Functional Diversification of the Dihydroflavonol 4-Reductase from Camellia nitidissima Chi. in the Control of Polyphenol Biosynthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Cloning of CnDFR
2.3. Sequence Alignment and Phylogenetic Analysis
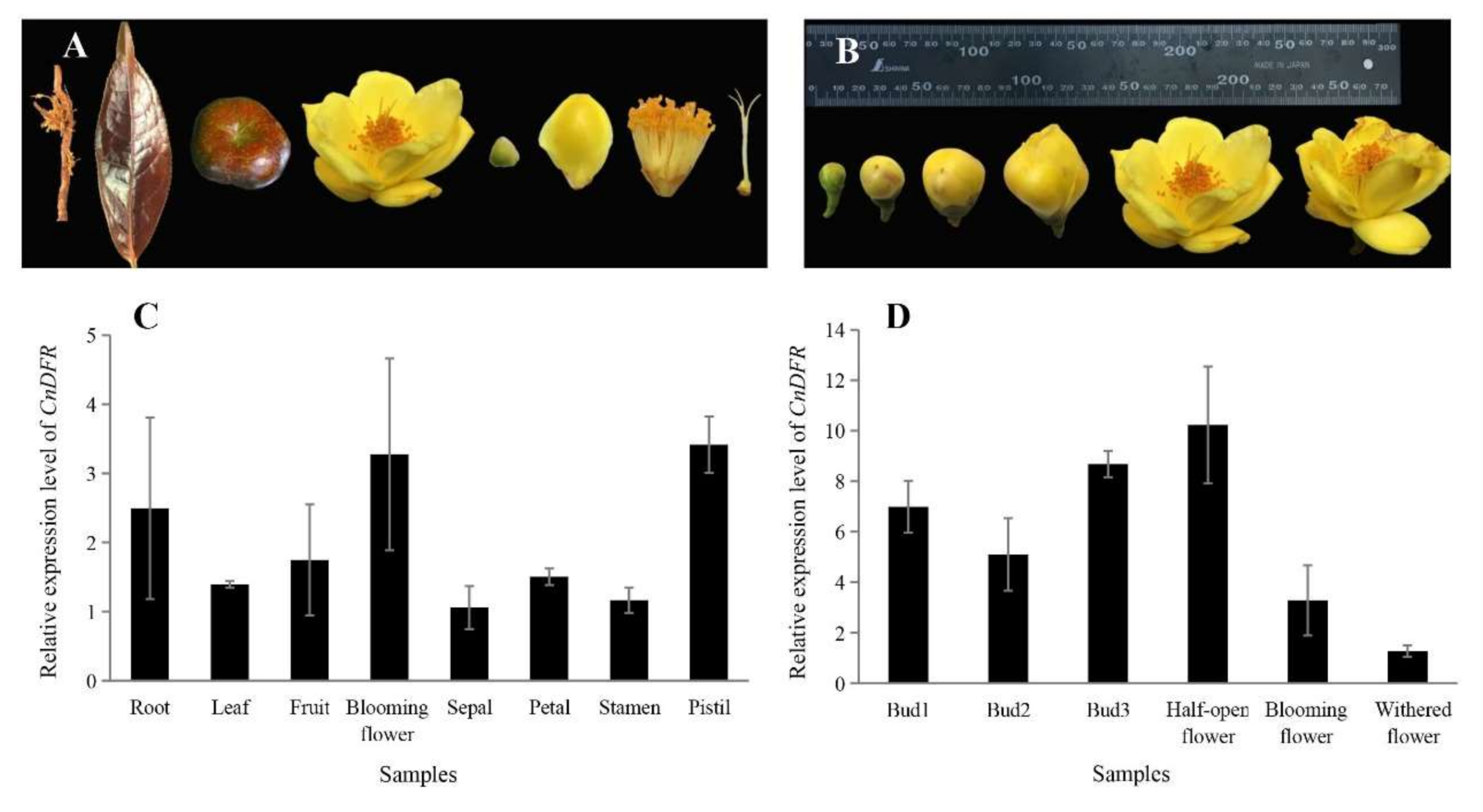
2.4. Quantitative PCR Analysis of CnDFR
2.5. High-Performance Liquid Chromatography Analysis
2.6. Agroinfiltration-Based Transient CnDFR-EGFP Gene Expression in Nicotiana Benthamiana Domin for Subcellular Localization of CnDFR
2.7. Tobacco Transformation Analysis of CnDFR
3. Results
3.1. Molecular Characterization of CnDFR Reveals Lineage-Specific Amino Acid Sites
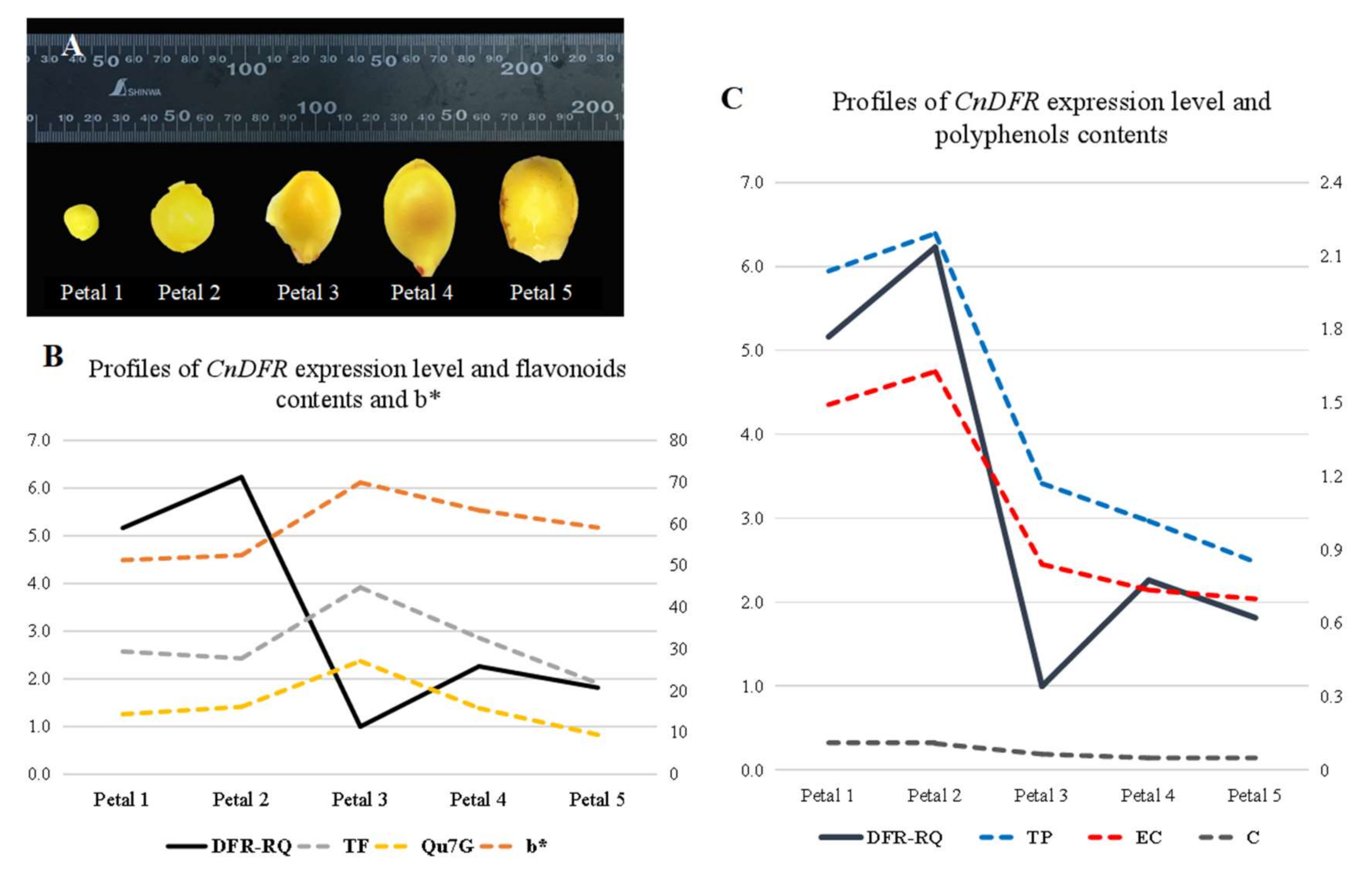
3.2. Expression of CnDFR in C. nitidissima Chi. Is Positively Correlated with Polyphenols Contents
3.3. Subcellular Localization of CnDFR Was in the Nucleus and Cell Membrane
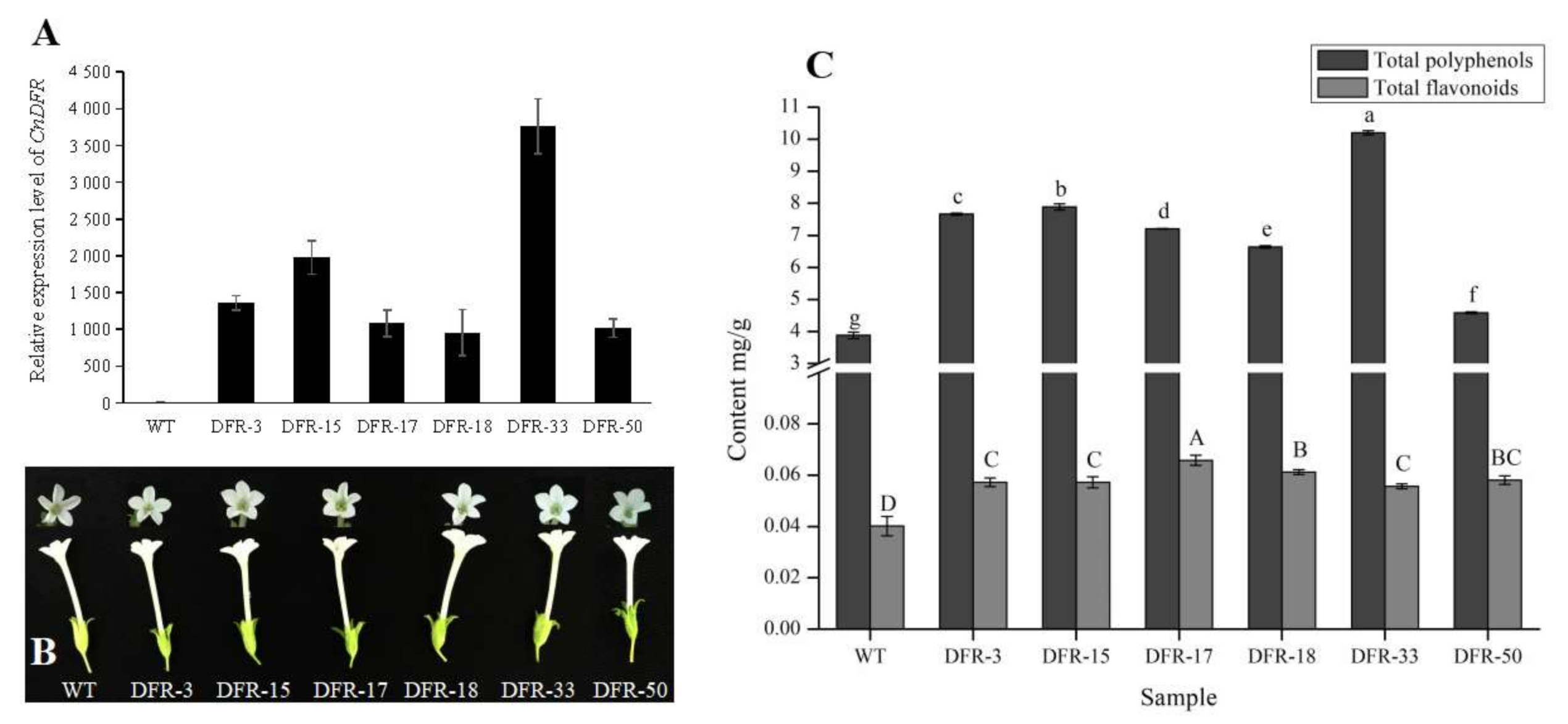
3.4. Overexpression of CnDFR Enhanced the Biosynthesis of Polyphenols But Not Anthocyanins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Bohorquez-Restrepo, A.; Irani, N.G.; Grotewold, E. Anthocyanin biosynthesis, regulation, and transport: New insights from model species. Recent Adv. Polyphen. Res. 2012, 3, 143–160. [Google Scholar] [CrossRef]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef] [PubMed]
- Ramiro-Puig, E.; Casadesús, G.; Lee, H.-G.; Zhu, X.; McShea, A.; Perry, G.; Pérez-Cano, F.J.; Smith, M.A.; Castell, M. Neuroprotective effect of cocoa flavonids on in vitro oxidative stress. Eur. J. Nutr. 2009, 48, 54–61. [Google Scholar] [CrossRef]
- Tsuda, T. Recent progress in anti-obesity and anti-diabetes effect of berries. Antioxidants 2016, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, W.; Zhang, P.; Pan, Q.; Zhan, J.; Huang, W. Gene transcript accumulation, tissue and subcellular localization of anthocyanidin synthase (ANS) in developing grape berries. Plant Sci. 2010, 179, 103–113. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. Acolorfulmodel forgenetics, biochemistry, cellbiology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant. Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Hassani, D.; Liu, H.; Chen, Y.; Wan, Z.; Zhuge, Q.; Li, S. Analysis of biochemical compounds and differentially expressed genes of the anthocyanin biosynthetic pathway in variegated peach flowers. Genet. Mol. Res. 2015, 14, 13425–13436. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; He, J. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.-M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Chen, M.; Zhang, J.; Li, K.; Song, T.; Zhang, X.; Yao, Y. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 2017, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, E.T.; Yi, H.; Shin, B.; Oh, B.J.; Cheong, H.; Choi, G. Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 1999, 19, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef] [Green Version]
- Gould, K.; Davies, K.M.; Winefield, C. Anthocyanins: Biosynthesis, Functions, and Applications; Springer Science & Business Media: New York, NY, USA, 2008; p. 167. [Google Scholar]
- Guo, N.; Han, S.; Zong, M.; Wang, G.; Zheng, S.; Liu, F. Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale. BMC Genom. 2019, 20, 564. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Andersen, M.; Jordheim, M. Anthocyanins. Encyclopedia of Life Sciences (ELS); John Wiley and Sons Ltd.: Chichester, UK, 2010. [Google Scholar] [CrossRef]
- Otani, M.; Kanemaki, Y.; Oba, F.; Shibuya, M.; Funayama, Y.; Nakano, M. Comprehensive isolation and expression analysis of the flavonoid biosynthesis-related genes in Tricyrtis spp. Biol. Plant 2018, 62, 684–692. [Google Scholar] [CrossRef]
- Provenzano, S.; Spelt, C.; Hosokawa, S.; Nakamura, N.; Brugliera, F.; Demelis, L.; Geerke, D.P.; Schubert, A.; Tanaka, Y.; Quattrocchio, F. Genetic control and evolution of anthocyanin methylation. Plant Physiol. 2014, 165, 962–977. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.; Teeri, T.; Forkmann, G. Heterologous expression of dihydroflavonol 4-reductases from various plants. FEBS Lett. 2002, 531, 453–458. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. B 2013, 368, 20120432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.H.; You, M.K.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Ha, S.H. RNAi-mediated suppression of dihydroflavonol 4-reductase in tobacco allows fine-tuning of flower color and flux through the flavonoid biosynthetic pathway. Plant Physiol. Biochem. 2016, 109, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, A.; Izumi, Y.; Yamagishi, M. Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 2003, 165, 759–767. [Google Scholar] [CrossRef]
- Polashock, J.J.; Griesbach, R.J.; Sullivan, R.F.; Vorsa, N. Cloning of a cDNA encoding the cranberry dihydroflavonol-4-reductase (DFR) and expression in transgenic tobacco. Plant Sci. 2002, 163, 241–251. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Yadav, S.K.; Ahuja, P.S. Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant Biotechnol. Rep. 2009, 3, 95–101. [Google Scholar] [CrossRef]
- Pitakdantham, W.; Sutabutra, T.; Pitaksutheepong, C.; Chiemsombat, P. Isolation and characterization of dihydroflavonol 4-reductase gene from Dendrobium Sonia’earsakul’. J. Issaas 2011, 17, 673–675. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Qiu, J.; Chen, F.; Lv, X.; Fu, C.; Zhao, D.; Hua, X.; Zhao, Q. Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene in Saussurea medusa. Mol. Biol. Rep. 2012, 39, 2991–2999. [Google Scholar] [CrossRef]
- Mori, S.; Otani, M.; Kobayashi, H.; Nakano, M. Isolation and characterization of the dihydroflavonol 4-reductase gene in the monocotyledonous ornamental Agapanthus praecox ssp. orientalis (Leighton) Leighton. Sci. Hortic. 2014, 166, 24–30. [Google Scholar] [CrossRef]
- Zhu, Y.; Peng, Q.; Li, K.; Xie, D.-Y. Molecular cloning and functional characterization of a dihydroflavonol 4-Reductase from Vitis bellula. Molecules 2018, 23, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Kobayashi, A.; Endo, M.; Sage-Ono, K.; Toki, S.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Nadda, G.; Kumar, S.; Yadav, S.K. Transgenic tobacco overexpressing tea cDNA encoding dihydroflavonol 4-reductase and anthocyanidin reductase induces early flowering and provides biotic stress tolerance. PLoS ONE 2013, 8, e65535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, Y.-J.; Yoshikawa, K.; Miyajima, I.; Okubo, H. Flower colors and pigments in hybrids with Camellia chrysantha. Sci. Hortic 1992, 51, 251–259. [Google Scholar] [CrossRef]
- Scogin, R. Floral pigments of the yellow Camellia, Camellia chrysantha (Theaceae). Aliso 1986, 11, 387–392. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Li, J.; Yin, H.; Fan, Z.; Li, X. Cloning of Camellia nitidissima flavonol synthase cDNA and construction of sense, RNA interference expression vectors. Bull. Bot. Res. 2013, 33, 58–65. [Google Scholar]
- Jiang, L.; Li, J.; Tong, R.; He, L.; Zhang, L.; Li, Z.; Huang, X. Relationship between flower color and important cellular environment elemental factors in yellow camellia. Guihaia 2019, 39, 1605–1612. [Google Scholar] [CrossRef]
- Tanikawa, N.; Kashiwabara, T.; Hokura, A.; Abe, T.; Shibata, M.; Nakayama, M. A peculiar yellow flower coloration of camellia using aluminum-flavonoid interaction. J. Jpn. Soc. Hortic. Sci. 2008, 77, 402–407. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Fan, Z.; Guo, H.; Ye, N.; Lyu, T.; Yang, W.; Wang, J.; Wang, J.-T.; Wu, B.; Li, J. Comparative genomics analysis reveals gene family expansion and changes of expression patterns associated with natural adaptations of flowering time and secondary metabolism in yellow Camellia. Funct. Intergr. Genom. 2018, 18, 659–671. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Zhu, Y.; Ni, S.; Chen, J.; Feng, X.; Zhang, Y.; Li, S.; Zhu, H.; Wen, Y. De novo assembly of the Camellia nitidissima transcriptome reveals key genes of flower pigment biosynthesis. Front. Plant Sci. 2017, 8, 1545. [Google Scholar] [CrossRef] [Green Version]
- Aida, R.; Yoshida, K.; Kondo, T.; Kishimoto, S.; Shibata, M. Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Sci. 2000, 160, 49–56. [Google Scholar] [CrossRef]
- Tian, J.; Han, Z.-y.; Zhang, J.; Hu, Y.; Song, T.; Yao, Y. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32 (Suppl. 2), W20–W25. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007, 36 (Suppl. 1), D13–D21. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Gooley, A.A.; Herbert, B.R.; Molloy, M.P.; Binz, P.-A.; Ou, K.; Sanchez, J.-C.; Bairoch, A.; Williams, K.L. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 1999, 289, 645–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gelvin, S.B. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Lukasik, E.; Gawehns, F.; Takken, F.L. The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 2012, 835, 61–74. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Sasaki, T.; Sivaguru, M.; Yamamoto, Y.; Osawa, H.; Ahn, S.J.; Matsumoto, H. Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol. 2005, 46, 812–816. [Google Scholar] [CrossRef] [Green Version]
- American Association for the Advancement of Science A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [CrossRef] [PubMed]
- Shimada, S.; Takahashi, K.; Sato, Y.; Sakuta, M. Dihydroflavonol 4-reductase cDNA from non-anthocyanin-producing species in the Caryophyllales. Plant Cell Physiol. 2004, 45, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, S.; Fukui, Y.; Nakamura, N.; Katsumoto, Y.; Yonekura-Sakakibara, K.; Fukuchi-Mizutani, M.; Ohira, K.; Ueyama, Y.; Ohkawa, H.; Holton, T.A. Flower color modification of Petunia hybrida commercial varieties by metabolic engineering. Plant Biotechnol. 2004, 21, 377–386. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Fan, Z.; Tong, R.; Zhou, X.; Li, J.; Yin, H. Functional Diversification of the Dihydroflavonol 4-Reductase from Camellia nitidissima Chi. in the Control of Polyphenol Biosynthesis. Genes 2020, 11, 1341. https://doi.org/10.3390/genes11111341
Jiang L, Fan Z, Tong R, Zhou X, Li J, Yin H. Functional Diversification of the Dihydroflavonol 4-Reductase from Camellia nitidissima Chi. in the Control of Polyphenol Biosynthesis. Genes. 2020; 11(11):1341. https://doi.org/10.3390/genes11111341
Chicago/Turabian StyleJiang, Lina, Zhengqi Fan, Ran Tong, Xingwen Zhou, Jiyuan Li, and Hengfu Yin. 2020. "Functional Diversification of the Dihydroflavonol 4-Reductase from Camellia nitidissima Chi. in the Control of Polyphenol Biosynthesis" Genes 11, no. 11: 1341. https://doi.org/10.3390/genes11111341




