Identification and Selection of Reference Genes for Quantitative Transcript Analysis in Corydalis yanhusuo
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Growth Environments
2.2. RNA Isolation and Complementary DNA Synthesis
2.3. Selection of Candidate Reference Genes and Primers Design
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.5. Statistical Analysis of Gene Expression Stability
2.6. Comprehensive Analysis and Validation of Selected Reference Genes
3. Results
3.1. Evaluation of Amplification Specificity and PCR Efficiency in C. yanhusuo
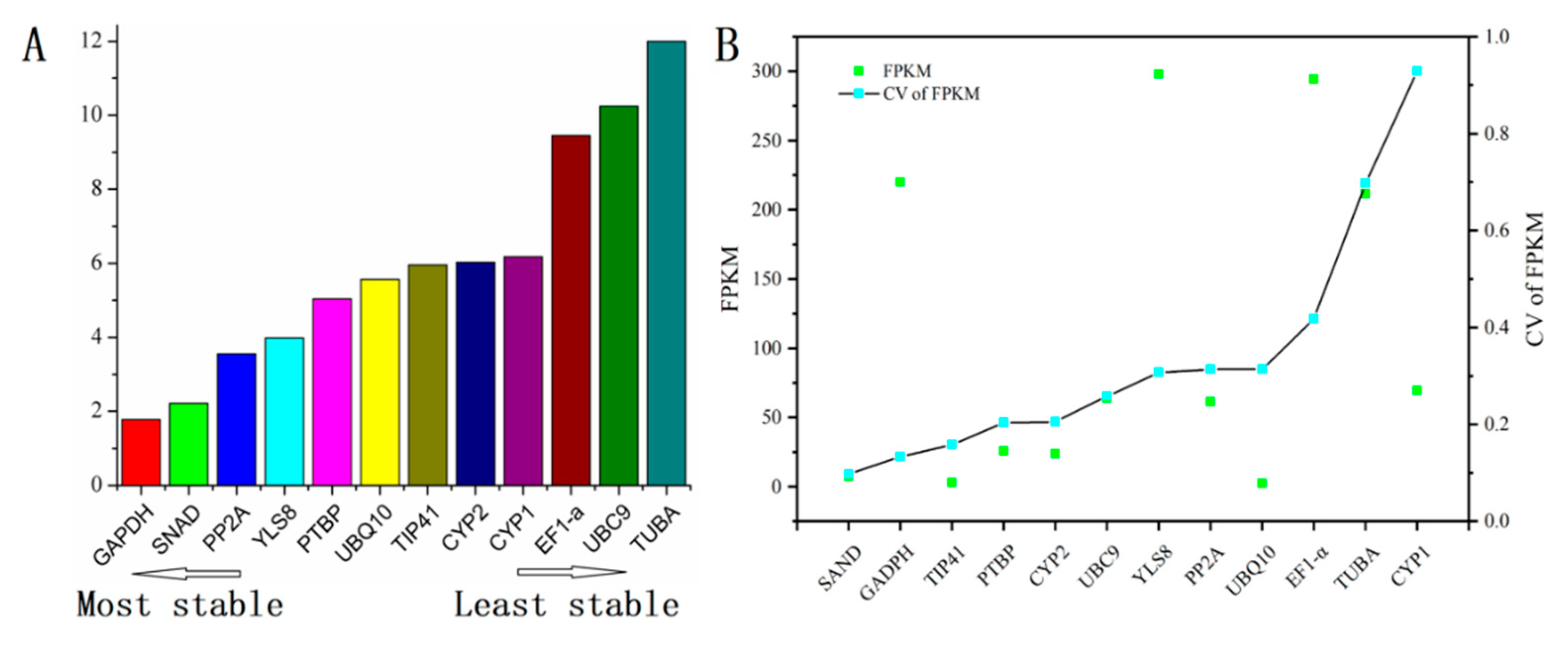
3.2. Expression Profiles of Reference Genes
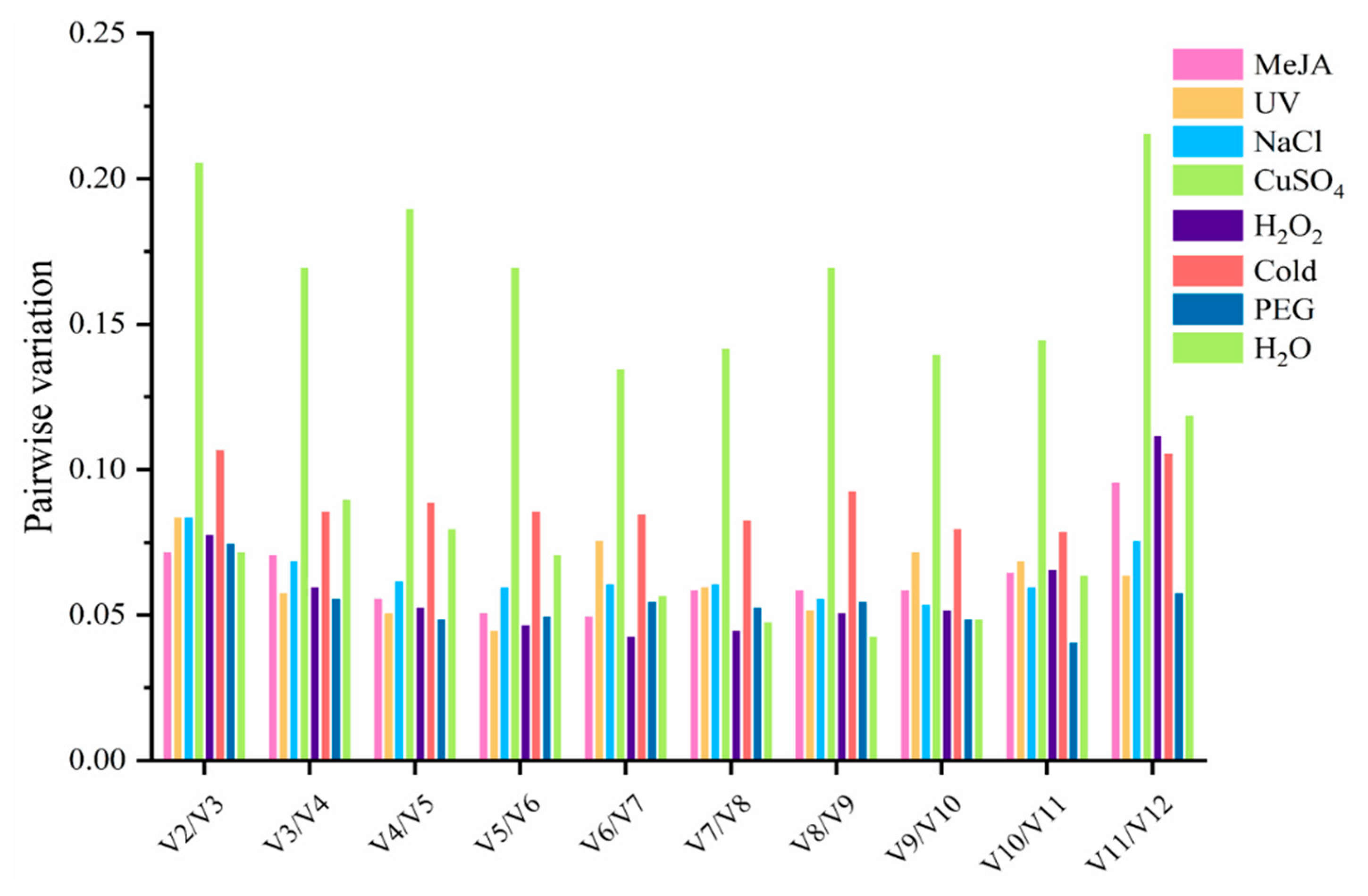
3.3. The Analysis of Expression Stability of Candidate Reference Genes
3.3.1. geNorm Analysis
3.3.2. NormFinder Analysis
3.3.3. BestKeeper Analysis
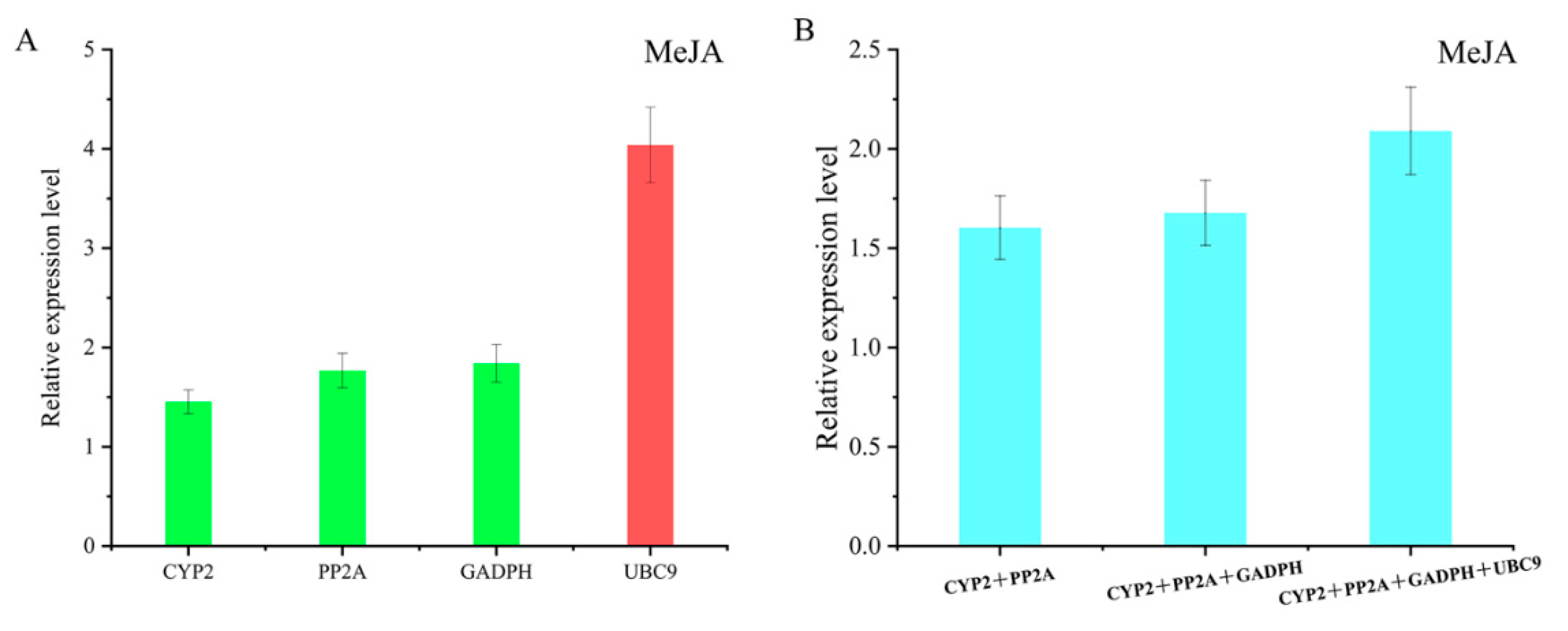
3.4. Comprehensive Analysis and Validation of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.B.; Mantsch, J.R. l-tetrahydropalamatine: A potential new medication for the treatment of cocaine addiction. Future Med. Chem. 2012, 4, 177–186. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lim, H.S.; Kim, Y.; Lee, J.; Kim, B.Y.; Jeong, S.J. Neuroprotective effect of Corydalis ternata extract and its phytochemical quantitative analysis. Chem. Pharm. Bull. (Tokyo) 2017, 65, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.L.; Shi, J.M.; Lee, S.M.; Zhang, Q.W.; Wang, Y.T. Angiogenic pathway inhibition of Corydalis yanhusuo and berberine in human umbilical vein endothelial cells. Oncol. Res. 2009, 17, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Wang, Z.; Gong, N.; Kweon, T.D.; Vo, B.; Wang, C.; Zhang, X.; Chung, J.Y.; Alachkar, A.; et al. The antinociceptive properties of the Corydalis yanhusuo extract. PLoS ONE 2016, 11, e0162875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, C.; Wang, L.; Parks, G.S.; Zhang, X.; Guo, Z.; Ke, Y.; Li, K.W.; Kim, M.K.; Vo, B.; et al. A novel analgesic isolated from a traditional Chinese medicine. Curr. Biol. 2014, 24, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, W.; Tang, Y.; Bai, W.; Yang, F.; Xie, L.; Li, X.; Zhou, S.; Pan, S.; Chen, Q.; et al. l-Tetrahydropalmatine, an active component of Corydalis yanhusuo W.T. Wang, protects against myocardial ischaemia-reperfusion injury in rats. PLoS ONE 2012, 7, e38627. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, Y.; Wu, W.; Huang, L.; Guo, D.; Liu, C. Approaches to establish Q-markers for the quality standards of traditional Chinese medicines. Acta Pharm. Sin. B 2017, 7, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, J.; Liu, Y.; Chen, J.; Lin, H.; Huang, Y.; Bian, X.; Zhao, Y. Selection and validation of appropriate reference genes for real-time quantitative PCR analysis in Momordica charantia. Phytochemistry 2019, 164, 1–11. [Google Scholar] [CrossRef]
- De Magalhães, J.P.; Finch, C.E.; Janssens, G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010, 9, 315–323. [Google Scholar]
- Stone, J.D.; Storchova, H. The application of RNA-seq to the comprehensive analysis of plant mitochondrial transcriptomes. Mol. Genet. Genom. 2015, 290, 1–9. [Google Scholar] [CrossRef]
- Bustin, S. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, J.; Li, Y.; Reid, N.; Langridge, P.; Shi, B.-J.; Tricker, P.J. Identification of reference genes for quantitative expression analysis of microRNAs and mRNAs in barley under various stress conditions. PLoS ONE 2015, 10, e0118503. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Goidin, D.; Mamessier, A.; Staquet, M.-J.; Schmitt, D.; Berthier-Vergnes, O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001, 295, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-j.; Altmann, S.W. mRNA and 18S–RNA coapplication–reverse transcription for quantitative gene expression analysis. Anal. Biochem. 2005, 345, 102–109. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. research0034.1-0034.11. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Meng, Y.; Moscou, M.J.; Wise, R.P. Blufensin1 negatively impacts basal defense in response to barley powdery mildew. Plant Physiol. 2009, 149, 271–285. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 2012, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef] [PubMed]
- Maroufi, A.; Van Bockstaele, E.; De Loose, M. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol. Biol. 2010, 11, 15. [Google Scholar] [CrossRef]
- Hoogewijs, D.; Houthoofd, K.; Matthijssens, F.; Vandesompele, J.; Vanfleteren, J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, H.M.; Simpson, R.S.; Casu, R.E.; Bonnett, G.D.; Maclean, D.J.; Manners, J.M. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol. Biol. Rep. 2004, 22, 325–337. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Q.-w.; Zhao, W.-J.; Du, Q.-Y.; Chang, Z.-J. Selection of suitable candidate genes for miRNA expression normalization in Yellow River Carp (Cyprinus carpio. var). Sci. Rep. 2019, 9, 8691. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Gen. 2014, 15, 648. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Eamens, A.L.; Grof, C.P. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods 2018, 14, 24. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, X.; Sun, M.; Cong, H.; Qiao, F. Evaluation of reference genes for normalizing RT-qPCR in leaves and suspension cells of Cephalotaxus hainanensis under various stimuli. Plant Methods 2019, 15, 31. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.; Ramakers, C.; Hoogaars, W.; Karlen, Y.; Bakker, O.; Van den Hoff, M.; Moorman, A. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucl. Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Ramadoss, N.; Gupta, D.; Vaidya, B.N.; Joshee, N.; Basu, C. Functional characterization of 1-aminocyclopropane-1-carboxylic acid oxidase gene in Arabidopsis thaliana and its potential in providing flood tolerance. Biochem. Biophys. Res. Commun. 2018, 503, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, T.; Gan, S.; Ren, X.; Fang, L.; Karungo, S.K.; Wang, Y.; Chen, L.; Li, S.; Xin, H. Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 2016, 6, 24066. [Google Scholar] [CrossRef]
- Tian, C.; Jiang, Q.; Wang, F.; Wang, G.-L.; Xu, Z.-S.; Xiong, A.-S. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 2015, 10, e0117569. [Google Scholar] [CrossRef]
- Gu, C.; Chen, S.; Liu, Z.; Shan, H.; Luo, H.; Guan, Z.; Chen, F. Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Mol. Biotechnol. 2011, 49, 192. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Han, X.; Zhang, J.; Sun, P.; Lu, M.; Hu, J. Selection of reliable reference genes for gene expression analysis under abiotic stresses in the desert biomass willow, Salix psammophila. Front. Plant Sci. 2016, 7, 1505. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mauriat, M.; Guénin, S.; Pelloux, J.; Lefebvre, J.F.; Louvet, R.; Rusterucci, C.; Moritz, T.; Guerineau, F.; Bellini, C. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008, 6, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Wang, F.; Jiang, Q.; Ma, J.; Xiong, A.-S. Identification of SSRs and differentially expressed genes in two cultivars of celery (Apium graveolens L.) by deep transcriptome sequencing. Horticult. Res. 2014, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Wang, P.; Jia, C.; Sun, P.; Qi, J.; Zhou, L.; Li, X.E. Identification and developmental expression profiling of putative alkaloid biosynthetic genes in Corydalis yanhusuo bulbs. Sci. Rep. 2016, 6, 19460. [Google Scholar] [CrossRef] [PubMed]
- Chandna, R.; Augustine, R.; Bisht, N.C. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE 2012, 7, e36918. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Ohl, F.; Jung, M.; Xu, C.; Stephan, C.; Rabien, A.; Burkhardt, M.; Nitsche, A.; Kristiansen, G.; Loening, S.A.; Radonić, A. Gene expression studies in prostate cancer tissue: Which reference gene should be selected for normalization? J. Mol. Med. 2005, 83, 1014–1024. [Google Scholar] [CrossRef]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 2015, 5, 788. [Google Scholar] [CrossRef]
- Borowski, J.M.; Galli, V.; da Silva Messias, R.; Perin, E.C.; Buss, J.H.; e Silva, S.D.d.A.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Galli, V.; da Silva Messias, R.; e Silva, S.D.d.A.; Rombaldi, C.V. Selection of reliable reference genes for quantitative real-time polymerase chain reaction studies in maize grains. Plant Cell Rep. 2013, 32, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Veazey, K.J.; Golding, M.C. Selection of stable reference genes for quantitative RT-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS ONE 2011, 6, e27592. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Gen. 2009, 10, 57. [Google Scholar] [CrossRef]


| Gene symbol | Description | Gene ID | Arabidopsis Homolog Locus | Primer Sequence Forward/Reverse (5′–3′) | Length (bp) | PCR Efficiency | R2 |
|---|---|---|---|---|---|---|---|
| CYP2 | Cyclophilin 2 | XP_008340167.1 | AT4G33060 | F: TGGTGCATCACTTGCTATGG R: GTTGTTTGGCTCCACCACTA | 164 | 1.848 | 0.960 |
| EF1-α | Elongation factor 1-α | XP_018856763.1 | AT1G07920 | F: CTGCCCCTTCAGGATGTTTA R: GCCTCGTGATGCATTTCAAC | 152 | 1.803 | 0.881 |
| PP2A | Serine/threonine-protein phosphatase PP2A | OVA18136.1 | ATG59830 | F: TCCCCATCTATCGAGACCCT R: GTCCTGGCCAAATGTGTATC | 124 | 1.828 | 0.960 |
| PTBP | Polypyrimidine tract-binding protein | OVA06588.1 | AT3G01150 | F: AGCCAGGGCAGTTGCTTATC R: CCAGGACAGTGCATCTTTCG | 134 | 1.799 | 0.841 |
| SAND | SAND family protein | XP_010260994.1 | AT2G28390 | F: AGATGGTGGCCTACGTGTTG R: GCCAATGTCAGCTTCCTTGA | 130 | 1.858 | 1.000 |
| TIP41 | TIP41-like protein | XP_010260049.1 | AT4G34270 | F: GTCATGCCGAGTTGTTGGTT R: AAATGTGGCTTCTCTCCAGC | 153 | 1.796 | 0.841 |
| UBC9 | Ubiquitin-conjugating enzyme 9 | OVA15929.1 | AT4G27960 | F: TGGCAAGCAACAATTATGGG R: GCAGATGCTTCCATTGCTGT | 159 | 1.788 | 0.841 |
| UBQ10 | Ubiquitin-conjugating enzyme 10 | XP_010261482.1 | AT4G05320 | F: CATCCAGAAGGAGTCTACCC R: AGCTTTCACGTTATCAATCG | 140 | 1.815 | 0.960 |
| CYP1 | Cyclophilin 1 | AAN31845.1 | AT2G16600 | F: TTCCAAAGTTTCAGAGTCCC R: CATGTGCTTGGGATTCAATC | 136 | 1.747 | 0.907 |
| TUBA | Tubulin beta | OVA16215.1 | AT5G12250 | F: TTGACCTCTGCTTAGACCGC R: GTGAACCCAATCCAGAACCA | 111 | 1.598 | 0.676 |
| YLS8 | Mitosis protein | KJB77370.1 | AT5G08290 | F: ACTTGTCGTAATTCGGTTCG R: CAACAAGGTAGATCACCGCA | 124 | 1.765 | 0.815 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | XP_010941981.2 | AT1G42970 | F: CAAGGTCATCAACGACAGGT R: TGCTGCTGGGAATGATGTTG | 149 | 1.860 | 1.000 |
| Rank | MeJA | UV | NaCl | CuSO4 | H2O2 | Cold | PEG | Control |
|---|---|---|---|---|---|---|---|---|
| 1 | UBQ10 0.024 | GAPDH 0.147 | PP2A 0.042 | TIP41 0.185 | CYP2 0.039 | PP2A 0.097 | PTBP 0.100 | UBQ10 0.168 |
| 2 | PTBP 0.054 | EF1-α 0.173 | EF1-α 0.063 | SAND 0.187 | GAPDH 0.055 | GAPDH 0.134 | PP2A 0.114 | SAND 0.183 |
| 3 | GAPDH 0.101 | UBC9 0.199 | CYP2 0.200 | GAPDH 0.207 | PP2A 0.076 | YLS8 0.182 | GAPDH 0.130 | PP2A 0.190 |
| 4 | EF1-α 0.156 | TIP41 0.207 | SAND 0.210 | EF1-α 0.382 | EF1-α 0.140 | SAND 0.267 | SAND 0.166 | PTBP 0.205 |
| 5 | PP2A 0.230 | UBQ10 0.220 | TIP41 0.217 | CYP2 0.531 | SAND 0.183 | UBQ10 0.277 | CYP2 0.196 | YLS8 0.216 |
| 6 | UBC9 0.240 | CYP2 0.278 | GAPDH 0.246 | YLS8 0.555 | TIP41 0.193 | TIP41 0.293 | EF1-α 0.229 | CYP2 0.230 |
| 7 | TIP41 0.251 | SAND 0.280 | YLS8 0.267 | UBC9 0.781 | PTBP 0.218 | CYP2 0.346 | YLS8 0.264 | EF1-α 0.236 |
| 8 | CYP1 0.306 | PP2A 0.317 | PTBP 0.308 | UBQ10 0.798 | YLS8 0.276 | UBC9 0.515 | CYP1 0.274 | GAPDH 0.270 |
| 9 | SAND 0.339 | PTBP 0.351 | UBC9 0.371 | PTBP 0.961 | UBQ10 0.285 | PTBP 0.557 | UBQ10 0.287 | UBC9 0.313 |
| 10 | CYP2 0.403 | YLS8 0.406 | UBQ10 0.372 | PP2A 0.969 | UBC9 0.339 | EF1-α 0.559 | TIP41 0.307 | TIP41 0.373 |
| 11 | YLS8 0.502 | CYP1 0.503 | CYP1 0.430 | CYP1 1.213 | CYP1 0.526 | CYP1 0.605 | UBC9 0.330 | CYP1 0.420 |
| 12 | TUBA 0.797 | TUBA 0.527 | TUBA 0.626 | TUBA 1.782 | TUBA 0.925 | TUBA 0.871 | TUBA 0.476 | TUBA 0.985 |
| Rank | MeJA | UV | NaCl | CuSO4 | H2O2 | Cold | PEG | Control (H2O) |
|---|---|---|---|---|---|---|---|---|
| 1 | UBC9 | YLS8 | SAND | YLS8 | SAND | YLS8 | PP2A | TIP41 |
| CV ± SD | 1.77 ± 0.38 | 1.50 ± 0.32 | 1.09 ± 0.23 | 2.25 ± 0.43 | 0.85 ± 0.18 | 0.86 ± 0.19 | 1.26 ± 0.24 | 6.15 ± 1.40 |
| 2 | CYP1 | UBC9 | YLS8 | GAPDH | YLS8 | TIP41 | TIP41 | TUBA |
| CV ± SD | 1.95 ± 0.35 | 1.50 ± 0.36 | 1.10 ± 0.22 | 2.28 ± 0.40 | 0.94 ± 0.19 | 1.29 ± 0.31 | 1.40 ± 0.33 | 6.64 ± 1.88 |
| 3 | PTBP | CYP1 | PP2A | SAND | EF1-α | UBC9 | PTBP | EF1-α |
| CV ± SD | 2.08 ± 0.41 | 1.55 ± 0.28 | 1.47 ± 0.26 | 2.82 ± 0.58 | 1.32 ± 0.30 | 1.96 ± 0.48 | 1.44 ± 0.30 | 7.06 ± 1.67 |
| 4 | YLS8 | UBQ10 | EF1-α | TIP41 | TIP41 | GAPDH | SAND | UBC9 |
| CV ± SD | 2.12 ± 0.43 | 2.22 ± 0.42 | 1.48 ± 0.32 | 2.82 ± 0.59 | 1.50 ± 0.32 | 2.14 ± 0.43 | 1.50 ± 0.32 | 7.56 ± 1.68 |
| 5 | TIP41 | TIP41 | TIP41 | UBC9 | PTBP | PP2A | CYP1 | GAPDH |
| CV ± SD | 2.21 ± 0.48 | 2.31 ± 0.5 | 1.61 ± 0.35 | 3.80 ± 0.75 | 1.61 ± 0.31 | 2.17 ± 0.46 | 1.65 ± 0.28 | 7.97 ± 1.63 |
| 6 | GAPDH | CYP2 | UBQ10 | EF1-α | CYP2 | PTBP | YLS8 | PTBP |
| CV ± SD | 2.24 ± 0.41 | 2.32 ± 0.41 | 1.74 ± 0.33 | 3.94 ± 0.83 | 1.76 ± 0.32 | 2.28 ± 0.52 | 1.84 ± 0.38 | 8.20 ± 1.70 |
| 7 | EF1-α | TUBA | PTBP | UBQ10 | PP2A | TUBA | EF1-α | SAND |
| CV ± SD | 2.24 ± 0.50 | 2.42 ± 0.54 | 2.38 ± 0.44 | 5.10 ± 0.92 | 2.05 ± 0.36 | 2.74 ± 0.74 | 2.11 ± 0.44 | 8.27 ± 1.86 |
| 8 | UBQ10 | EF1-α | CYP1 | PTBP | CYP1 | SAND | UBQ10 | UBQ10 |
| CV ± SD | 2.43 ± 0.47 | 2.73 ± 0.54 | 2.43 ± 0.40 | 5.72 ± 1.10 | 2.09 ± 0.36 | 2.75 ± 0.63 | 2.13 ± 0.38 | 8.29 ± 1.68 |
| 9 | SAND | GAPDH | GAPDH | TUBA | GAPDH | CYP2 | TUBA | CYP1 |
| CV ± SD | 3.18 ± 0.66 | 2.82 ± 0.52 | 2.60 ± 0.47 | 5.89 ± 1.49 | 2.37 ± 0.42 | 2.91 ± 0.62 | 2.24 ± 0.62 | 8.67 ± 1.63 |
| 10 | PP2A | SAND | CYP2 | CYP1 | UBQ10 | UBQ10 | GAPDH | YLS8 |
| CV ± SD | 3.40 ± 0.60 | 3.13 ± 0.69 | 2.66 ± 0.46 | 6.00 ± 1.02 | 3.35 ± 0.61 | 3.16 ± 0.65 | 2.28 ± 0.42 | 9.21 ± 2.01 |
| 11 | CYP2 | PTBP | TUBA | CYP2 | UBC9 | EF1-α | UBC9 | PP2A |
| CV ± SD | 3.50 ± 0.61 | 3.42 ± 0.70 | 2.68 ± 0.73 | 6.00 ± 1.06 | 3.36 ± 0.70 | 4.15 ± 0.85 | 2.32 ± 0.47 | 9.90 ± 1.96 |
| 12 | TUBA | PP2A | UBC9 | PP2A | TUBA | CYP1 | CYP2 | CYP2 |
| CV ± SD | 4.23 ± 1.02 | 3.59 ± 0.66 | 3.18 ± 0.66 | 6.93 ± 1.24 | 4.51 ± 1.23 | 4.49 ± 0.80 | 2.66 ± 0.49 | 10.26 ± 2.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, Z.; Zhang, K.; Lin, H.; Li, C.; Zhao, X.; Wu, J.; Nian, S. Identification and Selection of Reference Genes for Quantitative Transcript Analysis in Corydalis yanhusuo. Genes 2020, 11, 130. https://doi.org/10.3390/genes11020130
Bao Z, Zhang K, Lin H, Li C, Zhao X, Wu J, Nian S. Identification and Selection of Reference Genes for Quantitative Transcript Analysis in Corydalis yanhusuo. Genes. 2020; 11(2):130. https://doi.org/10.3390/genes11020130
Chicago/Turabian StyleBao, Zhenzhen, Kaidi Zhang, Hanfeng Lin, Changjian Li, Xiurong Zhao, Jie Wu, and Sihui Nian. 2020. "Identification and Selection of Reference Genes for Quantitative Transcript Analysis in Corydalis yanhusuo" Genes 11, no. 2: 130. https://doi.org/10.3390/genes11020130
APA StyleBao, Z., Zhang, K., Lin, H., Li, C., Zhao, X., Wu, J., & Nian, S. (2020). Identification and Selection of Reference Genes for Quantitative Transcript Analysis in Corydalis yanhusuo. Genes, 11(2), 130. https://doi.org/10.3390/genes11020130




