Distribution of Medically Relevant Antibiotic Resistance Genes and Mobile Genetic Elements in Soils of Temperate Forests and Grasslands Varying in Land Use
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, Soil Characteristics and DNA Extraction
2.2. Soil Fungal Diversity
2.3. Quantification of 16S rRNA Genes, IncP-1 Plasmids and Class 1 Integrons
2.4. Detection of Antibiotic Resistance Genes via qPCR Array
2.5. Quantification of ARGs in Soils Derived from 300 Study Plots
2.6. Statistical Analysis
3. Results
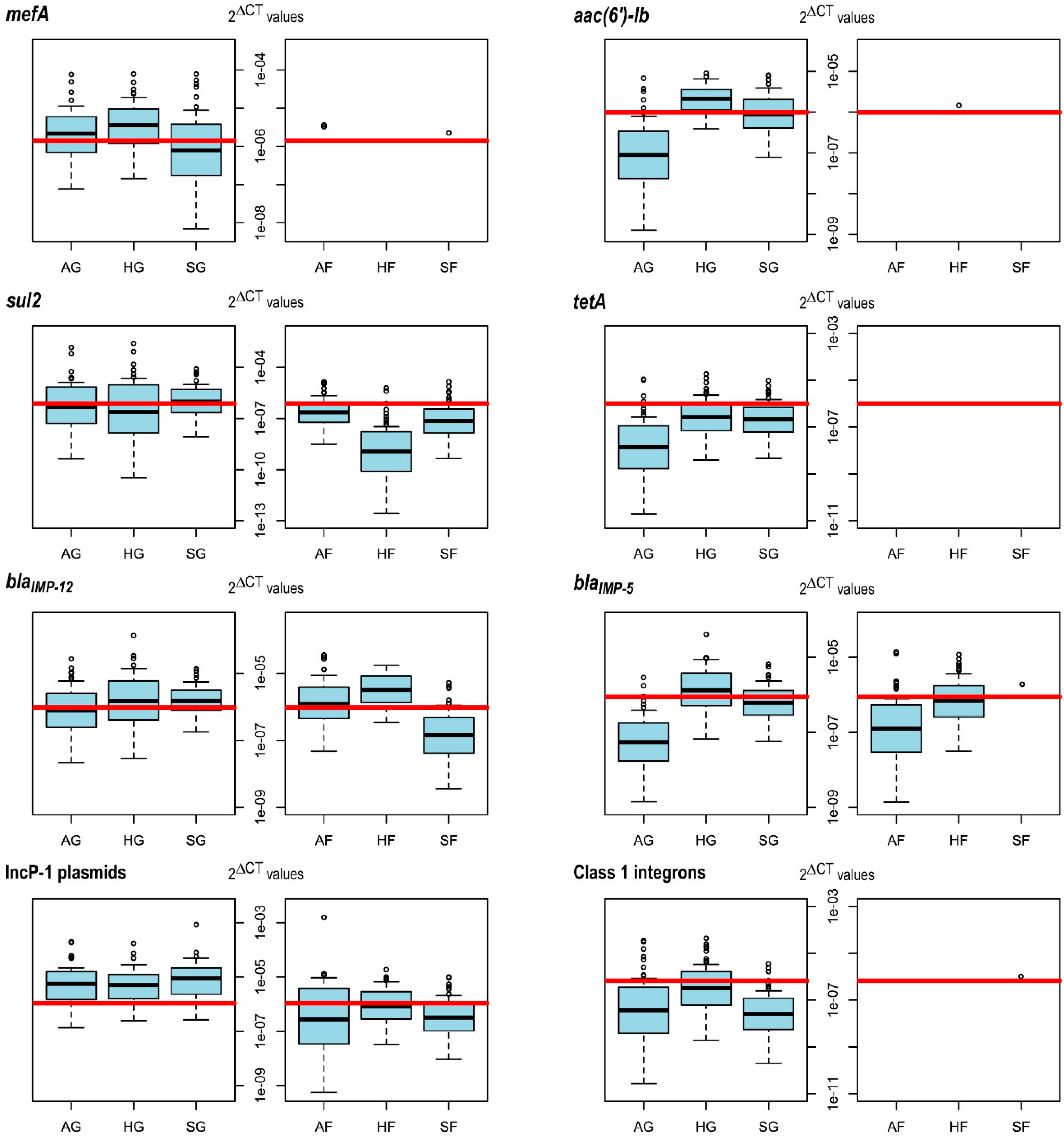
3.1. Selection of Targets for ARG Quantification in Forest and Grassland Soils
3.2. IncP-1 Plasmids, mefA and sul2 Are More Abundant in Grassland than in Forest Soils
3.3. Land Use Practices in Grassland Affect the Abundances of aac(6′)-lb, mefA and sul2
3.4. The Abundance of blaIMP-12 Increases with Fungal Diversity in Forest Soil
4. Discussion
4.1. sul2 and mefA
4.2. aac(6′)-lb
4.3. blaIMP-12, blaIMP-5 and MGEs
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review on Antimicrobial Resistance: London, UK, 2016. [Google Scholar]
- Auta, A.; Hadi, M.A.; Oga, E.; Adewuyi, E.O.; Abdu-Aguye, S.N.; Adeloye, D.; Strickland-Hodge, B.; Morgan, D.J. Global access to antibiotics without prescription in community pharmacies: A systematic review and meta-analysis. J. Infect. 2019, 78, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Versporten, A.; Bolokhovets, G.; Ghazaryan, L.; Abilova, V.; Pyshnik, G.; Spasojevic, T.; Korinteli, I.; Raka, L.; Kambaralieva, B.; Cizmovic, L.; et al. Antibiotic use in eastern Europe: A cross-national database study in coordination with the WHO regional office for Europe. Lancet Infect. Dis. 2014, 14, 381–387. [Google Scholar] [CrossRef]
- WHO Regional Office for Europe. Assessing Non-Prescription and Inappropriate Use of Antibiotics: Report on Survey; WHO: Copenhagen, Denmark, 2019. [Google Scholar]
- Anderson, M.; Clift, C.; Schulze, K.; Sagan, A.; Nahrgang, S.; Ait Ouakrim, D.; Mossialos, E. Averting the AMR crisis: What are the avenues for policy action for countries in Europe? In European Observatory Policy Briefs; European Observatory on Health Systems and Policies: Copenhagen, Denmark, 2019. [Google Scholar]
- World Health Organization. Critically Important Antibacterial Agents for Human Medicine for Risk Management Strategies of Non-Human Use: Report of a WHO Working Group Consultation; World Health Organization: Canberra, Australia, 2005. [Google Scholar]
- Van Doorn, H.R. Emerging infectious diseases. Medicine 2014, 42, 60–63. [Google Scholar] [CrossRef]
- World Health Organization. Of All Human Diseases, 60% Originate in Animals—“One Health” Is the Only Way to Keep Antibiotics Working. Available online: http://www.euro.who.int/en/health-topics/disease-prevention/food-safety/news/news/2018/11/of-all-human-diseases,-60-originate-in-animals-one-health-is-the-only-way-to-keep-antibiotics-working (accessed on 28 August 2019).
- European Union. Ban on Antibiotics as Growth Promoters in Animal Feed Enters into Effect. Available online: https://europa.eu/rapid/press-release_IP-05-1687_en.htm (accessed on 22 November 2019).
- Anomaly, J. What’s wrong with factory farming? Public Health Ethics 2015, 8, 246–254. [Google Scholar] [CrossRef]
- Pluhar, E.B. Meat and morality: Alternatives to factory farming. J. Agric. Environ. Ethics 2010, 23, 455–468. [Google Scholar] [CrossRef]
- EMA. Antimicrobial Resistance in the Environment: Considerations for Current and Future Risk Assessment of Veterinary Medicinal Products; European Medicines Agency: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
- Canteón, R. Antibiotic resistance genes from the environment: A perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 2009, 15, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Surette, M.D.; Wright, G.D. Lessons from the environmental antibiotic resistome. Annu. Rev. Microbiol. 2017, 71, 309–329. [Google Scholar] [CrossRef]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Knapp, C.W.; Christensen, B.T.; McCluskey, S.; Dolfing, J. Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 2016, 6, 21550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Xu, M.; Stedtfeld, R.D.; Sheng, H.; Fan, J.; Liu, M.; Chai, B.; Soares de Carvalho, T.; Li, H.; Li, Z.; et al. Long-term effect of different fertilization and cropping systems on the soil antibiotic resistome. Environ. Sci. Technol. 2018, 52, 13037–13046. [Google Scholar] [CrossRef] [PubMed]
- DeFrancesco, K.A.; Cobbold, R.N.; Rice, D.H.; Besser, T.E.; Hancock, D.D. Antimicrobial resistance of commensal Escherichia coli from dairy cattle associated with recent multi-resistant salmonellosis outbreaks. Vet. Microbiol. 2004, 98, 55–61. [Google Scholar] [CrossRef]
- Berendsen, B.J.A.; Wegh, R.S.; Memelink, J.; Zuidema, T.; Stolker, L.A.M. The analysis of animal faeces as a tool to monitor antibiotic usage. Talanta 2015, 132, 258–268. [Google Scholar] [CrossRef]
- Guo, T.; Lou, C.; Zhai, W.; Tang, X.; Hashmi, M.Z.; Murtaza, R.; Li, Y.; Liu, X.; Xu, J. Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Total Environ. 2018, 635, 995–1003. [Google Scholar] [CrossRef]
- Peng, S.; Feng, Y.; Wang, Y.; Guo, X.; Chu, H.; Lin, X. Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. J. Hazard. Mater. 2017, 340, 16–25. [Google Scholar] [CrossRef]
- Jechalke, S.; Schreiter, S.; Wolters, B.; Dealtry, S.; Heuer, H.; Smalla, K. Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis. Front. Microbiol. 2014, 4, 420. [Google Scholar] [CrossRef]
- Hu, H.-W.; Wang, J.-T.; Singh, B.K.; Liu, Y.-R.; Chen, Y.-L.; Zhang, Y.-J.; He, J.-Z. Diversity of herbaceous plants and bacterial communities regulates soil resistome across forest biomes. Environ. Microbiol. 2018, 20, 3186–3200. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.; Currie, C. The natural history of antibiotics. Curr. Biol. 2009, 11, R437–R441. [Google Scholar] [CrossRef] [Green Version]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Nacke, H.; Goldmann, K.; Schöning, I.; Pfeiffer, B.; Kaiser, K.; Castillo-Villamizar, G.A.; Schrumpf, M.; Buscot, F.; Daniel, R.; Wubet, T. Fine spatial scale variation of soil microbial communities under European beech and Norway spruce. Front. Microbiol. 2016, 7, 2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M.; Bossdorf, O.; Gockel, S.; Hänsel, F.; Hemp, A.; Hessenmöller, D.; Korte, G.; Nieschulze, J.; Pfeiffer, S.; Prati, D.; et al. Implementing large-scale and long-term functional biodiversity research: The biodiversity exploratories. Basic Appl. Ecol. 2010, 11, 473–485. [Google Scholar] [CrossRef]
- Solly, E.F.; Schöning, I.; Boch, S.; Kandeler, E.; Marhan, S.; Michalzik, B.; Müller, J.; Zscheischler, J.; Trumbore, S.E.; Schrumpf, M. Factors controlling decomposition rates of fine root litter in temperate forests and grasslands. Plant Soil 2014, 382, 203–218. [Google Scholar] [CrossRef] [Green Version]
- Vogt, J.; Klaus, V.H.; Both, S.; Fürstenau, C.; Gockel, S.; Gossner, M.M.; Heinze, J.; Hemp, A.; Hölzel, N.; Jung, K.; et al. Eleven years’ data of grassland management in Germany. Biodivers. Data J. 2019, 7, e36387. [Google Scholar] [CrossRef]
- Blüthgen, N.; Dormann, C.F.; Prati, D.; Klaus, V.H.; Kleinebecker, T.; Hölzel, N.; Alt, F.; Boch, S.; Gockel, S.; Hemp, A.; et al. A quantitative index of land-use intensity in grasslands: Integrating mowing, grazing and fertilization. Basic Appl. Ecol. 2012, 13, 207–220. [Google Scholar] [CrossRef]
- Dean, R.B.; Dixon, W.J. Simplified statistics for small numbers of observations. Anal. Chem. 1951, 23, 636–638. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Boyer, F.; Mercier, C.; Bonin, A.; Le Bras, Y.; Taberlet, P.; Coissac, E. obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 176–182. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Michin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef] [Green Version]
- Jechalke, S.; Dealtry, S.; Smalla, K.; Heuer, H. Quantification of IncP-1 plasmid prevalence in environmental Samples. Appl. Environ. Microbiol. 2013, 79, 1410–1413. [Google Scholar] [CrossRef] [Green Version]
- Pansegrau, W.; Lanka, E.; Barth, P.T.; Figurski, D.H.; Guiney, D.G.; Haas, D.; Helinski, D.R.; Schwab, H.; Stanisich, V.A.; Thomas, C.M. Complete nucleotide sequence of Birmingham IncPα plasmids. J. Mol. Biol. 1994, 239, 623–663. [Google Scholar] [CrossRef]
- Barraud, O.; Baclet, M.C.; Denis, F.; Ploy, M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010, 65, 1642–1645. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.-W.; Han, X.-M.; Shi, X.-Z.; Wang, J.-T.; Han, L.-L.; Chen, D.; He, J.-Z. Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 2016, 92, fiv169. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Wang, J.; Han, Y.; Chen, J.; Liu, G.; Lu, H.; Yan, B.; Chen, S. Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 2017, 220, 909–918. [Google Scholar] [CrossRef]
- Kleiber, C.; Zeileis, A. Applied Econometrics with R; Springer New York: New York, NY, USA, 2008; ISBN 978-0-387-77316-2. [Google Scholar]
- We, T.; Simko, V. R Package “corrplot”: Visualization of a Correlation Matrix; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Lopaka, L. NADA: Nondetects and Data Analysis for Environmental Data; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Mcfadden, D. Conditional logit analysis of qualitative choice behavior. In Froniers in Econometrics; Zarembka, P., Ed.; Academic Press: New York, NY, USA, 1974; pp. 105–142. [Google Scholar]
- Kaiser, K.; Wemheuer, B.; Korolkow, V.; Wemheuer, F.; Nacke, H.; Schöning, I.; Schrumpf, M.; Daniel, R. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci. Rep. 2016, 6, 33696. [Google Scholar] [CrossRef] [Green Version]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014, 509, 612–616. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Sköld, O. Resistance to trimethoprim and sulfonamides. Vet. Res. 2001, 32, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, D.E.; Tietze, E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 2001, 65, 481–496. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [Green Version]
- Seiffert, S.N.; Marschall, J.; Perreten, V.; Carattoli, A.; Furrer, H.; Endimiani, A. Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. Int. J. Antimicrob. Agents 2014, 44, 260–262. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Schroeter, A.; Malorny, B.; Lehmann, S.; Helmuth, R. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 2003, 52, 489–492. [Google Scholar] [CrossRef]
- Leclercq, R.; Courvalin, P. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2002, 46, 2727–2734. [Google Scholar] [CrossRef] [Green Version]
- Griffith, F. The serological classification of Streptococcus pyogenes. Epidemiol. Infect. 1934, 34, 542–584. [Google Scholar] [CrossRef] [Green Version]
- Oster, P.; Zanchi, A.; Cresti, S.; Lattanzi, M.; Montagnani, F.; Cellesi, C.; Rossolini, G.M. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob. Agents Chemother. 1999, 43, 2510–2512. [Google Scholar] [CrossRef] [Green Version]
- Santagati, M.; Iannelli, F.; Cascone, C.; Campanile, F.; Oggioni, M.R.; Stefani, S.; Pozzi, G. The novel conjugative transposon Tn 1207.3 carries the macrolide efflux gene mef (A) in Streptococcus pyogenes. Microb. Drug Resist. 2003, 9, 243–247. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Bertin, S.; Leblond, P.; Madec, J.-Y.; Payot, S. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl. Environ. Microbiol. 2010, 76, 7957–7965. [Google Scholar] [CrossRef] [Green Version]
- Collignon, P.; Powers, J.H.; Chiller, T.M.; Aidara-Kane, A.; Aarestrup, F.M. World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 2009, 49, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Giguère, S. Antimicrobial Therapy in Veterinary Medicine; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Bartram, D.J.; Moyaert, H.; Vanimisetti, B.H.; Ramage, C.P.; Reddick, D.; Stegemann, M.R. Comparative efficacy of tulathromycin and tildipirosin for the treatment of experimental Mycoplasma bovis infection in calves. Vet. Med. Sci. 2016, 2, 170–178. [Google Scholar] [CrossRef] [Green Version]
- Baggot, J.D.; Gingerich, D.A. Pharmacokinetic interpretation of erythromycin and tylosin activity in serum after intravenous administration of a single dose to cows. Res. Vet. Sci. 1976, 21, 318–323. [Google Scholar] [CrossRef]
- Blondeau, J. Differential impact of macrolide compounds in the selection of macrolide nonsusceptible Streptococcus pneumoniae. Therapy 2005, 2, 813–818. [Google Scholar] [CrossRef]
- Kim, M.; Park, T.; Yu, Z. Metagenomic investigation of gastrointestinal microbiome in cattle. Asian Australasian J. Anim. Sci. 2017, 30, 1515. [Google Scholar] [CrossRef]
- Holman, D.B.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- United States Pharmacopeial Convention. The United States Pharmacopeia 31: The National Formulary 26; United States Pharmacopeial Convention: Rockville, MD, USA, 2007. [Google Scholar]
- Ramirez, M.S.; Nikolaidis, N.; Tolmasky, M.E. Rise and dissemination of aminoglycoside resistance: The aac(6′)-Ib paradigm. Front. Microbiol. 2013, 4, 121. [Google Scholar] [CrossRef] [Green Version]
- Lambert, T.; Ploy, M.-C.; Courvalin, P. A spontaneous point mutation i the aac(6′)-lb′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 1994, 115, 297–303. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Hye Park, C.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Wallmann, J. Erfahrungen und Schlussfolgerungen aus der Antibiotikaabgabeerfassung in der Veterinärmedizin; BVL: Berlin, Germany, 2014. [Google Scholar]
- Aidara-Kane, A.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Collignon, P.J.; WHO guideline development group. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control. 2018, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Ruegg, P.L. Responsible Use of Antibiotics for Treatment of Clinical Mastitis—DAIReXNET. Available online: https://dairy-cattle.extension.org/responsible-use-of-antibiotics-for-treatment-of-clinical-mastitis/ (accessed on 10 November 2019).
- Persson, Y.; Katholm, J.; Landin, H.; Mörk, M.J. Efficacy of enrofloxacin for the treatment of acute clinical mastitis caused by Escherichia coli in dairy cows. Vet. Rec. 2015, 176, 673. [Google Scholar] [CrossRef]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Thiele-Bruhn, S. Pharmaceutical antibiotic compounds in soils—A review. J. Plant Nutr. Soil Sci. 2003, 166, 145–167. [Google Scholar] [CrossRef]
- Schulz, J.; Kemper, N.; Hartung, J.; Janusch, F.; Mohring, S.A.I.; Hamscher, G. Analysis of fluoroquinolones in dusts from intensive livestock farming and the co-occurrence of fluoroquinolone-resistant Escherichia coli. Sci. Rep. 2019, 9, 5117. [Google Scholar] [CrossRef]
- Lillenberg, M.; Litvin, S.V.; Nei, L.; Roasto, M.; Sepp, K.I. Enrofloxacin and ciprofloxacin uptake by plants from soil. Agron. Res. 2010, 8, 807–814. [Google Scholar]
- Hu, X.; Zhou, Q.; Luo, Y. Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 2010, 158, 2992–2998. [Google Scholar] [CrossRef]
- Alonso, A.; Sanchez, P.; Martinez, J.L. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 2001, 3, 1–9. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Maynard, C.; St-Arnaud, M.; Greer, C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014, 8, 344–358. [Google Scholar] [CrossRef]
- Diene, S.M.; Rolain, J.-M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef] [Green Version]
- Cornaglia, G.; Giamarellou, H.; Rossolini, G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011, 11, 381–393. [Google Scholar] [CrossRef]
- Docquier, J.-D.; Riccio, M.L.; Mugnaioli, C.; Luzzaro, F.; Endimiani, A.; Toniolo, A.; Amicosante, G.; Rossolini, G.M. IMP-12, a new plasmid-encoded metallo-beta-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 2003, 47, 1522–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.-H.; Chen, G.; Ito, R.; Kimura, S.; Hu, Z.-Q. Identification of a plasmid-borne blaIMP-11 gene in clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Med. Microbiol. 2012, 61, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, K.; Xiao, S.; Xie, L.; Gu, F.; Li, X.; Ni, Y.; Sun, J.; Han, L. A multidrug resistance Plasmid pIMP26, carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in Enterobacter cloacae. Sci. Rep. 2019, 9, 10212. [Google Scholar] [CrossRef] [Green Version]
- Braschi, I.; Blasioli, S.; Fellet, C.; Lorenzini, R.; Garelli, A.; Pori, M.; Giacomini, D. Persistence and degradation of new β-lactam antibiotics in the soil and water environment. Chemosphere 2013, 93, 152–159. [Google Scholar] [CrossRef]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014, 9, 536–545. [Google Scholar] [CrossRef]
- Chen, S.; Larsson, M.; Robinson, R.C.; Chen, S.L. Direct and convenient measurement of plasmid stability in lab and clinical isolates of E. coli. Sci. Rep. 2017, 7, 4788. [Google Scholar] [CrossRef]
- Da Silva, G.; Correia, M.; Vital, C.; Ribeiro, G.; Sousa, J.C.; Leitão, R.; Peixe, L.; Duarte, A. Molecular characterization of blaIMP-5, a new integrons-borne metallo-β-lactamase gene from an Acinetobacter baumanii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 2002, 215, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Goldmann, K.; Schöning, I.; Buscot, F.; Wubet, T. Forest management type influences diversity and community composition of soil fungi across temperate forest ecosystems. Front. Microbiol. 2015, 6, 1300. [Google Scholar] [CrossRef] [Green Version]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2011, 2, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, H.S.; Abraham, E.P. Isolation of antibiotics from a species of Cephalosporium. Cephalosporins P1, P2, P3, P4 and P5. Biochem. J. 1951, 50, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Pöggeler, S.; Hoff, B.; Kück, U. Asexual cephalosporin C producer Acremonium chrysogenum carries a functional mating type locus. Appl. Environ. Microbiol. 2008, 74, 6006–6016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuer, H.; Smalla, K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012, 36, 1083–1104. [Google Scholar] [CrossRef] [Green Version]
- Popowska, M.; Krawczyk-Balska, A. Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 2013, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Sen, D.; Van der Auwera, G.A.; Rogers, L.M.; Thomas, C.M.; Brown, C.J.; Top, E.M. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 2011, 77, 7975–7983. [Google Scholar] [CrossRef] [Green Version]
| A | B | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p | Est. | R2 | Df | Null/Resid. | p | Est. | R2 | Df | Null/Resid. | |
| aac(6′)-lb | 0.21 | 143 | 191.3/152 | 0.12 | 142 | 333.7/292.7 | ||||
| Intercept | 3.8 × 10−7 | −12.29 | <2 × 10−16 | −31.49 | ||||||
| Mowing | 0.03 | 0.45 | 4.4 × 10−2 | 0.37 | ||||||
| pH | 7.4 × 10−7 | 1.74 | 1.5 × 10−7 | 1.65 | ||||||
| mefA | 0.24 | 140 | 198.6/163 | 0.11 | 141 | 198.6/178.5 | ||||
| Intercept | 0.02 | 0.52 | <2 × 10−16 | −18.81 | ||||||
| Mowing | 0.13 | 0.33 | 8.1 × 10−2 | 0.41 | ||||||
| Org. N | 9.4 × 10−4 | 1.14 | 3.9 × 10−3 | 0.64 | ||||||
| Moisture | 3.0 × 10−3 | −0.68 | 9.2 × 10−5 | −1.01 | ||||||
| sul2 | 0.1 | 140 | 182.5/171.6 | 0.08 | 140 | 182.5/177.9 | ||||
| Intercept | 9.3 × 10−4 | −0.62 | <2 × 10−16 | −22.18 | ||||||
| Org. N | 0.01 | 0.53 | 2.2 × 10−3 | 1.60 | ||||||
| Moisture | 0.10 | −0.37 | 8.9 × 10−2 | −1.14 | ||||||
| blaIMP-12 | 0.17 | 133 | 188.5/155.6 | 0.09 | 132 | 417.1/380.7 | ||||
| Intercept | 8.4 × 10−4 | −1.64 | <2 × 10−16 | −21.93 | ||||||
| Beech | 7.0 × 10−5 | 2.13 | 7.4 × 10−6 | 2.75 | ||||||
| Shan-H | 4.6 × 10−3 | 0.65 | 2.4 × 10−3 | 0.73 |
| Target | Occurrence and Relative Abundance Significantly Increased in Grassland Compared to Forest Soil | Factors Significantly Influencing Target Occurrence and Relative Abundance in Grassland Soil | Factors Significantly Influencing Target Occurrence and Relative Abundance in Forest Soil |
|---|---|---|---|
| IncP-1 | Yes | - | - |
| aac(6′)-lb | Further analysis required * | Mowing frequency and pH | - |
| mefA | Yes | Organic nitrogen input | - |
| sul2 | Yes | Organic nitrogen input and soil moisture | - |
| blaIMP-12 | No | - | Fungal diversity and beech as dominant tree species |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willms, I.M.; Yuan, J.; Penone, C.; Goldmann, K.; Vogt, J.; Wubet, T.; Schöning, I.; Schrumpf, M.; Buscot, F.; Nacke, H. Distribution of Medically Relevant Antibiotic Resistance Genes and Mobile Genetic Elements in Soils of Temperate Forests and Grasslands Varying in Land Use. Genes 2020, 11, 150. https://doi.org/10.3390/genes11020150
Willms IM, Yuan J, Penone C, Goldmann K, Vogt J, Wubet T, Schöning I, Schrumpf M, Buscot F, Nacke H. Distribution of Medically Relevant Antibiotic Resistance Genes and Mobile Genetic Elements in Soils of Temperate Forests and Grasslands Varying in Land Use. Genes. 2020; 11(2):150. https://doi.org/10.3390/genes11020150
Chicago/Turabian StyleWillms, Inka M., Jingyue Yuan, Caterina Penone, Kezia Goldmann, Juliane Vogt, Tesfaye Wubet, Ingo Schöning, Marion Schrumpf, François Buscot, and Heiko Nacke. 2020. "Distribution of Medically Relevant Antibiotic Resistance Genes and Mobile Genetic Elements in Soils of Temperate Forests and Grasslands Varying in Land Use" Genes 11, no. 2: 150. https://doi.org/10.3390/genes11020150
APA StyleWillms, I. M., Yuan, J., Penone, C., Goldmann, K., Vogt, J., Wubet, T., Schöning, I., Schrumpf, M., Buscot, F., & Nacke, H. (2020). Distribution of Medically Relevant Antibiotic Resistance Genes and Mobile Genetic Elements in Soils of Temperate Forests and Grasslands Varying in Land Use. Genes, 11(2), 150. https://doi.org/10.3390/genes11020150






