DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation
Abstract
1. Introduction
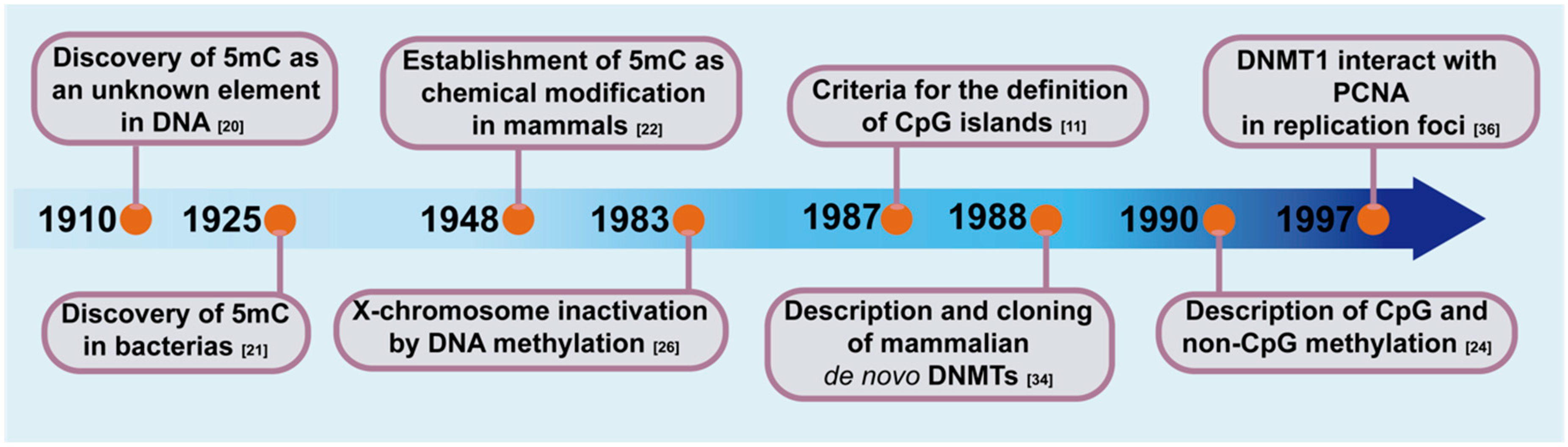
2. Discovery of 5-Methylcytosine and Its Function in the Genome
3. DNA Methyltransferases (DNMTs)
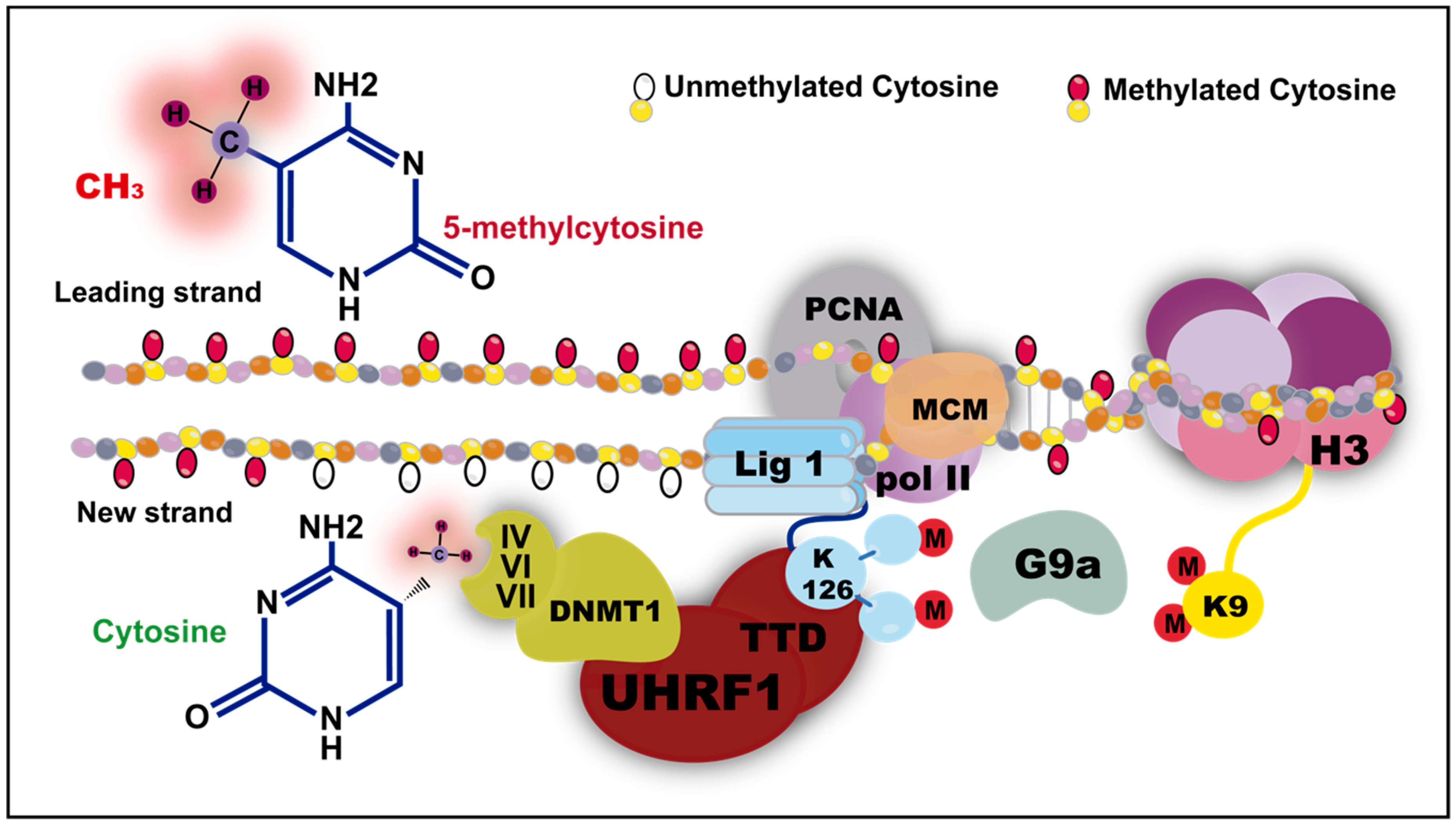
3.1. Characteristics and Function of DNMT1 (Maintenance Methylation)
3.2. Characteristics and Function of DNMT3A and DNMT3B (De Novo Methylation)
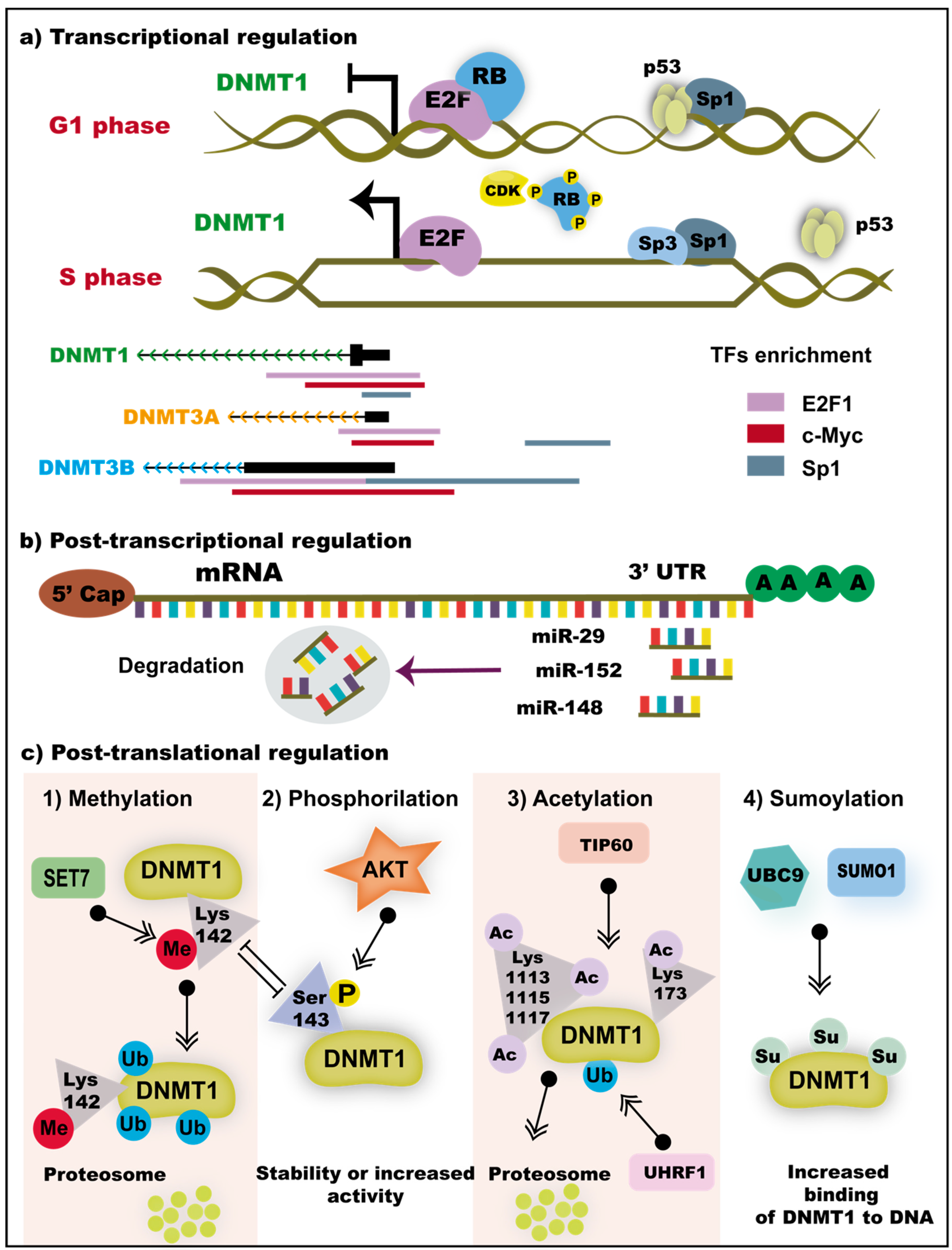
3.3. Transcriptional and Post-Transcriptional Regulation of DNMTs
4. Maintenance of the Methylation Machinery
5. Elements that Influence DNA Methylation
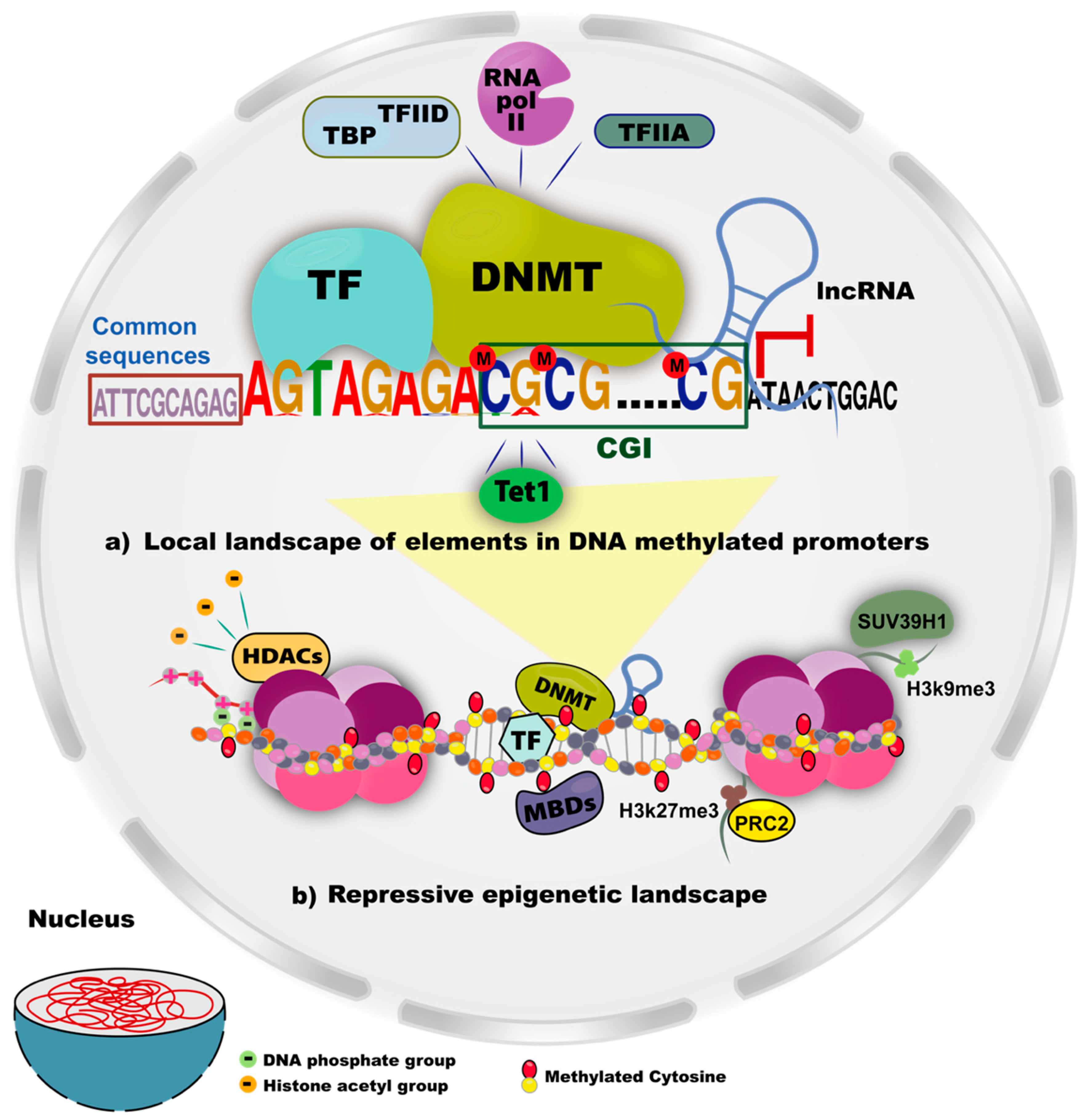
5.1. CpGs Content in Promoters
5.2. Transcription Factors Involved in DNA Methylation
5.3. Common Sequences in Methylated Genes
5.4. Long Non-Coding RNAs
| DNMT1 | DNMT3A | DNMT3B | |
|---|---|---|---|
| (a) Transcription factors | STAT3 [92] YY1 [96] Sp1 [96] SALL4 [97] PML-RAR [91] P53 [96] | SALL4 [97] MYC [95] PU.1 [93] | SALL4 [97] PU.1 [93] ZHX1 [94] MYC [98] PML-RAR [91] |
| (b) Common motifs | 5′TAAAAATACAAAAA3′ [100] 5′ATTAGCCGGG3′ [100] | 5′TNCpGNC3′ [101] 5′-TTGCCGGGCT3′ [100] 5′CACATCTGGACAGATGTGGGCG3′ [102] | 5′CTTGCGCAAG3′ [99] 5′NTCpGGN3′ [101] 5′CpGpG3′ [54] 5′CANAGCTG3′ [13] 5′GCAGCCGGCAT3′ [100] |
| (c) LncRNA | DALI [114] NEAT1 [15] DACOR [106]. Dum [14] LincRNA-p21 [111] EcCEBP [114] Kcnq1ot1 [112] HOTAIR [109,110] PARTICLE [108] | HOTAIR [109,110] Dum [14] | HOTAIR [109,110] Dum [14] |
5.5. Histone Methylation Patterns and DNMTs
6. Perspectives (De Novo DNMTs and Methylation Editing)
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schübeler, D. Function and information content of DNA methylation. Nat. Cell Biol. 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Stricker, S.H.; Köferle, A.; Beck, A.K.S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017, 18, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hore, T.A.; Reik, W. Reprogramming the Methylome: Erasing Memory and Creating Diversity. Cell Stem Cell 2014, 14, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Cedar, H.; Razin, A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nat. Cell Biol. 1982, 295, 620–622. [Google Scholar] [CrossRef] [PubMed]
- Rhee, I.; Bachman, K.E.; Park, B.H.; Jair, K.-W.; Yen, R.-W.C.; Schuebel, K.E.; Cui, H.; Feinberg, A.P.; Lengauer, C.; Kinzler, K.W.; et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nat. Cell Biol. 2002, 416, 552–556. [Google Scholar] [CrossRef]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef]
- Riethoven, J.-J.M. Regulatory Regions in DNA: Promoters, Enhancers, Silencers, and Insulators. Methods Mol. Biol. 2010, 674, 33–42. [Google Scholar] [CrossRef]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef]
- Shibata, M.; Gulden, F.O.; Sestan, N. From trans to cis: Transcriptional regulatory networks in neocortical development. Trends Genet. 2015, 31, 77–87. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG Islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Lienert, F.; Wirbelauer, C.; Som, I.; Dean, A.; Mohn, F.; Schübeler, D. Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 2011, 43, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Kamei, Y.; Ehara, T.; Yuan, X.; Suganami, T.; Takai-Igarashi, T.; Hatada, I.; Ogawa, Y. Analysis of DNA methylation change induced by Dnmt3b in mouse hepatocytes. Biochem. Biophys. Res. Commun. 2013, 434, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.-Y.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A.; et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am. J. Cancer Res. 2018, 8, 81–90. [Google Scholar]
- Schlesinger, Y.; Straussman, R.; Keshet, I.; Farkash, S.; Hecht, M.; Zimmerman, J.; Eden, E.; Yakhini, Z.; Ben-Shushan, E.; Reubinoff, B.E.; et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2006, 39, 232–236. [Google Scholar] [CrossRef]
- Dhayalan, A.; Rajavelu, A.; Rathert, P.; Tamas, R.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. The Dnmt3a PWWP Domain Reads Histone 3 Lysine 36 Trimethylation and Guides DNA Methylation. J. Biol. Chem. 2010, 285, 26114–26120. [Google Scholar] [CrossRef]
- Wheeler, H.L. Yale University. Sheffield Scientific School, Johnson TB. Papers on Pyrimidines [Internet]. New Haven, Conn; 1910. 744p. Available online: http://archive.org/details/papersonpyrimidi00wheerich (accessed on 29 August 2018).
- Johnson, T.B.; Coghill, R.D. researches on pyrimidines. C111. The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J. Am. Chem. Soc. 1925, 47, 2838–2844. [Google Scholar] [CrossRef]
- Hotchkiss, R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948, 175, 315–322. [Google Scholar] [PubMed]
- Just, T.; Waddington, C.H. An Introduction to Modern Genetics. Am. Midl. Nat. 1939, 22, 754. [Google Scholar] [CrossRef]
- Bestor, T.H. DNA methylation: Evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990, 326, 179–187. [Google Scholar] [PubMed]
- Dugaiczyk, A.; Hedgpeth, J.; Boyer, H.W.; Goodman, H.M. Physical identity of the SV40 deoxyribonucleic acid sequence recognized by the Eco RI restriction endonuclease and modification methylase. Biochemistry 1974, 13, 503–512. [Google Scholar] [CrossRef]
- Riggs, A. X inactivation, differentiation, and DNA methylation. Cytogenet. Genome Res. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Vanyushin, B.F.; Tkacheva, S.G.; Belozersky, A.N. Rare Bases in Animal DNA. Nat. Cell Biol. 1970, 225, 948–949. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Van Der Ploeg, L.H.; Flavell, R.A. DNA methylation in the human γ delta β -globin locus in erythroid and nonerythroid tissues. Cell 1980, 19, 947–958. [Google Scholar] [CrossRef]
- Jones, P.A.; Taylor, S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 1980, 20, 85–93. [Google Scholar] [CrossRef]
- Razin, A.; Riggs, A.D. DNA methylation and gene function. Science 1980, 210, 604–610. [Google Scholar] [CrossRef]
- Ye, F.; Kong, X.; Zhang, H.; Liu, Y.; Shao, Z.; Jin, J.; Cai, Y.; Zhang, R.; Li, L.; Zhang, Y.W.; et al. Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1. ACS Chem. Biol. 2018, 13, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Jurkowska, R.Z. Allosteric control of mammalian DNA methyltransferases—A new regulatory paradigm. Nucleic Acids Res. 2016, 44, 8556–8575. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.; Estève, P.-O.; Chin, H.G.; Samaranayke, M.; Kim, G.-D.; Pradhan, S. CXXC Domain of Human DNMT1 Is Essential for Enzymatic Activity. Biochemistry 2008, 47, 10000–10009. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.-H.; Ian, H.-I.; Koh, T.-W.; Ng, H.-H.; Xu, G.; Li, B.F.L. Human DNA-(Cytosine-5) Methyltransferase-PCNA Complex as a Target for p21WAF1. Science 1997, 277, 1996–2000. [Google Scholar] [CrossRef]
- Berkyurek, A.C.; Suetake, I.; Arita, K.; Takeshita, K.; Nakagawa, A.; Shirakawa, M.; Tajima, S. The DNA Methyltransferase Dnmt1 Directly Interacts with the SET and RING Finger-associated (SRA) Domain of the Multifunctional Protein Uhrf1 to Facilitate Accession of the Catalytic Center to Hemi-methylated DNA. J. Biol. Chem. 2014, 289, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Gifford, C.A.; Ziller, M.J.; Gu, H.; Trapnell, C.; Donaghey, J.; Tsankov, A.; Shalek, A.K.; Kelley, D.R.; Shishkin, A.A.; Issner, R.; et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell 2013, 153, 1149–1163. [Google Scholar] [CrossRef]
- Vilkaitis, G.; Suetake, I.; Klimašauskas, S.; Tajima, S. Processive Methylation of Hemimethylated CpG Sites by Mouse Dnmt1 DNA Methyltransferase. J. Biol. Chem. 2004, 280, 64–72. [Google Scholar] [CrossRef]
- Jeltsch, A. On the enzymatic properties of Dnmt1: Specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics 2006, 1, 63–66. [Google Scholar] [CrossRef]
- Bashtrykov, P.; Jankevicius, G.; Smarandache, A.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. Specificity of Dnmt1 for Methylation of Hemimethylated CpG Sites Resides in Its Catalytic Domain. Chem. Biol. 2012, 19, 572–578. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation, methyltransferases, and cancer. Oncogene 2001, 20, 3139–3155. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Bachman, K.E.; Herman, J.G.; Baylin, S.B. DNA methylation, chromatin inheritance, and cancer. Oncogene 2001, 20, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Gowher, H.; Loutchanwoot, P.; Vorobjeva, O.; Handa, V.; Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Mutational Analysis of the Catalytic Domain of the Murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 2006, 357, 928–941. [Google Scholar] [CrossRef]
- Norvil, A.B.; Petell, C.J.; Alabdi, L.; Wu, L.; Rossie, S.; Gowher, H. Dnmt3b Methylates DNA by a Noncooperative Mechanism, and Its Activity Is Unaffected by Manipulations at the Predicted Dimer Interface. Biochemistry 2016, 57, 4312–4324. [Google Scholar] [CrossRef]
- Rinaldi, L.; Datta, D.; Serrat, J.; Morey, L.; Solanas, G.; Avgustinova, A.; Blanco, E.; Pons, J.I.; Matallanas, D.; Von Kriegsheim, A.; et al. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell 2016, 19, 491–501. [Google Scholar] [CrossRef]
- Qiu, C.; Sawada, K.; Zhang, X.; Cheng, X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Genet. 2002, 9, 217–224. [Google Scholar] [CrossRef]
- Chen, T.; Tsujimoto, N.; Li, E. The PWWP Domain of Dnmt3a and Dnmt3b Is Required for Directing DNA Methylation to the Major Satellite Repeats at Pericentric Heterochromatin. Mol. Cell. Biol. 2004, 24, 9048–9058. [Google Scholar] [CrossRef]
- Jia, D.; Jurkowska, R.Z.; Zhang, X.; Jeltsch, A.; Cheng, X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nat. Cell Biol. 2007, 449, 248–251. [Google Scholar] [CrossRef]
- Ishida, C.; Ura, K.; Hirao, A.; Sasaki, H.; Toyoda, A.; Sakaki, Y.; Niwa, H.; Li, E.; Kaneda, Y. Genomic organization and promoter analysis of the Dnmt3b gene. Gene 2003, 310, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Wang, Z.; Okano, M.; Nogami, M.; Li, Y.; He, W.-W.; Okumura, K.; Li, E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 1999, 236, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.-L.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nat. Cell Biol. 2018, 554, 387–391. [Google Scholar] [CrossRef]
- Lin, C.-C.; Chen, Y.-P.; Yang, W.-Z.; Shen, J.C.K.; Yuan, H.S. Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B. Nucleic Acids Res. 2020, 48, 3949–3961. [Google Scholar] [CrossRef] [PubMed]
- Kinney, S.R.M.; Pradhan, S. Regulation of Expression and Activity of DNA (Cytosine-5) Methyltransferases in Mammalian Cells. Prog. Mol. Biol. Transl. Sci. 2011, 101, 311–333. [Google Scholar] [CrossRef] [PubMed]
- Bigey, P.; Ramchandani, S.; Theberge, J.; Araujo, F.D.; Szyf, M. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene 2000, 242, 407–418. [Google Scholar] [CrossRef]
- Robertson, K.D. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G0/G1 to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000, 28, 2108–2113. [Google Scholar] [CrossRef]
- Margot, J.B.; Cardoso, M.C.; Leonhardt, H. Mammalian DNA methyltransferases show different subnuclear distributions. J. Cell. Biochem. 2001, 83, 373–379. [Google Scholar] [CrossRef]
- McCabe, M.T.; Cabioglu, N.; Summy, J.; Miller, C.; Parikh, N.U.; Sahin, A.A.; Tuzlali, S.; Pumiglia, K.; Gallick, G.; Price, J.E. Regulation of DNA Methyltransferase 1 by the pRb/E2F1 Pathway. Cancer Res. 2005, 65, 3624–3632. [Google Scholar] [CrossRef]
- Lin, R.-K.; Wu, C.-Y.; Chang, J.-W.; Juan, L.-J.; Hsu, H.-S.; Chen, C.-Y.; Lu, Y.Y.; Tang, Y.A.; Yang, Y.C.; Yang, P.C.; et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010, 70, 5807–5817. [Google Scholar]
- Peterson, E.J.; Bögler, O.; Taylor, S.M. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res. 2003, 63, 6579–6582. [Google Scholar] [PubMed]
- Jinawath, A.; Miyake, S.; Yanagisawa, Y.; Akiyama, Y.; Yuasa, Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem. J. 2005, 385, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Huang, N.; Patel, T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010, 51, 881–890. [Google Scholar] [CrossRef]
- Huang, J.; Wang, Y.; Guo, Y.; Sun, S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase. Hepatology 2010, 52, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, Y.; Liang, Y.; Zhao, M.; Long, H.; Ding, S.; Yin, H.; Lu, Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase. Arthritis Rheum. 2011, 63, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, H.; Wang, Z.; Zhang, B.; Liu, W.; Lu, H.; Xiao, L.; Liu, X.; Wang, R.; Li, X.; et al. MiR-185 Targets the DNA Methyltransferases 1 and Regulates Global DNA Methylation in human glioma. Mol. Cancer 2011, 10, 124. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Jeong, W.; Lim, W.; Lim, C.-H.; Bae, S.-M.; Kim, J.; Bazer, F.W.; Song, G. Hypermethylation and Post-Transcriptional Regulation of DNA Methyltransferases in the Ovarian Carcinomas of the Laying Hen. PLoS ONE 2013, 8, e61658. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Duursma, A.M.; Kedde, M.; Schrier, M.; Le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 2008, 14, 872–877. [Google Scholar] [CrossRef]
- Sandhu, R.; Rivenbark, A.G.; Coleman, W.B. Loss of post-transcriptional regulation of DNMT3b by microRNAs: A possible molecular mechanism for the hypermethylation defect observed in a subset of breast cancer cell lines. Int. J. Oncol. 2012, 41, 721–732. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robaina, M.C.; Mazzoccoli, L.; Arruda, V.O.; Reis, F.R.D.S.; Apa, A.G.; De Rezende, L.M.M.; Klumb, E.M. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015, 98, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Yu, G.; Zhao, Y. MicroRNA-30a-3p inhibits the progression of lung cancer via the PI3K/AKT by targeting DNA methyltransferase 3a. OncoTargets Ther. 2019, 12, 7015–7024. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.; Chang, H.; Xu, G.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009, 41, 125–129. [Google Scholar] [CrossRef]
- Estève, P.-O.; Chin, H.G.; Benner, J.; Feehery, G.R.; Samaranayake, M.; Horwitz, G.A.; Jacobsen, S.E.; Pradhan, S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA 2009, 106, 5076–5081. [Google Scholar] [CrossRef]
- Estève, P.-O.; Chang, Y.; Samaranayake, M.; Upadhyay, A.K.; Horton, J.R.; Feehery, G.R.; Cheng, X.; Pradhan, S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat. Struct. Mol. Biol. 2011, 18, 42–48. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Du, Z.; Song, J.; Wang, Y.; Zhao, Y.; Guda, K.; Yang, S.; Kao, H.-Y.; Xu, Y.; Willis, J.; Markowitz, S.D.; et al. DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination. Sci. Signal. 2010, 3, ra80. [Google Scholar] [CrossRef]
- Lee, B.; Muller, M.T. SUMOylation enhances DNA methyltransferase 1 activity. Biochem. J. 2009, 421, 449–461. [Google Scholar] [CrossRef]
- Melchior, F. SUMO—Nonclassical Ubiquitin. Annu. Rev. Cell Dev. Biol. 2000, 16, 591–626. [Google Scholar] [CrossRef]
- Ferry, L.; Fournier, A.; Tsusaka, T.; Adelmant, G.; Shimazu, T.; Matano, S.; Kirsh, O.; Amouroux, R.; Dohmae, N.; Suzuki, T.; et al. Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol. Cell 2017, 67, 550–565.e5. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Mulholland, C.B.; Bultmann, S.; Kori, S.; Endo, A.; Saeki, Y.; Qin, W.; Trummer, C.; Chiba, Y.; Yokoyama, H.; et al. Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nat. Cell Biol. 2007, 450, 908–912. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Sci. 2007, 317, 1760–1764. [Google Scholar] [CrossRef]
- Rothbart, S.B.; Krajewski, K.; Nady, N.; Tempel, W.; Xue, S.; Badeaux, A.I.; Barsyte-Lovejoy, D.; Martinez, J.Y.; Bedford, M.T.; Fuchs, S.M.; et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012, 19, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Elango, N.; Yi, S.V. DNA Methylation and Structural and Functional Bimodality of Vertebrate Promoters. Mol. Biol. Evol. 2008, 25, 1602–1608. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.-H.; LeProust, E.; Park, I.-H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale methylomics reveals gene body signatures in human cell lines. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [PubMed]
- Landolin, J.M.; Johnson, D.S.; Trinklein, N.D.; Aldred, S.F.; Medina, C.; Shulha, H.; Weng, Z.; Myers, R.M. Sequence features that drive human promoter function and tissue specificity. Genome Res. 2010, 20, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Di Croce, L.; Raker, V.A.; Corsaro, M.; Fazi, F.; Fanelli, M.; Faretta, M.; Fuks, F.; Coco, F.L.; Kouzarides, T.; Nervi, C.; et al. Methyltransferase Recruitment and DNA Hypermethylation of Target Promoters by an Oncogenic Transcription Factor. Science 2002, 295, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.Y.; Marzec, M.; Raghunath, P.N.; Nagasawa, T.; Wasik, M.A. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl. Acad. Sci. USA 2005, 102, 6948–6953. [Google Scholar] [CrossRef]
- Suzuki, M.; Yamada, T.; KiharaNegishi, F.; Sakurai, T.; Hara, E.; Tenen, D.G.; Hozumi, N.; Oikawa, T. Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene 2006, 25, 2477–2488. [Google Scholar] [CrossRef]
- Kim, S.-H.; Park, J.; Choi, M.-C.; Kim, H.-P.; Park, J.-H.; Jung, Y.; Lee, J.-H.; Oh, -Y.; Im, S.-A.; Bang, Y.-J.; et al. Zinc-fingers and homeoboxes 1 (ZHX1) binds DNA methyltransferase (DNMT) 3B to enhance DNMT3B-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 2007, 355, 318–323. [Google Scholar] [CrossRef]
- Hervouet, E.; Vallette, F.M.; Cartron, P.-F. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics 2009, 4, 487–499. [Google Scholar] [CrossRef]
- Hervouet, E.; Vallette, F.M.; Cartron, P.-F. Dnmt1/Transcription Factor Interactions. Genes Cancer 2010, 1, 434–443. [Google Scholar] [CrossRef]
- Yang, J.; Corsello, T.R.; Ma, Y. Stem Cell Gene SALL4 Suppresses Transcription through Recruitment of DNA Methyltransferases. J. Biol. Chem. 2011, 287, 1996–2005. [Google Scholar] [CrossRef]
- Palakurthy, R.K.; Wajapeyee, N.; Santra, M.K.; Gazin, C.; Lin, L.; Gobeil, S.; Green, M.R. Epigenetic Silencing of the RASSF1A Tumor Suppressor Gene through HOXB3-Mediated Induction of DNMT3B Expression. Mol. Cell 2009, 36, 219–230. [Google Scholar] [CrossRef]
- Handa, V.; Jeltsch, A. Profound Flanking Sequence Preference of Dnmt3a and Dnmt3b Mammalian DNA Methyltransferases Shape the Human Epigenome. J. Mol. Biol. 2005, 348, 1103–1112. [Google Scholar] [CrossRef]
- Fan, H.; Zhao, Z.; Cheng, Y.; Cui, H.; Qiao, F.; Wang, L.; Hu, J.; Wu, H.; Song, W. Genome-wide profiling of DNA methylation reveals preferred sequences of DNMTs in hepatocellular carcinoma cells. Tumor Biol. 2015, 37, 877–885. [Google Scholar] [CrossRef]
- Wienholz, B.L.; Kareta, M.S.; Moarefi, A.H.; Gordon, C.A.; Ginno, P.A.; Chédin, F. DNMT3L Modulates Significant and Distinct Flanking Sequence Preference for DNA Methylation by DNMT3A and DNMT3B In Vivo. PLoS Genet. 2010, 6, e1001106. [Google Scholar] [CrossRef]
- Hu, S.; Xie, Z.; Onishi, A.; Yu, X.; Jiang, L.; Lin, J.; Rho, H.; Woodard, C.; Wang, H.; Jeong, J.-S.; et al. Profiling the Human Protein-DNA Interactome Reveals MAPK1 as a Transcriptional Repressor of Interferon Signalling. Cell 2009, 139, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.-H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.-M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, V.B.; Hain, S.; Maugg, D.; Smida, J.; Azimzadeh, O.; Tapio, S.; Ovsepian, S.V.; Atkinson, M.J. Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. 2017, 7, 1790. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Gao, H.; Tong, Y.; Yang, J.; Tang, R.; Niu, Y.; Li, M.; Guo, L. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab. Investig. 2015, 96, 60–68. [Google Scholar] [CrossRef]
- Cheng, D.; Deng, J.; Zhang, B.; He, X.; Meng, Z.; Li, G.; Ye, H.; Zheng, S.; Wei, L.; Deng, X.; et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 2018, 36, 159–170. [Google Scholar] [CrossRef]
- Baoming, Q.; Wu, H.; Zhu, X.; Guo, X.; Hutchins, A.P.; Luo, Z.; Song, H.; Chen, Y.; Lai, K.; Yin, M.; et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015, 25, 80–92. [Google Scholar] [CrossRef]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; Pontes, L.L.D.F.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nat. Cell Biol. 2013, 503, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife 2014, 3, e04530. [Google Scholar] [CrossRef] [PubMed]
- Viré, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.-M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nat. Cell Biol. 2006, 439, 871–874. [Google Scholar] [CrossRef]
- Zhang, Y.; Charlton, J.; Karnik, R.; Beerman, I.; Smith, Z.D.; Gu, H.; Weiderpass, E.; Mi, X.; Clement, M.K.; Pop, R.; et al. Targets and genomic constraints of ectopic Dnmt3b expression. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.J.; Carpenter, P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef]
- Lehnertz, B.; Ueda, Y.; Derijck, A.A.; Braunschweig, U.; Perez-Burgos, L.; Kubicek, S.; Chen, T.; Li, E.; Jenuwein, T.; Peters, A.H. Suv39h-Mediated Histone H3 Lysine 9 Methylation Directs DNA Methylation to Major Satellite Repeats at Pericentric Heterochromatin. Curr. Biol. 2003, 13, 1192–1200. [Google Scholar] [CrossRef]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nat. Cell Biol. 2007, 448, 714–717. [Google Scholar] [CrossRef]
- Kundaje, A.; Roadmap Epigenomics Consortium; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nat. Cell Biol. 2015, 518, 317–330. [Google Scholar] [CrossRef]
- Dawson, M.A. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science 2017, 355, 1147–1152. [Google Scholar] [CrossRef]
- Hwang, J.-Y.; Aromolaran, K.A.; Zukin, R.S. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 2017, 18, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C.O.; Peisach, E.; Grant, R.A. Design and Selection of Novel Cys2His2Zinc Finger Proteins. Annu. Rev. Biochem. 2001, 70, 313–340. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA Methylation in the Mammalian Genome. Cell 2016, 167, 233–247.e17. [Google Scholar] [CrossRef]
- Vojta, A.; Dobrinić, P.; Tadić, V.; Bočkor, L.; Korać, P.; Julg, B.; Klasić, M.; Zoldoš, V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loaeza-Loaeza, J.; Beltran, A.S.; Hernández-Sotelo, D. DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation. Genes 2020, 11, 1336. https://doi.org/10.3390/genes11111336
Loaeza-Loaeza J, Beltran AS, Hernández-Sotelo D. DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation. Genes. 2020; 11(11):1336. https://doi.org/10.3390/genes11111336
Chicago/Turabian StyleLoaeza-Loaeza, Jaqueline, Adriana S. Beltran, and Daniel Hernández-Sotelo. 2020. "DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation" Genes 11, no. 11: 1336. https://doi.org/10.3390/genes11111336
APA StyleLoaeza-Loaeza, J., Beltran, A. S., & Hernández-Sotelo, D. (2020). DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation. Genes, 11(11), 1336. https://doi.org/10.3390/genes11111336






