Transcriptional Analysis of Carotenoids Accumulation and Metabolism in a Pink-Fleshed Lemon Mutant
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Carotenoid Extraction and Quantification by HPLC-PDA
2.3. Gene Expression Analysis by Quantitative Real-Time PCR
2.4. Statistical Analysis
3. Results
3.1. Phenotypic Characteristics of the Pink Lemon Fruit
3.2. Carotenoids Content and Composition in Pink Lemon Fruit
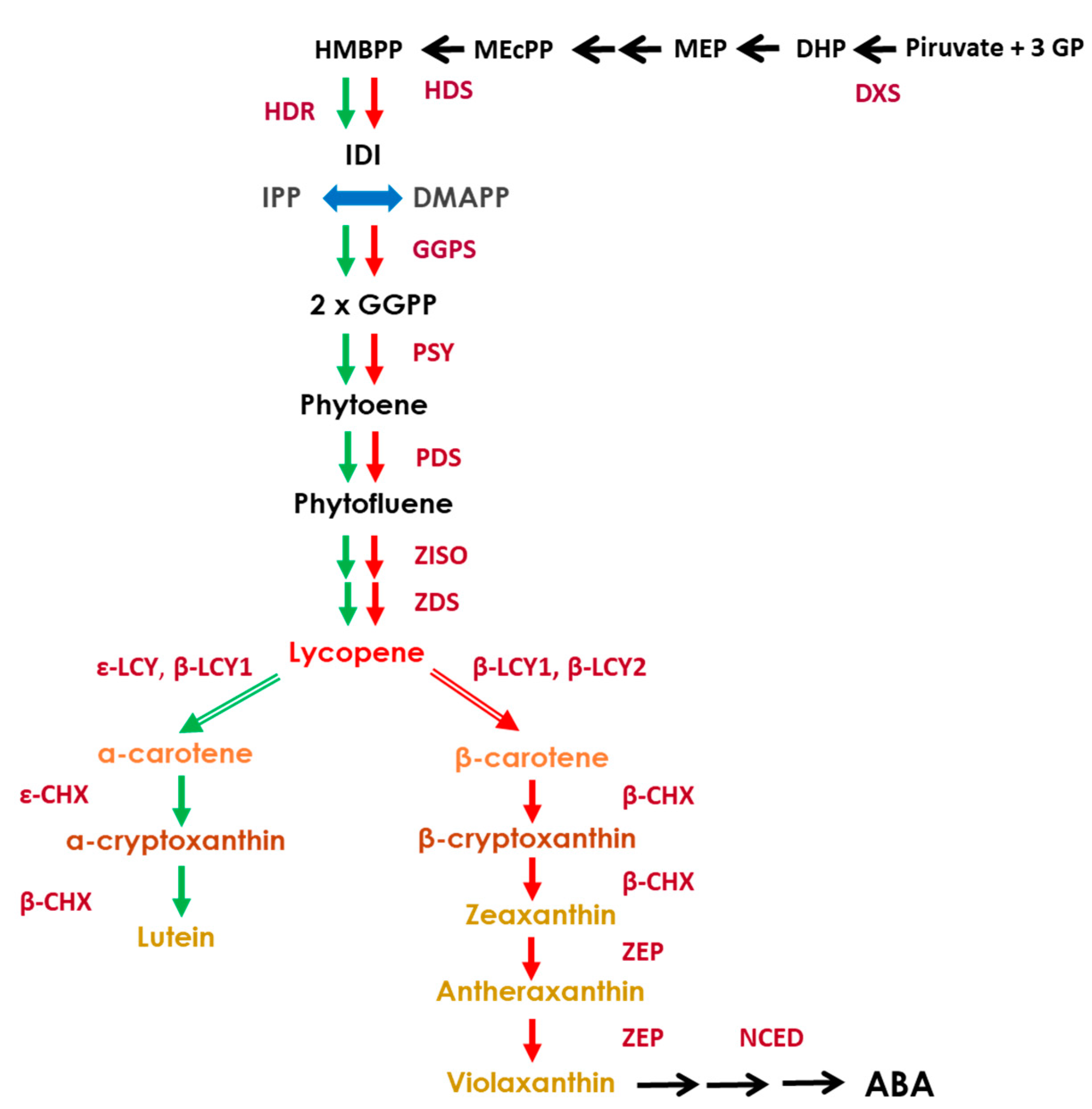
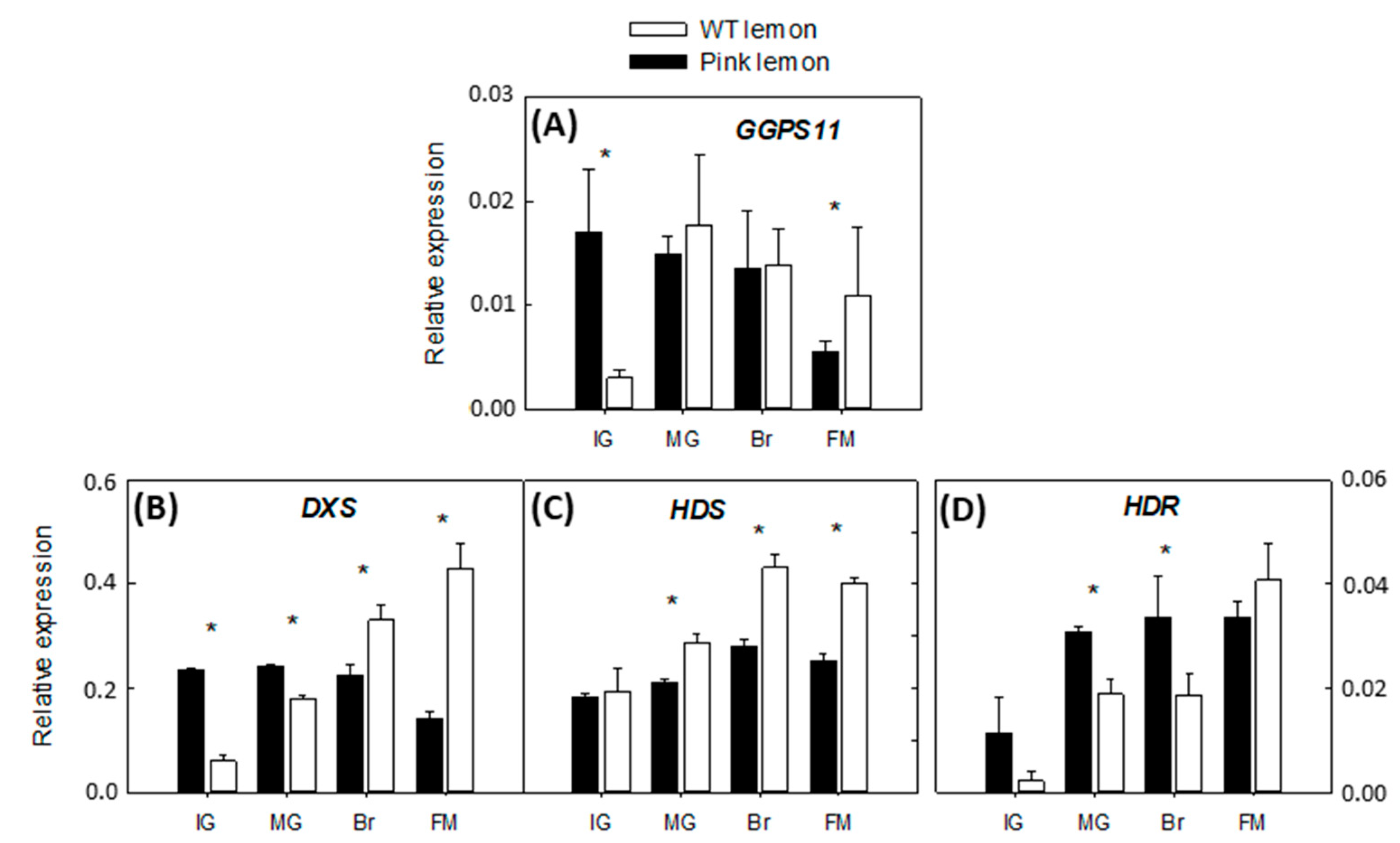
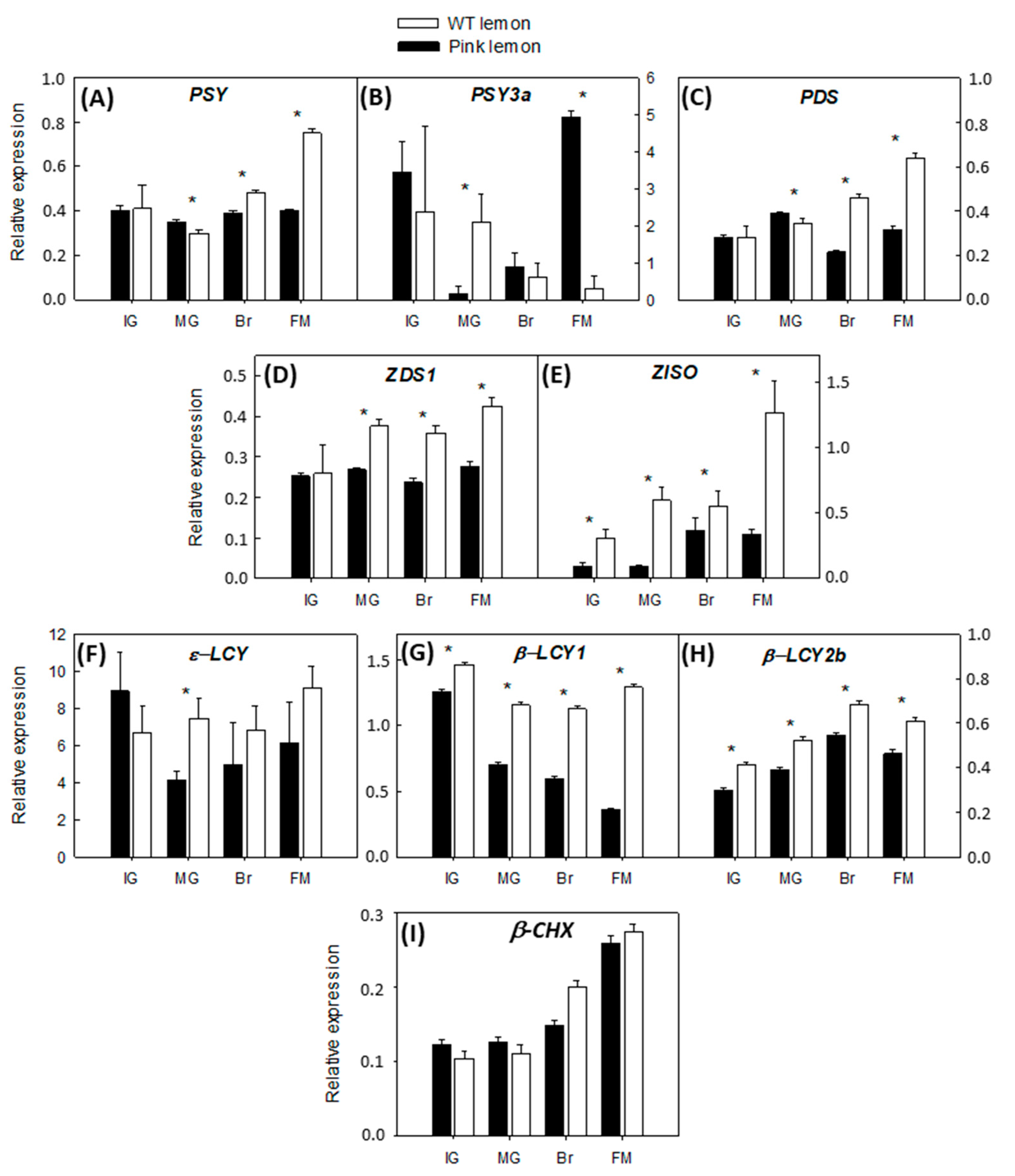
3.3. Expression of the Genes Involved in the Biochemical Pathway of Carotenoids
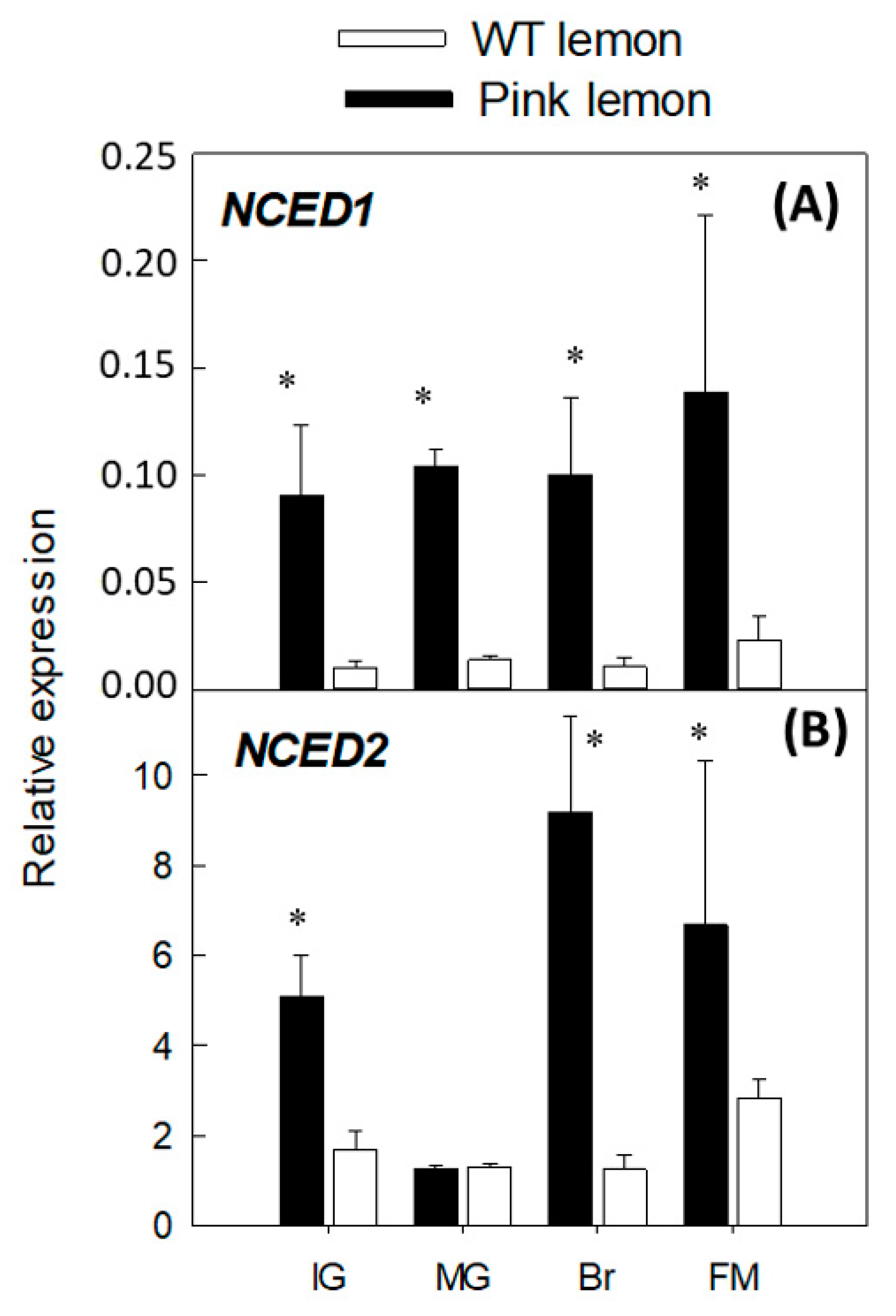
3.4. Expression of Genes Involved in the Biosynthesis of Abscisic Acid
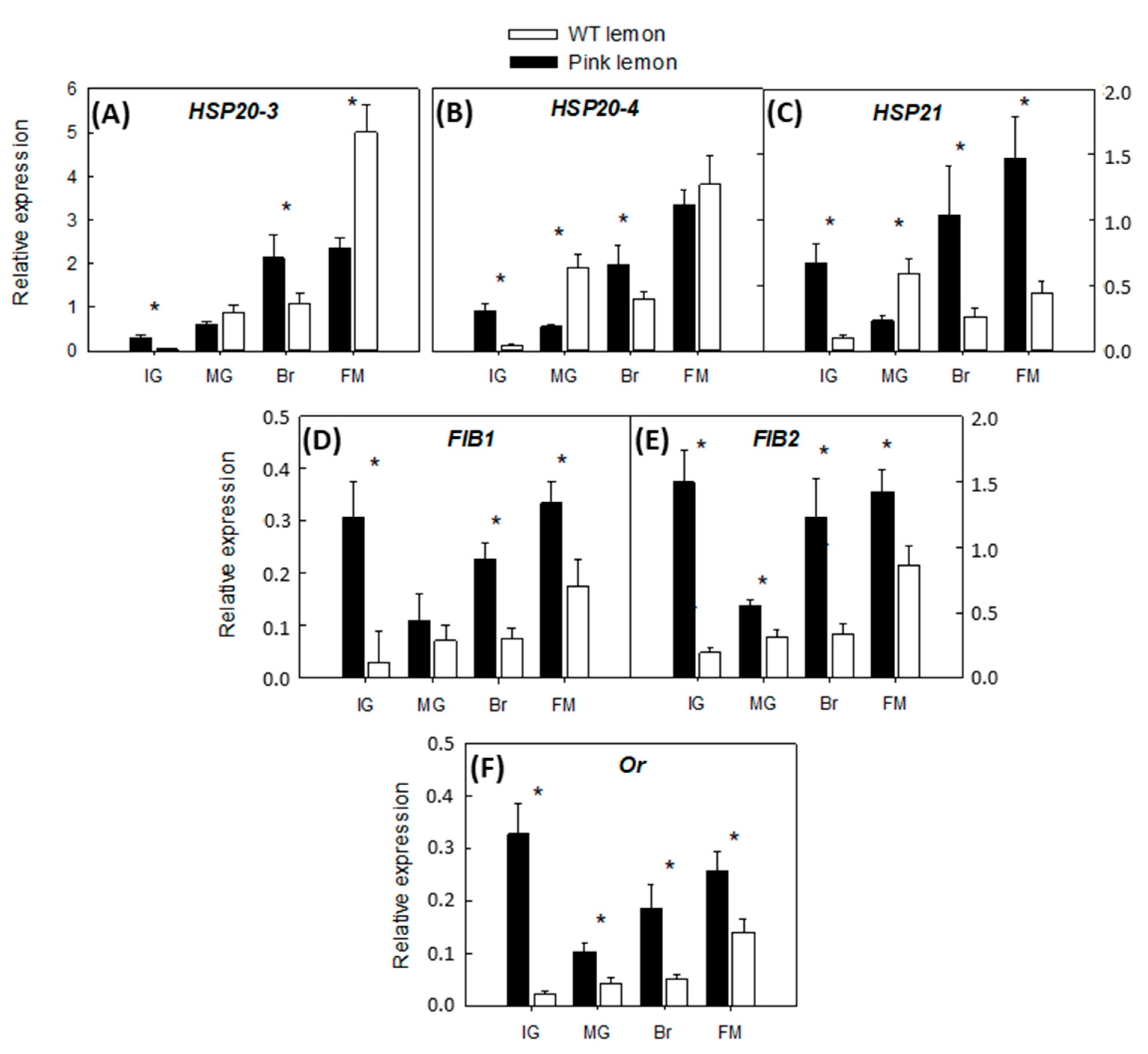
3.5. Expression of Accessory Genes Involved in the Accumulation of Carotenoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Woodside, J.V.; McGrath, A.J.; Lyner, N.; McKinley, M.C. Carotenoids and health in older people. Maturitas 2015, 80, 63–68. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquézar, B.; Alós, E.; Lado, J.; Zacarías, L. Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci. Hortic. 2013, 163, 46–62. [Google Scholar] [CrossRef]
- Lado, J.; Cronje, P.; Alquézar, B.; Page, A.; Manzi, M.; Gómez-Cadenas, A.; Stead, A.D.; Zacarías, L.; Rodrigo, M.J. Fruit shading enhances peel color, carotenes accumulation and chromoplast differentiation in red grapefruit. Physiol. Plant. 2015, 154, 469–484. [Google Scholar] [CrossRef]
- Tadeo, F.R.; Terol, J.; Rodrigo, M.J.; Licciardello, C.; Sadka, A. Fruit growth and development. In The Citrus Book; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 245–270. [Google Scholar]
- Ma, G.; Zhang, L.; Sugiura, M.; Kato, M. Citrus and health. In The Citrus Book; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 495–511. [Google Scholar]
- Gross, J. Carotenoids: Pigments in Fruits; London Academic Press: London, UK, 1987. [Google Scholar]
- Rodríguez-Concepción, M. Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 2010, 504, 118–122. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2. [Google Scholar] [CrossRef]
- Ahrazem, O.; Gómez-Gómez, L.; Rodrigo, M.J.; Avalos, J.; Limón, M.C. Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: Features and functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef]
- Sun, T.; Li, L. Toward the ‘golden’ era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2020, 290. [Google Scholar] [CrossRef]
- Tatmala, N.; Ma, G.; Zhang, L.; Kato, M.; Kaewsuksaeng, S. Characterization of carotenoid accumulation and carotenogenic gene expression during fruit ripening in red colored pulp of ‘siam red ruby’ pumelo (Citrus grandis) cultivated in thailand. Hortic. J. 2020, 89, 237–243. [Google Scholar] [CrossRef]
- Ikoma, Y.; Matsumoto, H.; Kato, M. Diversity in the carotenoid profiles and the expression of genes related to carotenoid accumulation among citrus genotypes. Breed. Sci. 2016, 66, 139–147. [Google Scholar] [CrossRef]
- Mendes, A.F.S.; Chen, C.; Gmitter, F.G.; Moore, G.A.; Costa, M.G.C. Expression and phylogenetic analysis of two new lycopene β-cyclases from Citrus paradisi. Physiol. Plant. 2011, 141, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alquézar, B.; Zacarías, L.; Rodrigo, M.J. Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J. Exp. Bot. 2009, 60, 1783–1797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, G.; Shirai, Y.; Kato, M.; Yamawaki, K.; Ikoma, Y.; Matsumoto, H. Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta 2012, 236, 1315–1325. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Yungyuen, W.; Tsukamoto, I.; Iijima, N.; Oikawa, M.; Yamawaki, K.; Yahata, M.; Kato, M. Expression and functional analysis of citrus carotene hydroxylases: Unravelling the xanthophyll biosynthesis in citrus fruits. BMC Plant Biol. 2016, 16, 1–12. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Matsuta, A.; Matsutani, K.; Yamawaki, K.; Yahata, M.; Wahyudi, A.; Motohashi, R.; Kato, M. Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 2013, 163, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, F.; Zheng, X.; Xie, Z.; Ye, J.; Cheng, Y.; Deng, X.; Zeng, Y. Investigation of chromoplast ultrastructure and tissue-specific accumulation of carotenoids in citrus flesh. Sci. Hortic. 2019, 256. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquezar, B.; Zacarías, L. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 2006, 57, 633–643. [Google Scholar] [CrossRef]
- Agustí, J.; Zapater, M.; Iglesias, D.J.; Cercós, M.; Tadeo, F.R.; Talón, M. Differential expression of putative 9-cis-epoxycarotenoid dioxygenases and abscisic acid accumulation in water stressed vegetative and reproductive tissues of citrus. Plant Sci. 2007, 172, 85–94. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef]
- Neta-sharir, I.; Isaacson, T.; Lurie, S.; Weiss, D. Dual Role for Tomato Heat Shock Protein 21: Protecting Photosystem II from Oxidative Stress and Promoting Color Changes during Fruit Maturation. Plant Cell 2005, 17, 1829–1838. [Google Scholar] [CrossRef]
- Simkin, A.J.; Gaffé, J.; Alcaraz, J.P.; Carde, J.P.; Bramley, P.M.; Fraser, P.D.; Kuntz, M. Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 2007, 68, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Zhou, X.; Yuan, H.; Álvarez, D.; Sun, T.; Schlossarek, D.; Yang, Y.; Shen, G.; Zhang, H.; Rodriguez-Concepcion, M.; et al. Clp Protease and OR Directly Control the Proteostasis of Phytoene Synthase, the Crucial Enzyme for Carotenoid Biosynthesis in Arabidopsis. Mol. Plant 2018, 11, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Ikoma, Y.; Matsumoto, H.; Sugiura, M.; Hyodo, H.; Yano, M. Accumulation of Carotenoids and Expression of Carotenoid Biosynthetic Genes during Maturation in Citrus Fruit. Plant Physiol. 2004, 134, 824–837. [Google Scholar] [CrossRef]
- Alquezar, B.; Rodrigo, M.J.; Zacarías, L. Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 2008, 69, 1997–2007. [Google Scholar] [CrossRef]
- Lu, P.J.; Wang, C.Y.; Yin, T.T.; Zhong, S.L.; Grierson, D.; Chen, K.S.; Xu, C.J. Cytological and molecular characterization of carotenoid accumulation in normal and high-lycopene mutant oranges. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Xu, Q.; Yu, K.; Zhu, A.; Ye, J.; Liu, Q.; Zhang, J.; Deng, X. Comparative transcripts profiling reveals new insight into molecular processes regulating lycopene accumulation in a sweet orange (Citrus sinensis) red-flesh mutant. BMC Genom. 2009, 10, 1–15. [Google Scholar] [CrossRef]
- Yu, K.; Xu, Q.; Da, X.; Guo, F.; Ding, Y.; Deng, X. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genom. 2012, 13. [Google Scholar] [CrossRef]
- Lu, S.; Van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Conlin, B.J.; Paolillo, D.J.; Garvin, D.F.; Vrebalov, J.; et al. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef] [PubMed]
- Alquezar, B.; Rodrigo, M.J.; Lado, J.; Zacarías, L. A comparative physiological and transcriptional study of carotenoid biosynthesis in white and red grapefruit (Citrus paradisi Macf.). Tree Genet. Genomes 2013, 9, 1257–1269. [Google Scholar] [CrossRef]
- Liu, W.; Ye, Q.; Jin, X.; Han, F.; Huang, X.; Cai, S.; Yang, L. A spontaneous bud mutant that causes lycopene and β-carotene accumulation in the juice sacs of the parental Guanxi pummelo fruits (Citrus grandis (L.) Osbeck). Sci. Hortic. 2016, 198, 379–384. [Google Scholar] [CrossRef]
- Yan, F.; Shi, M.; He, Z.; Wu, L.; Xu, X.; He, M.; Chen, J.; Deng, X.; Cheng, Y.; Xu, J. Largely different carotenogenesis in two pummelo fruits with different flesh colors. PLoS ONE 2018, 13, e0200320. [Google Scholar] [CrossRef] [PubMed]
- Promkaew, P.; Pongprasert, N.; Wongs-Aree, C.; Kaewsuksaeng, S.; Opio, P.; Kondo, S.; Srilaong, V. Carotenoids accumulation and carotenoids biosynthesis gene expression during fruit development in pulp of Tubtim-Siam pummelo fruit. Sci. Hortic. 2020, 260, 108870. [Google Scholar] [CrossRef]
- Shamel, A.D. A pink-fruited lemon. In Citrus-Fruit Improvement: A Study of Bud Variation in the Eureka Lemon; US Department of Agriculture: Washington, DC, USA, 1932; Volume 23, pp. 23–27. [Google Scholar]
- Stewart, I.; Wheaton, T.A. Carotenoids in Citrus: Their Accumulation Induced by Ethylene. J. Agric. Food Chem. 1972, 20, 448–449. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Cilla, A.; Barberá, R.; Zacarías, L. Carotenoid bioaccessibility in pulp and fresh juice from carotenoid-rich sweet oranges and mandarins. Food Funct. 2015, 6, 1950–1959. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Gurrea, A.; Page, A.; Stead, A.; Rodrigo, M.J. Exploring the diversity in Citrus fruit colouration to decipher the relationship between plastid ultrastructure and carotenoid composition. Planta 2015, 242, 645–661. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquézar, B.; Alós, E.; Medina, V.; Carmona, L.; Bruno, M.; Al-Babili, S.; Zacarías, L. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013, 64, 4461–4478. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfel, L. Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR; Oxford University Press: Oxford, UK, 2002; Volume 30, pp. 9–36. [Google Scholar]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Differential transcriptional regulation of l-ascorbic acid content in peel and pulp of citrus fruits during development and maturation. Planta 2014, 239, 1113–1128. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Lado, J.; Alós, E.; Alquézar, B.; Dery, O.; Hirschberg, J.; Zacarías, L. A mutant allele of ζ-carotene isomerase (Z-ISO) is associated with the yellow pigmentation of the “pinalate” sweet orange mutant and reveals new insights into its role in fruit carotenogenesis. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Rodrigo, M.J. Regulation of Carotenoid Biosynthesis during Fruit Development. In Carotenoids in Nature; Springer International Publishing: Cham, Switzerland, 2016; pp. 161–198. [Google Scholar]
- Xu, C.J.; Fraser, P.D.; Wang, W.J.; Bramley, P.M. Differences in the carotenoid content of ordinary Citrus and lycopene-accumulating mutants. J. Agric. Food Chem. 2006, 54, 5474–5481. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xu, J.; Liu, Y.; Zhao, X.; Deng, X.; Guo, L.; Gu, J. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 2007, 58, 4161–4171. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Kato, M.; Matsumoto, H.; Ikoma, Y.; Okuda, H.; Yano, M. The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. J. Exp. Bot. 2006, 57, 2153–2164. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Seo, M.; Marion-Poll, A. Abscisic acid metabolism and transport. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2019; Volume 92, pp. 1–49. [Google Scholar] [CrossRef]
- van Wijk, K.; Kessler, F. Plastoglobuli: Plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef]
- Wurtzel, E.T. Changing form and function through carotenoids and synthetic biology. Plant Physiol. 2019, 179, 830–843. [Google Scholar] [CrossRef]
- Singh, D.K.; McNellis, T.W. Fibrillin protein function: The tip of the iceberg? Trends Plant Sci. 2011, 16, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Kilcrease, J.; Rodriguez-Uribe, L.; Richins, R.D.; Arcos, J.M.G.; Victorino, J.; O’Connell, M.A. Correlations of carotenoid content and transcript abundances for fibrillin and carotenogenic enzymes in Capsicum annum fruit pericarp. Plant Sci. 2015, 232, 57–66. [Google Scholar] [CrossRef]
- Osorio, C.E. The Role of Orange Gene in Carotenoid Accumulation: Manipulating Chromoplasts toward a Colored Future. Front. Plant Sci. 2019, 10, 1235. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, M.; Sun, Z.; Yuan, H.; Zeng, S.; Thannhauser, T.W.; Vrebalov, J.; Ma, Q.; Xu, Y.; Fei, Z.; Van Eck, J.; et al. Ectopic expression of ORANGE promotes carotenoid accumulation and fruit development in tomato. Plant Biotechnol. J. 2019, 17, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, Y.; Xu, Q.; Owsiany, K.; Welsch, R.; Chitchumroonchokchai, C.; Lu, S.; Van Eck, J.; Deng, X.X.; Failla, M.; et al. The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol. Plant 2012, 5, 339–352. [Google Scholar] [CrossRef] [PubMed]

| Pink Lemon | Wild Type | |||||||
|---|---|---|---|---|---|---|---|---|
| Carotenoids (µg/g FW) | IG | MG | BR | FM | IM | MG | BR | FM |
| Phytoene | 17.59 ± 1.12 | 44.01 ± 0.90 | 4.98 ± 0.30 | 8.81 ± 0.07 | tr. | tr. | 0.04 ± 0.01 | 0.04 ± 0.01 |
| Phytofluene | 2.27 ± 0.17 | 8.93 ± 1.75 | 1.00 ± 0.01 | 1.79 ± 0.01 | nd | nd | nd | tr. |
| ζ-carotene | nd | 0.05 ± 0.01 | nd | nd | nd | nd | nd | nd |
| Neurosporene | 0.06 ± 0.01 | 0.24 ± 0.03 | nd | nd | nd | nd | nd | nd |
| Lycopene | 0.24 ± 0.01 | 0.49 ± 0.13 | tr. | 0.02±0.01 | nd | nd | nd | nd |
| δ-carotene | tr. | 0.04 ± 0.01 | nd | nd | nd | nd | nd | nd |
| Lutein | 0.06 ± 0.01 | nd | nd | nd | tr. | tr. | tr. | tr. |
| β-carotene | nd | nd | nd | nd | nd | tr | nd | tr. |
| β-cryptoxanthin | 0.02 ± 0.01 | nd | 0.02 ± 0.01 | nd | nd | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| Anteraxanthin | nd | nd | nd | nd | nd | nd | tr. | tr. |
| Violaxanthin | nd | tr | nd | nd | nd | tr. | nd | tr. |
| Total carotenoids α | 20.24 ± 1.54 | 53.33 ± 2.17 | 6.01 ± 0.29 | 10.61 ± 0.03 | tr. | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.08 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lana, G.; Zacarias-Garcia, J.; Distefano, G.; Gentile, A.; Rodrigo, M.J.; Zacarias, L. Transcriptional Analysis of Carotenoids Accumulation and Metabolism in a Pink-Fleshed Lemon Mutant. Genes 2020, 11, 1294. https://doi.org/10.3390/genes11111294
Lana G, Zacarias-Garcia J, Distefano G, Gentile A, Rodrigo MJ, Zacarias L. Transcriptional Analysis of Carotenoids Accumulation and Metabolism in a Pink-Fleshed Lemon Mutant. Genes. 2020; 11(11):1294. https://doi.org/10.3390/genes11111294
Chicago/Turabian StyleLana, Giuseppe, Jaime Zacarias-Garcia, Gaetano Distefano, Alessandra Gentile, María J. Rodrigo, and Lorenzo Zacarias. 2020. "Transcriptional Analysis of Carotenoids Accumulation and Metabolism in a Pink-Fleshed Lemon Mutant" Genes 11, no. 11: 1294. https://doi.org/10.3390/genes11111294
APA StyleLana, G., Zacarias-Garcia, J., Distefano, G., Gentile, A., Rodrigo, M. J., & Zacarias, L. (2020). Transcriptional Analysis of Carotenoids Accumulation and Metabolism in a Pink-Fleshed Lemon Mutant. Genes, 11(11), 1294. https://doi.org/10.3390/genes11111294







