadruplex DNA Structures in Vitro and Are Unwound by the Pfh1 DNA Helicase. Nucleic Acids Res. 2016, 44, 6213–6231. [Google Scholar] [CrossRef]
- Bedard, L.G.; Dronamraju, R.; Kerschner, L.J.; Hunter, O.G.; Axley, D.E.; Boyd, K.A.; Strahl, D.B.; Mosley, L.A. Quantitative Analysis of Dynamic Protein Interactions During Transcription Reveals a Role for Casein Kinase Ii in Polymerase-Associated Factor (Paf) Complex Phosphorylation and Regulation of Histone H2b Monoubiquitylation. J. Biol. Chem. 2016, 291, 13410–13420. [Google Scholar] [CrossRef]
- Ohya, T.; Arai, H.; Kubota, Y.; Shinagawa, H.; Hishida, T. A Sumo-Like Domain Protein, Esc2, Is Required for Genome Integrity and Sister Chromatid Cohesion in Saccharomyces cerevisiae. Genetics 2008, 180, 41–50. [Google Scholar] [CrossRef]
- Ouenzar, F.; Lalonde, M.; Laprade, H.; Morin, G.; Gallardo, F.; Tremblay-Belzile, S.; Chartrand, P. Cell Cycle-Dependent Spatial Segregation of Telomerase from Sites of DNA Damage. J. Cell Biol. 2017, 216, 2355–2371. [Google Scholar] [CrossRef]
- Dehghani-Tafti, S.; Levdikov, V.; Antson, A.A.; Bax, B.; Sanders, C.M. Structural and Functional Analysis of the Nucleotide and DNA Binding Activities of the Human Pif1 Helicase. Nucleic Acids Res. 2019, 47, 3208–3222. [Google Scholar] [CrossRef]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.P.; Foiani, M.; Nicolas, A. G-Quadruplex-Induced Instability During Leading-Strand Replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef]
- Sabouri, N.; Capra, J.A.; Zakian, A.V. The Essential Schizosaccharomyces Pombe Pfh1 DNA Helicase Promotes Fork Movement Past G-Quadruplex Motifs to Prevent DNA Damage. BMC Biol. 2014, 12, 101. [Google Scholar] [CrossRef]
- Budd, M.E.; Reis, C.C.; Smith, S.; Myung, K.; Campbell, L.J. Evidence Suggesting That Pif1 Helicase Functions in DNA Replication with the Dna2 Helicase/Nuclease and DNA Polymerase δ. Mol. Cell Biol. 2006, 26, 2490–2500. [Google Scholar] [CrossRef]
- Ryu, G.H.; Tanaka, H.; Kim, H.D.; Kim, H.J.; Bae, H.S.; Kwon, N.Y.; Rhee, S.J.; MacNeill, A.S.; Seo, S.Y. Genetic and Biochemical Analyses of Pfh1 DNA Helicase Function in Fission Yeast. Nucleic Acids Res. 2004, 32, 4205–4216. [Google Scholar] [CrossRef][Green Version]
- Hiraga, S.; Botsios, S.; Donaldson, D.A. Histone H3 Lysine 56 Acetylation by Rtt109 Is Crucial for Chromosome Positioning. J. Cell Biol. 2008, 183, 641–651. [Google Scholar] [CrossRef]
- Munoz-Galvan, S.; Garcia-Rubio, M.; Ortega, P.; Ruiz, F.J.; Jimeno, S.; Pardo, B.; Gomez-Gonzalez, B.; Aguilera, A. A New Role for Rrm3 in Repair of Replication-Born DNA Breakage by Sister Chromatid Recombination. PLoS Genet. 2017, 13, e1006781. [Google Scholar] [CrossRef]
- Wilson, M.A.; Kwon, Y.; Xu, Y.; Chung, H.W.; Chi, P.; Niu, H.; Mayle, R.; Chen, X.; Malkova, A.; Sung, P.; et al. Pif1 Helicase and Pol δ Promote Recombination-Coupled DNA Synthesis Via Bubble Migration. Nature 2013, 502, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Baxter, J.; Bazán, M.A.O.; Yeeles, J.T.; Labib, K.P. Pif1-Family Helicases Support Fork Convergence During DNA Replication Termination in Eukaryotes. Mol. Cell 2019, 74, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Steinacher, R.; Osman, F.; Dalgaard, J.Z.; Lorenz, A.; Whitby, M.C. The DNA Helicase Pfh1 Promotes Fork Merging at Replication Termination Sites to Ensure Genome Stability. Genes Dev. 2012, 26, 594–602. [Google Scholar] [CrossRef]
- Garcia-Rodriguez, N.; Wong, R.P.; Ulrich, D.H. The Helicase Pif1 Functions in the Template Switching Pathway of DNA Damage Bypass. Nucleic Acids Res. 2018, 46, 8347–8356. [Google Scholar] [CrossRef]
- Scholes, D.T.; Banerjee, M.; Bowen, B.; Curcio, J.M. Multiple Regulators of Ty1 Transposition in Saccharomyces cerevisiae Have Conserved Roles in Genome Maintenance. Genetics 2001, 159, 1449–1465. [Google Scholar]
- Stamenova, R.; Maxwell, P.H.; Kenny, A.E.; Curcio, J.M. Rrm3 Protects the Saccharomyces cerevisiae Genome from Instability at Nascent Sites of Retrotransposition. Genetics 2009, 182, 711–723. [Google Scholar] [CrossRef]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and Mms21-Mediated Sumoylation Counteracts Recombinogenic Events at Damaged Replication Forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef]
- Bessler, J.B.; Zakian, V.A. The Amino Terminus of the Saccharomyces cerevisiae DNA Helicase Rrm3p Modulates Protein Function Altering Replication and Checkpoint Activity. Genetics 2004, 168, 1205–1218. [Google Scholar] [CrossRef]
- Xu, H.; Boone, C.; Klein, L.H. Mrc1 Is Required for Sister Chromatid Cohesion to Aid in Recombination Repair of Spontaneous Damage. Mol. Cell Biol. 2004, 24, 7082–7090. [Google Scholar] [CrossRef][Green Version]
- Weitao, T.; Budd, M.; Hoopes, L.L.; Campbell, L.J. Dna2 Helicase/Nuclease Causes Replicative Fork Stalling and Double-Strand Breaks in the Ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 22513–22522. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.Z.; Schnakenberg, S.L.; Zakian, V.A. Saccharomyces cerevisiae Rrm3p DNA Helicase Promotes Genome Integrity by Preventing Replication Fork Stalling: Viability of Rrm3 Cells Requires the Intra-S-Phase Checkpoint and Fork Restart Activities. Mol. Cell Biol. 2004, 24, 3198–3212. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.; Evangelista, M.; Parsons, B.A.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic Genetic Analysis with Ordered Arrays of Yeast Deletion Mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef]
- Chang, M.; Bellaoui, M.; Zhang, C.; Desai, R.; Morozov, P.; Delgado-Cruzata, L.; Rothstein, R.; Freyer, G.A.; Boone, C.; Brown, G.W. Rmi1/Nce4, a Suppressor of Genome Instability, Encodes a Member of the Recq Helicase/Topo Iii Complex. EMBO J. 2005, 24, 2024–2033. [Google Scholar] [CrossRef]
- Keogh, M.C.; Kim, J.A.; Downey, M.; Fillingham, J.; Chowdhury, D.; Harrison, J.C.; Onishi, M.; Datta, N.; Galicia, S.; Emili, A.; et al. A Phosphatase Complex That Dephosphorylates Gammah2ax Regulates DNA Damage Checkpoint Recovery. Nature 2006, 439, 497–501. [Google Scholar] [CrossRef]
- Luciano, P.; Dehe, P.M.; Audebert, S.; Geli, V.; Corda, Y. Replisome Function During Replicative Stress Is Modulated by Histone H3 Lysine 56 Acetylation through Ctf4. Genetics 2015, 199, 1047–1063. [Google Scholar] [CrossRef]
- Mirzaei, H.; Syed, S.; Kennedy, J.; Schmidt, H.K. Sgs1 Truncations Induce Genome Rearrangements but Suppress Detrimental Effects of Blm Overexpression in Saccharomyces cerevisiae. J. Mol. Biol. 2011, 405, 877–891. [Google Scholar] [CrossRef]
- Morohashi, H.; Maculins, T.; Labib, K. The Amino-Terminal Tpr Domain of Dia2 Tethers Scf(Dia2) to the Replisome Progression Complex. Curr. Biol. 2009, 19, 1943–1949. [Google Scholar] [CrossRef]
- Pan, X.; Ye, P.; Yuan, S.D.; Wang, X.; Bader, J.S.; Boeke, J.D. A DNA Integrity Network in the Yeast Saccharomyces cerevisiae. Cell 2006, 124, 1069–1081. [Google Scholar] [CrossRef]
- Putnam, C.D.; Hayes, T.K.; Kolodner, D.R. Post-Replication Repair Suppresses Duplication-Mediated Genome Instability. PLoS Genet. 2010, 6, e1000933. [Google Scholar] [CrossRef]
- Sacher, M.; Pfander, B.; Hoege, C.; Jentsch, S. Control of Rad52 Recombination Activity by Double-Strand Break-Induced Sumo Modification. Nat. Cell Biol. 2006, 8, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Deshpande, R.A.; Budd, M.; Campbell, J.; Revere, A.; Zhang, X.; Schmidt, K.H.; Paull, T.T. Genetic Separation of Sae2 Nuclease Activity from Mre11 Nuclease Functions in Budding Yeast. Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Smith, S.; Myung, K. Suppression of Gross Chromosomal Rearrangements by Yku70-Yku80 Heterodimer through DNA Damage Checkpoints. Proc. Natl. Acad. Sci. USA 2006, 103, 1816–1821. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, D.; Villa, M.; Gobbini, E.; Cassani, C.; Tedeschi, G.; Longhese, M.P. Escape of Sgs1 from Rad9 Inhibition Reduces the Requirement for Sae2 and Functional Mrx in DNA End Resection. EMBO Rep. 2015, 16, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Lydall, D. Pif1- and Exo1-Dependent Nucleases Coordinate Checkpoint Activation Following Telomere Uncapping. EMBO J. 2010, 29, 4020–4034. [Google Scholar] [CrossRef]
- Garbacz, M.A.; Lujan, S.A.; Burkholder, A.B.; Cox, P.B.; Wu, Q.; Zhou, X.Z.; Haber, E.J.; Kunkel, A.T. Evidence That DNA Polymerase δ Contributes to Initiating Leading Strand DNA Replication in Saccharomyces cerevisiae. Nat. Commun. 2018, 9, 858. [Google Scholar] [CrossRef]
- Stundon, J.L.; Zakian, V.A. Identification of Saccharomyces cerevisiae Genes Whose Deletion Causes Synthetic Effects in Cells with Reduced Levels of the Nuclear Pif1 DNA Helicase. G3 Genes Genomes Genet. 2015, 5, 2913–2918. [Google Scholar]
- Vega, L.R.; Phillips, J.A.; Thornton, B.R.; Benanti, J.A.; Onigbanjo, M.T.; Toczyski, D.P.; Zakian, A.V. Sensitivity of Yeast Strains with Long G-Tails to Levels of Telomere-Bound Telomerase. PLoS Genet. 2007, 3, e105. [Google Scholar] [CrossRef]
- Wagner, M.; Price, G.; Rothstein, R. The Absence of Top3 Reveals an Interaction between the Sgs1 and Pif1 DNA Helicases in Saccharomyces cerevisiae. Genetics 2006, 174, 555–573. [Google Scholar] [CrossRef]
- Gibson, D.G.; Aparicio, J.G.; Hu, F.; Aparicio, M.O. Diminished S-Phase Cyclin-Dependent Kinase Function Elicits Vital Rad53-Dependent Checkpoint Responses in Saccharomyces cerevisiae. Mol. Cell Biol. 2004, 24, 10208–10222. [Google Scholar] [CrossRef]
- Menolfi, D.; Delamarre, A.; Lengronne, A.; Pasero, P.; Branzei, D. Essential Roles of the Smc5/6 Complex in Replication through Natural Pausing Sites and Endogenous DNA Damage Tolerance. Mol. Cell 2015, 60, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Ciftci-Yilmaz, S.; Au, W.C.; Mishra, P.K.; Eisenstatt, J.R.; Chang, J.; Dawson, R.A.; Zhu, I.; Rahman, M.; Bilke, S.; Costanzo, M.; et al. A Genome-Wide Screen Reveals a Role for the Hir Histone Chaperone Complex in Preventing Mislocalization of Budding Yeast Cenp-A. Genetics 2018, 210, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.H.; Kolodner, R.D. Requirement of Rrm3 Helicase for Repair of Spontaneous DNA Lesions in Cells Lacking Srs2 or Sgs1 Helicase. Mol. Cell Biol. 2004, 24, 3213–3226. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.J.; Gonzalez-Barrera, S.; Sunjevaric, I.; Alvaro, D.; Ciccone, S.; Wagner, M.; Rothstein, R. Selective Ploidy Ablation, a High-Throughput Plasmid Transfer Protocol, Identifies New Genes Affecting Topoisomerase I-Induced DNA Damage. Genome Res. 2011, 21, 477–486. [Google Scholar] [CrossRef]
- Srikumar, T.; Lewicki, M.C.; Costanzo, M.; Tkach, M.J.; van Bakel, H.; Tsui, K.; Johnson, E.S.; Brown, G.W.; Andrews, B.J.; Boone, C.; et al. Global Analysis of Sumo Chain Function Reveals Multiple Roles in Chromatin Regulation. J. Cell Biol. 2013, 201, 145–163. [Google Scholar] [CrossRef]
- Addinall, S.G.; Holstein, E.M.; Lawless, C.; Yu, M.; Chapman, K.; Banks, A.P.; Ngo, H.P.; Maringele, L.; Taschuk, M.; Young, A.; et al. Quantitative Fitness Analysis Shows That Nmd Proteins and Many Other Protein Complexes Suppress or Enhance Distinct Telomere Cap Defects. PLoS Genet. 2011, 7, e1001362. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Qi, Y.; Lu, Y.J.; Pan, X.; Yuan, D.S.; Zhao, Y.; Bader, J.S.; Boeke, J.D. A Comprehensive Synthetic Genetic Interaction Network Governing Yeast Histone Acetylation and Deacetylation. Genes Dev. 2008, 22, 2062–2074. [Google Scholar] [CrossRef]
- Moriel-Carretero, M.; Aguilera, A. Postincision-Deficient Tfiih Causes Replication Fork Breakage and Uncovers Alternative Rad51- or Pol32-Mediated Restart Mechanisms. Mol. Cell 2010, 37, 690–701. [Google Scholar] [CrossRef]
- Osman, C.; Haag, M.; Potting, C.; Rodenfels, J.; Dip, P.V.; Wieland, F.T.; Brugger, B.; Westermann, B.; Langer, T. The Genetic Interactome of Prohibitins: Coordinated Control of Cardiolipin and Phosphatidylethanolamine by Conserved Regulators in Mitochondria. J. Cell Biol. 2009, 184, 583–596. [Google Scholar] [CrossRef]
- Ye, P.; Peyser, B.D.; Pan, X.; Boeke, D.J.; Spencer, A.F.; Bader, S.J. Gene Function Prediction from Congruent Synthetic Lethal Interactions in Yeast. Mol. Syst. Biol. 2005, 1, 2005.0026. [Google Scholar] [CrossRef]
- Zhang, W.; Durocher, D. De Novo Telomere Formation Is Suppressed by the Mec1-Dependent Inhibition of Cdc13 Accumulation at DNA Breaks. Genes Dev. 2010, 24, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Audry, J.; Maestroni, L.; Delagoutte, E.; Gauthier, T.; Nakamura, T.M.; Gachet, Y.; Saintome, C.; Geli, V.; Coulon, S. Rpa Prevents G-Rich Structure Formation at Lagging-Strand Telomeres to Allow Maintenance of Chromosome Ends. EMBO J. 2015, 34, 1942–1958. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.B.; Yang, C.P.; Li, R.X.; Zeng, R.; Zhou, Q.J. Def1p Is Involved in Telomere Maintenance in Budding Yeast. J. Biol. Chem. 2005, 280, 24784–24791. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.L.; Hu, Y.; Shen, N.; Tong, J.X.; Wang, J.; Ding, J.; Zhou, J.Q. Sua5p a Single-Stranded Telomeric DNA-Binding Protein Facilitates Telomere Replication. EMBO J. 2009, 28, 1466–1478. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Chan, A.; Zakian, A.V. Hrq1, a Homolog of the Human Recq4 Helicase, Acts Catalytically and Structurally to Promote Genome Integrity. Cell Rep. 2014, 6, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Adkins, C.J.; Cartwright, B.R.; Friedman, L.K. Yeast Est2p Affects Telomere Length by Influencing Association of Rap1p with Telomeric Chromatin. Mol. Cell Biol. 2008, 28, 2380–2390. [Google Scholar] [CrossRef][Green Version]
- Hwang, J.Y.; Smith, S.; Myung, K. The Rad1-Rad10 Complex Promotes the Production of Gross Chromosomal Rearrangements from Spontaneous DNA Damage in Saccharomyces cerevisiae. Genetics 2005, 169, 1927–1937. [Google Scholar] [CrossRef]
- Myung, K.; Smith, S.; Kolodner, D.R. Mitotic Checkpoint Function in the Formation of Gross Chromosomal Rearrangements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2004, 101, 15980–15985. [Google Scholar] [CrossRef]
- Nene, R.V.; Putnam, C.D.; Li, B.Z.; Nguyen, K.G.; Srivatsan, A.; Campbell, S.C.; Desai, A.; Kolodner, R.D. Cdc73 Suppresses Genome Instability by Mediating Telomere Homeostasis. PLoS Genet. 2018, 14, e1007170. [Google Scholar] [CrossRef]
- Banerjee, S.; Smith, S.; Oum, H.J.; Liaw, J.H.; Hwang, Y.J.; Sikdar, N.; Motegi, A.; Lee, S.E.; Myung, K. Mph1p Promotes Gross Chromosomal Rearrangement through Partial Inhibition of Homologous Recombination. J. Cell Biol. 2008, 181, 1083–1093. [Google Scholar] [CrossRef]
- Liang, J.; Li, B.Z.; Tan, A.P.; Kolodner, R.D.; Putnam, C.D.; Zhou, H. Sumo E3 Ligase Mms21 Prevents Spontaneous DNA Damage Induced Genome Rearrangements. PLoS Genet. 2018, 14, e1007250. [Google Scholar] [CrossRef] [PubMed]
- Putnam, C.D.; Pallis, K.; Hayes, K.T.; Kolodner, D.R. DNA Repair Pathway Selection Caused by Defects in Tel1, Sae2, De Novo Telomere Addition Generates Specific Chromosomal Rearrangement Signatures. PLoS Genet. 2014, 10, e1004277. [Google Scholar] [CrossRef] [PubMed]
- Sakofsky, C.J.; Ayyar, S.; Deem, K.A.; Chung, H.W.; Ira, G.; Malkova, A. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol. Cell 2015, 60, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Gupta, A.; Kolodner, D.R.; Myung, K. Suppression of Gross Chromosomal Rearrangements by the Multiple Functions of the Mre11-Rad50-Xrs2 Complex in Saccharomyces cerevisiae. DNA Repair 2005, 4, 606–617. [Google Scholar] [CrossRef]
- Myung, K.; Chen, C.; Kolodner, D.R. Multiple Pathways Cooperate in the Suppression of Genome Instability in Saccharomyces cerevisiae. Nature 2001, 411, 1073–1076. [Google Scholar] [CrossRef]
- Azvolinsky, A.; Dunaway, S.; Torres, Z.J.; Bessler, B.J.; Zakian, A.V. The S. cerevisiae Rrm3p DNA Helicase Moves with the Replication Fork and Affects Replication of All Yeast Chromosomes. Genes Dev. 2006, 20, 3104–3116. [Google Scholar] [CrossRef]
- Balakirev, M.Y.; Mullally, J.E.; Favier, A.; Assard, N.; Sulpice, E.; Lindsey, D.F.; Rulina, A.V.; Gidrol, X.; Wilkinson, K.D. Wss1 Metalloprotease Partners with Cdc48/Doa1 in Processing Genotoxic Sumo Conjugates. Elife 2015, 4, e06763. [Google Scholar] [CrossRef]
- Buser, R.; Kellner, V.; Melnik, A.; Wilson-Zbinden, C.; Schellhaas, R.; Kastner, L.; Piwko, W.; Dees, M.; Picotti, P.; Maric, M.; et al. The Replisome-Coupled E3 Ubiquitin Ligase Rtt101mms22 Counteracts Mrc1 Function to Tolerate Genotoxic Stress. PLoS Genet. 2016, 12, e1005843. [Google Scholar] [CrossRef]
- Kurat, C.F.; Lambert, J.P.; Petschnigg, J.; Friesen, H.; Pawson, T.; Rosebrock, A.; Gingras, A.C.; Fillingham, J.; Andrews, B. Cell Cycle-Regulated Oscillator Coordinates Core Histone Gene Transcription through Histone Acetylation. Proc. Natl. Acad. Sci. USA 2014, 111, 14124–14129. [Google Scholar] [CrossRef]
- Matsuda, K.; Makise, M.; Sueyasu, Y.; Takehara, M.; Asano, T.; Mizushima, T. Yeast Two-Hybrid Analysis of the Origin Recognition Complex of Saccharomyces cerevisiae: Interaction between Subunits and Identification of Binding Proteins. FEMS Yeast Res. 2007, 7, 1263–1269. [Google Scholar] [CrossRef][Green Version]
- Miller, J.E.; Zhang, L.; Jiang, H.; Li, Y.; Pugh, B.F.; Reese, J.C. Genome-Wide Mapping of Decay Factor-mRNA Interactions in Yeast Identifies Nutrient-Responsive Transcripts as Targets of the Deadenylase Ccr4. G3 Genes Genomes Genet. 2018, 8, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Oliete-Calvo, P.; Serrano-Quilez, J.; Nuno-Cabanes, C.; Perez-Martinez, E.M.; Soares, M.L.; Dichtl, B.; Buratowski, S.; Perez-Ortin, J.E.; Rodriguez-Navarro, S. A Role for Mog1 in H2bub1 and H3k4me3 Regulation Affecting RNAPII Transcription and mRNA Export. EMBO Rep. 2018, 19, e45992. [Google Scholar] [CrossRef] [PubMed]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global Analysis of Protein Phosphorylation in Yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Willmund, F.; del Alamo, M.; Pechmann, S.; Chen, T.; Albanèse, V.; Dammer, E.B.; Peng, J.; Frydman, J. The Cotranslational Function of Ribosome-Associated Hsp70 in Eukaryotic Protein Homeostasis. Cell 2013, 152, 196–209. [Google Scholar] [CrossRef]
- Telekawa, C.; Boisvert, F.M.; Bachand, F. Proteomic Profiling and Functional Characterization of Post-Translational Modifications of the Fission Yeast Rna Exosome. Nucleic Acids Res. 2018, 46, 11169–11183. [Google Scholar] [CrossRef]
- Zamir, L.; Zaretsky, M.; Fridman, Y.; Ner-Gaon, H.; Rubin, E.; Aharoni, A. Tight Coevolution of Proliferating Cell Nuclear Antigen (Pcna)-Partner Interaction Networks in Fungi Leads to Interspecies Network Incompatibility. Proc. Natl. Acad. Sci. USA 2012, 109, E406–E414. [Google Scholar] [CrossRef]
- She, R.; Chakravarty, A.K.; Layton, C.J.; Chircus, L.M.; Andreasson, J.O.; Damaraju, N.; McMahon, L.P.; Buenrostro, D.J.; Jarosz, F.D.; Greenleaf, J.W. Comprehensive and Quantitative Mapping of Rna-Protein Interactions across a Transcribed Eukaryotic Genome. Proc. Natl. Acad. Sci. USA 2017, 114, 3619–3624. [Google Scholar] [CrossRef]
- Ramanagoudr-Bhojappa, R.; Blair, L.P.; Tackett, A.J.; Raney, D.K. Physical and Functional Interaction between Yeast Pif1 Helicase and Rim1 Single-Stranded DNA Binding Protein. Nucleic Acids Res. 2013, 41, 1029–1046. [Google Scholar] [CrossRef]
- Lopez, C.R.; Singh, S.; Hambarde, S.; Griffin, C.W.; Gao, J.; Chib, S.; Yu, Y.; Ira, G.; Raney, K.D.; Kim, N. Yeast Sub1 and Human Pc4 Are G-Quadruplex Binding Proteins That Suppress Genome Instability at Co-Transcriptionally Formed G4 DNA. Nucleic Acids Res. 2017, 45, 5850–5862. [Google Scholar] [CrossRef]
- Lapointe, C.P.; Wilinski, D.; Saunders, A.H.; Wickens, M. Protein-Rna Networks Revealed through Covalent Rna Marks. Nat. Methods 2015, 12, 1163–1170. [Google Scholar] [CrossRef]
- Lakshminarasimhan, M.; Boanca, G.; Banks, A.C.; Hattem, L.G.; Gabriel, E.A.; Groppe, D.B.; Smoyer, C.; Malanowski, K.E.; Peak, A.; Florens, L.; et al. Proteomic and Genomic Analyses of the Rvb1 and Rvb2 Interaction Network Upon Deletion of R2tp Complex Components. Mol. Cell Proteom. 2016, 15, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, C.J.; Costello, J.L.; Talavera, D.; Rowe, W.; Castelli, M.L.; Sims, F.P.; Grant, M.C.; Ashe, P.M.; Hubbard, J.S.; Pavitt, D.G. Integrated Multi-Omics Analyses Reveal the Pleiotropic Nature of the Control of Gene Expression by Puf3p. Sci. Rep. 2015, 5, 15518. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.A.; Balakrishnan, L.; Ying-Lin, T.S.; Campbell, L.J.; Bambara, A.R. Components of the Secondary Pathway Stimulate the Primary Pathway of Eukaryotic Okazaki Fragment Processing. J. Biol. Chem. 2010, 285, 28496–28505. [Google Scholar] [CrossRef]
- Gilmore, J.M.; Sardiu, M.E.; Venkatesh, S.; Stutzman, B.; Peak, A.; Seidel, C.W.; Workman, J.L.; Florens, L.; Washburn, M.P. Characterization of a Highly Conserved Histone Related Protein, Ydl156w, Its Functional Associations Using Quantitative Proteomic Analyses. Mol. Cell Proteom. 2012, 11, M111.011544. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C.; Stirling, P.C.; McCormack, E.A.; Filmore, H.; Paul, A.; Brost, L.R.; Costanzo, M.; Boone, C.; Leroux, M.R.; Willison, K.R. The Interaction Network of the Chaperonin Cct. EMBO J. 2008, 27, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Elbaz-Alon, Y.; Rosenfeld-Gur, E.; Shinder, V.; Futerman, H.A.; Geiger, T.; Schuldiner, M. A Dynamic Interface between Vacuoles and Mitochondria in Yeast. Dev. Cell 2014, 30, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Batisse, J.; Batisse, C.; Budd, A.; Bottcher, B.; Hurt, E. Purification of Nuclear Poly(A)-Binding Protein Nab2 Reveals Association with the Yeast Transcriptome and a Messenger Ribonucleoprotein Core Structure. J. Biol. Chem. 2009, 284, 34911–34917. [Google Scholar] [CrossRef]
- Barranco-Medina, S.; Galletto, R. DNA Binding Induces Dimerization of Saccharomyces cerevisiae Pif1. Biochemistry 2010, 49, 8445–8454. [Google Scholar] [CrossRef]
- Babour, A.; Shen, Q.; Dos-Santos, J.; Murray, S.; Gay, A.; Challal, D.; Fasken, M.; Palancade, B.; Corbett, A.; Libri, D.; et al. The Chromatin Remodeler Isw1 Is a Quality Control Factor That Surveys Nuclear Mrnp Biogenesis. Cell 2016, 167, 1201–1214. [Google Scholar] [CrossRef][Green Version]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, A.J.; Asbury, L.C.; Biggins, S. Tension Directly Stabilizes Reconstituted Kinetochore-Microtubule Attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef]
- Sofueva, S.; Osman, F.; Lorenz, A.; Steinacher, R.; Castagnetti, S.; Ledesma, J.; Whitby, M.C. Ultrafine Anaphase Bridges, Broken DNA and Illegitimate Recombination Induced by a Replication Fork Barrier. Nucleic Acids Res. 2011, 39, 6568–6584. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gonzalez, B.; Garcia-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marin, A.; Foiani, M.; Aguilera, A. Genome-Wide Function of Tho/Trex in Active Genes Prevents R-Loop-Dependent Replication Obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef] [PubMed]
- Chib, S.; Byrd, A.K.; Raney, D.K. Yeast Helicase Pif1 Unwinds RNA:DNA Hybrids with Higher Processivity Than DNA:DNA Duplexes. J. Biol. Chem. 2016, 291, 5889–5901. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.; Aguilera, A. Impairment of Replication Fork Progression Mediates RNA PolII Transcription-Associated Recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef]
- Herrera-Moyano, E.; Mergui, X.; Garcia-Rubio, L.M.; Barroso, S.; Aguilera, A. The Yeast and Human Fact Chromatin-Reorganizing Complexes Solve R-Loop-Mediated Transcription-Replication Conflicts. Genes Dev. 2014, 28, 735–748. [Google Scholar] [CrossRef]
- Eugster, A.; Lanzuolo, C.; Bonneton, M.; Luciano, P.; Pollice, A.; Pulitzer, J.F.; Stegberg, E.; Berthiau, A.S.; Forstemann, K.; Corda, Y.; et al. The Finger Subdomain of Yeast Telomerase Cooperates with Pif1p to Limit Telomere Elongation. Nat. Struct. Mol. Biol. 2006, 13, 734–739. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Symington, L.S. Sae2, Exo1 and Sgs1 Collaborate in DNA Double-Strand Break Processing. Nature 2008, 455, 770–774. [Google Scholar] [CrossRef]
- Wei, X.B.; Zhang, B.; Bazeille, N.; Yu, Y.; Liu, N.N.; Rene, B.; Mauffret, O.; Xi, X.G. A 3′–5′ Exonuclease Activity Embedded in the Helicase Core Domain of Candida Albicans Pif1 Helicase. Sci. Rep. 2017, 7, 42865. [Google Scholar] [CrossRef]
- Le, S.; Moore, J.K.; Haber, J.E.; Greider, W.C. Rad50 and Rad51 Define Two Pathways That Collaborate to Maintain Telomeres in the Absence of Telomerase. Genetics 1999, 152, 143–152. [Google Scholar]
- McEachern, M.J.; Haber, J.E. Break-Induced Replication and Recombinational Telomere Elongation in Yeast. Annu. Rev. Biochem. 2006, 75, 111–135. [Google Scholar] [CrossRef]
- Hu, Y.; Tang, H.B.; Liu, N.N.; Tong, X.J.; Dang, W.; Duan, M.Y.; Fu, H.X.; Zhang, Y.; Peng, J.; Meng, F.L.; et al. Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators. PLoS Genet. 2013, 9, e1003208. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.M.; Wang, J.C.; Noguchi, H.; Imasaki, T.; Takagi, Y.; Bochman, M.L. Yeast Hrq1 Shares Structural and Functional Homology with the Disease-Linked Human Recq4 Helicase. Nucleic Acids Res. 2017, 45, 5217–5230. [Google Scholar] [CrossRef] [PubMed]
- Nickens, D.G.; Rogers, C.M.; Bochman, L.M. The Saccharomyces cerevisiae Hrq1 and Pif1 DNA Helicases Synergistically Modulate Telomerase Activity in Vitro. J. Biol. Chem. 2018, 293, 14481–14496. [Google Scholar] [CrossRef] [PubMed]
- Makovets, S.; Blackburn, E.H. DNA Damage Signalling Prevents Deleterious Telomere Addition at DNA Breaks. Nat. Cell Biol. 2009, 11, 1383–1386. [Google Scholar] [CrossRef]
- Vasianovich, Y.; Harrington, L.A.; Makovets, S. Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening. PLoS Genet. 2014, 10, e1004679. [Google Scholar] [CrossRef]
- Mangahas, J.L.; Alexander, M.K.; Sandell, L.L.; Zakian, A.V. Repair of Chromosome Ends after Telomere Loss in Saccharomyces. Mol. Biol. Cell 2001, 12, 4078–4089. [Google Scholar] [CrossRef][Green Version]
- White, C.I.; Haber, J.E. Intermediates of Recombination During Mating Type Switching in Saccharomyces cerevisiae. EMBO J. 1990, 9, 663–673. [Google Scholar] [CrossRef]
- Chang, M.; Luke, B.; Kraft, C.; Li, J.Z.; Peter, M.; Lingner, J.; Rothstein, R. Telomerase Is Essential to Alleviate Pif1-Induced Replication Stress at Telomeres. Genetics 2009, 183, 779–791. [Google Scholar] [CrossRef]
- Bambara, R.A.; Murante, R.S.; Henricksen, A.L. Enzymes and Reactions at the Eukaryotic DNA Replication Fork. J. Biol. Chem. 1997, 272, 4647–4650. [Google Scholar] [CrossRef]
- Harrington, J.J.; Lieber, M.R. The Characterization of a Mammalian DNA Structure-Specific Endonuclease. EMBO J. 1994, 13, 1235–1246. [Google Scholar] [CrossRef]
- Jin, Y.H.; Ayyagari, R.; Resnick, A.M.; Gordenin, A.D.; Burgers, M.P. Okazaki Fragment Maturation in Yeast. Ii. Cooperation between the Polymerase and 3′–5′-Exonuclease Activities of Pol δ in the Creation of a Ligatable Nick. J. Biol. Chem. 2003, 278, 1626–1633. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.L.; Bambara, R.A. Reconstituted Okazaki Fragment Processing Indicates Two Pathways of Primer Removal. J. Biol. Chem. 2006, 281, 26051–26061. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.H.; Bae, K.H.; Kim, J.A.; Seo, S.Y. Rpa Governs Endonuclease Switching During Processing of Okazaki Fragments in Eukaryotes. Nature 2001, 412, 456–461. [Google Scholar] [CrossRef]
- Kao, H.I.; Campbell, J.L.; Bambara, A.R. Dna2p Helicase/Nuclease Is a Tracking Protein, Like Fen1, for Flap Cleavage During Okazaki Fragment Maturation. J. Biol. Chem. 2004, 279, 50840–50849. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.I.; Veeraraghavan, J.; Polaczek, P.; Campbell, L.J.; Bambara, A.R. On the Roles of Saccharomyces cerevisiae Dna2p and Flap Endonuclease 1 in Okazaki Fragment Processing. J. Biol. Chem. 2004, 279, 15014–15024. [Google Scholar] [CrossRef] [PubMed]
- Zaher, M.S.; Rashid, F.; Song, B.; Joudeh, I.L.; Sobhy, A.M.; Tehseen, M.; Hingorani, M.M.; Hamdan, S.M. Missed Cleavage Opportunities by Fen1 Lead to Okazaki Fragment Maturation Via the Long-Flap Pathway. Nucleic Acids Res. 2018, 46, 2956–2974. [Google Scholar] [CrossRef]
- Stith, C.M.; Sterling, J.; Resnick, A.M.; Gordenin, A.D.; Burgers, M.P. Flexibility of Eukaryotic Okazaki Fragment Maturation through Regulated Strand Displacement Synthesis. J. Biol. Chem. 2008, 283, 34129–34140. [Google Scholar] [CrossRef]
- Pike, J.E.; Henry, R.A.; Burgers, P.M.; Campbell, J.L.; Bambara, A.R. An Alternative Pathway for Okazaki Fragment Processing: Resolution of Fold-Back Flaps by Pif1 Helicase. J. Biol. Chem. 2010, 285, 41712–41723. [Google Scholar] [CrossRef]
- Ribeyre, C.; Lopes, J.; Boule, B.J.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The Yeast Pif1 Helicase Prevents Genomic Instability Caused by G-Quadruplex-Forming Ceb1 Sequences in Vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef]
- Byrd, A.K.; Raney, K.D. A Parallel Quadruplex DNA Is Bound Tightly but Unfolded Slowly by Pif1 Helicase. J. Biol. Chem. 2015, 290, 6482–6494. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.M.; Wang, Y.R.; Xi, X.G.; Hou, M.X. DNA-Unwinding Activity of Saccharomyces cerevisiae Pif1 Is Modulated by Thermal Stability, Folding Conformation, Loop Lengths of G-Quadruplex DNA. J. Biol. Chem. 2018, 293, 18504–18513. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.K.; Bell, M.R.; Raney, D.K. Pif1 Helicase Unfolding of G-Quadruplex DNA Is Highly Dependent on Sequence and Reaction Conditions. J. Biol. Chem. 2018, 293, 17792–17802. [Google Scholar] [CrossRef]
- Galletto, R.; Tomko, E.J. Translocation of Saccharomyces cerevisiae Pif1 Helicase Monomers on Single-Stranded DNA. Nucleic Acids Res. 2013, 41, 4613–4627. [Google Scholar] [CrossRef] [PubMed]
- George, T.; Wen, Q.; Griffiths, R.; Ganesh, A.; Meuth, M.; Sanders, C.M. Human Pif1 Helicase Unwinds Synthetic DNA Structures Resembling Stalled DNA Replication Forks. Nucleic Acids Res. 2009, 37, 6491–6502. [Google Scholar] [CrossRef] [PubMed]
- Wanzek, K.; Schwindt, E.; Capra, A.J.; Paeschke, K. Mms1 Binds to G-Rich Regions in Saccharomyces cerevisiae and Influences Replication and Genome Stability. Nucleic Acids Res. 2017, 45, 7796–7806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Capra, J.A.; Paeschke, K.; Singh, M.; Zakian, A.V. G-Quadruplex DNA Sequences Are Evolutionarily Conserved and Associated with Distinct Genomic Features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010, 6, e1000861. [Google Scholar] [CrossRef]
- Wanrooij, P.H.; Uhler, J.P.; Shi, Y.H.; Westerlund, F.; Falkenberg, M.; Gustafsson, M.C. A Hybrid G-Quadruplex Structure Formed between Rna and DNA Explains the Extraordinary Stability of the Mitochondrial R-Loop. Nucleic Acids Res. 2012, 40, 10334–10344. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, W.Q.; Liu, N.N.; Duan, X.L.; Li, M.; Dou, X.S.; Hou, M.X.; Xi, G.X. G-Quadruplex and G-Rich Sequence Stimulate Pif1p-Catalyzed Downstream Duplex DNA Unwinding through Reducing Waiting Time at Ss/dsDNA Junction. Nucleic Acids Res. 2016, 44, 8385–8394. [Google Scholar] [CrossRef]
- Duan, X.L.; Liu, N.N.; Yang, Y.T.; Li, H.H.; Li, M.; Dou, X.S.; Xi, G.X. G-Quadruplexes Significantly Stimulate Pif1 Helicase-Catalyzed Duplex DNA Unwinding. J. Biol. Chem. 2015, 290, 7722–7735. [Google Scholar] [CrossRef]
- Li, J.R.; Lu, C.Y.; Lin, J.J.; Li, W.H. Multiple Pif1 Helicases Are Required to Sequentially Disrupt G-Quadruplex Structure and Unwind Duplex DNA. Biochem. Biophys. Res. Commun. 2016, 473, 1235–1239. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, J.; Bochman, L.M.; Zakian, A.V.; Ha, T. Periodic DNA Patrolling Underlies Diverse Functions of Pif1 on R-Loops and G-Rich DNA. Elife 2014, 3, e02190. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, L.K.; Kowalczykowski, C.S.; Zakian, A.V. Pif1 Family Helicases Suppress Genome Instability at G-Quadruplex Motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.L.; Shoemaker, D.D.; Boeke, D.J. DNA Helicase Gene Interaction Network Defined Using Synthetic Lethality Analyzed by Microarray. Nature Genet. 2003, 35, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Shah, S.S.; Wright, W.D.; Gore, S.K.; Koszul, R.; Heyer, D.W. Dynamic Processing of Displacement Loops During Recombinational DNA Repair. Mol. Cell 2019, 73, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Prakash, R.; Satory, D.; Dray, E.; Papusha, A.; Scheller, J.; Kramer, W.; Krejci, L.; Klein, H.; Haber, J.E.; Sung, P.; et al. Yeast Mph1 Helicase Dissociates Rad51-Made D-Loops: Implications for Crossover Control in Mitotic Recombination. Genes Dev. 2009, 23, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Quinet, A.; Lemacon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Hyun, K.; Kim, J.; Hohng, S. ATP Binding to Rad5 Initiates Replication Fork Reversal by Inducing the Unwinding of the Leading Arm and the Formation of the Holliday Junction. Cell Rep. 2018, 23, 1831–1839. [Google Scholar] [CrossRef] [PubMed]
- Hashash, N.; Johnson, A.L.; Cha, S.R. Regulation of Fragile Sites Expression in Budding Yeast by Mec1, Rrm3 and Hydroxyurea. J. Cell Sci. 2011, 124, 181–185. [Google Scholar] [CrossRef]
- Cheng, X.; Qin, Y.; Ivessa, S.A. Loss of Mitochondrial DNA under Genotoxic Stress Conditions in the Absence of the Yeast DNA Helicase Pif1p Occurs Independently of the DNA Helicase Rrm3p. Mol. Genet. Genom. 2009, 281, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ivessa, A.S. Association of the Yeast DNA Helicase Pif1p with Mitochondrial Membranes and Mitochondrial DNA. Eur J. Cell Biol. 2010, 89, 742–747. [Google Scholar] [CrossRef]
- Liu, Z.; Butow, R.A. Mitochondrial Retrograde Signaling. Annu. Rev. Genet. 2006, 40, 159–185. [Google Scholar] [CrossRef] [PubMed]
- Doudican, N.A.; Song, B.; Shadel, S.G.; Doetsch, W.P. Oxidative DNA Damage Causes Mitochondrial Genomic Instability in Saccharomyces cerevisiae. Mol. Cell Biol. 2005, 25, 5196–5204. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, T.W.; Doudican, N.A.; Mackereth, M.D.; Doetsch, P.W.; Shadel, S.G. Mitochondrial Dysfunction Due to Oxidative Mitochondrial DNA Damage Is Reduced through Cooperative Actions of Diverse Proteins. Mol. Cell. Biol. 2002, 22, 4086–4093. [Google Scholar] [CrossRef]
- O’Rourke, T.W.; Doudican, N.A.; Zhang, H.; Eaton, S.J.; Doetsch, W.P.; Shadel, S.G. Differential Involvement of the Related DNA Helicases Pif1p and Rrm3p in mtDNA Point Mutagenesis and Stability. Genes 2005, 354, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Zhang, H.; Eaton, S.J.; Rodeheffer, S.M.; Lebedeva, A.M.; O’Rourke, T.W.; Siede, W.; Shadel, G.S. The Conserved Mec1/Rad53 Nuclear Checkpoint Pathway Regulates Mitochondrial DNA Copy Number in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 3010–3018. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.M.; Wu, W.Q.; Duan, X.L.; Liu, N.N.; Li, H.H.; Fu, J.; Dou, X.S.; Li, M.; Xi, X.G. Molecular Mechanism of G-Quadruplex Unwinding Helicase: Sequential and Repetitive Unfolding of G-Quadruplex by Pif1 Helicase. Biochem. J. 2015, 466, 189–199. [Google Scholar] [CrossRef]
- Llorente, B.; Smith, C.E.; Symington, S.L. Break-Induced Replication: What Is It and What Is It For? Cell Cycle 2008, 7, 859–864. [Google Scholar] [CrossRef]
- Saini, N.; Ramakrishnan, S.; Elango, R.; Ayyar, S.; Zhang, Y.; Deem, A.; Ira, G.; Haber, J.E.; Lobachev, K.S.; Malkova, A. Migrating Bubble During Break-Induced Replication Drives Conservative DNA Synthesis. Nature 2013, 502, 389–392. [Google Scholar] [CrossRef]
- Li, J.H.; Lin, W.X.; Zhang, B.; Nong, G.D.; Ju, P.H.; Ma, B.J.; Xu, H.C.; Ye, F.F.; Xi, G.X.; Li, M.; et al. Pif1 Is a Force-Regulated Helicase. Nucleic Acids Res. 2016, 44, 4330–4339. [Google Scholar] [CrossRef]
- Bairwa, N.K.; Mohanty, B.K.; Stamenova, R.; Curcio, J.M.; Bastia, D. The Intra-S Phase Checkpoint Protein Tof1 Collaborates with the Helicase Rrm3 and the F-Box Protein Dia2 to Maintain Genome Stability in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 2445–2454. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Wu, J.; Kolodner, D.R. Control of Translocations between Highly Diverged Genes by Sgs1, the Saccharomyces cerevisiae Homolog of the Bloom’s Syndrome Protein. Mol. Cell Biol. 2006, 26, 5406–5420. [Google Scholar] [CrossRef] [PubMed]
- Cha, R.S.; Kleckner, N. Atr Homolog Mec1 Promotes Fork Progression, Thus Averting Breaks in Replication Slow Zones. Science 2002, 297, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, K.M.; Aubert, S.D.; Freese, K.P.; Zakian, V.A.; King, M.C.; Welcsh, L.P. A Genomewide Screen for Suppressors of Alu-Mediated Rearrangements Reveals a Role for Pif1. PLoS ONE 2012, 7, e30748. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jimeno, S.; Camarillo, R.; Mejias-Navarro, F.; Fernandez-Avila, J.M.; Soria-Bretones, I.; Prados-Carvajal, R.; Huertas, P. The Helicase Pif1 Facilitates Resection over Sequences Prone to Forming G4 Structures. Cell Rep. 2018, 25, 3543. [Google Scholar] [CrossRef]
- Chung, W.H. To Peep into Pif1 Helicase: Multifaceted All the Way from Genome Stability to Repair-Associated DNA Synthesis. J. Microbiol. 2014, 52, 89–98. [Google Scholar] [CrossRef]
| S. cerevesiae | S. pombe | ||||
|---|---|---|---|---|---|
| Rrm3 | Pif1 | Pfh1 | |||
| Essential | No | No | Yes | ||
| Nucleus | [28] | [28] | [28] | ||
| Mitochondria | [29] | [28] | [28] | ||
| Localization | Telomeres | [27] | [30,31] | [32] | |
| mtDNA | N.D. | [33] | [28] | ||
| rDNA | [16] | [16] | [16] | ||
| tRNA genes | [34,35] | [35,36] | [17] | ||
| Centromeres | [34,37] | [37] | N.D. | ||
| Highly transcribed genes | [38] | [39] | [40,41] | ||
| Active/Inactive DNA replication forks | [10] | [10] | N.D. | ||
| Transcription-replication conflicts | [38] | [38] | [17] | ||
| Rad52 DNA-damage foci | [28] | [28] | [28] | ||
| Origins of replication | [16,42] | [16] | N.D. | ||
| Function | Replication through telomeres | [27] | N.D.* | [32] | |
| Telomere anchoring | [43] | [44] | N.D. | ||
| Telomerase inhibition | [27] | [13] | [32] | ||
| G4 structures | [45] | [39,46] | [41,47] | ||
| Okazaki fragment maturation | [36] | [48] | [49] | ||
| Centromere Replication and Segregation | [37] | [37] | N.D. | ||
| Sister chromatid cohesion | [50] | [37] | N.D. | ||
| Sister chromatid exchange | [51] | [51] | N.D. | ||
| Break-induced replication | [11,52] | [11,52] | N.D. | ||
| DNA synthesis restriction (HU) | [42] | N.D. | N.D. | ||
| Fork convergence | [16,53] | [53] | [17,54] | ||
| Daughter-strand gap repair | [51] | [55] | N.D. | ||
| Silent mating type locus | [34] | N.D. | [17] | ||
| Fork progression through tRNA genes | [35,36] | [35,36] | [17] | ||
| Fork progression through rDNA | [16] | [16] | [17] | ||
| Repression of Ty1 mobility | [56,57] | N.D. | N.D. | ||
| Maintenance of mtDNA | [36] | [33,36] | [28] | ||
| H2AX/H2A phosphorylation | N.D. | N.D. | [17] | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muellner, J.; Schmidt, K.H. Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family. Genes 2020, 11, 224. https://doi.org/10.3390/genes11020224
Muellner J, Schmidt KH. Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family. Genes. 2020; 11(2):224. https://doi.org/10.3390/genes11020224
Chicago/Turabian StyleMuellner, Julius, and Kristina H. Schmidt. 2020. "Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family" Genes 11, no. 2: 224. https://doi.org/10.3390/genes11020224
APA StyleMuellner, J., & Schmidt, K. H. (2020). Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family. Genes, 11(2), 224. https://doi.org/10.3390/genes11020224




