Novel Regulators of Sugar-Mediated Lateral Root Development in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. β-glucuronidase (GUS) Staining and Microscopy
2.3. EMS Mutagenesis and Mutant Screening
2.4. Genetic Analyses
2.5. DNA Extraction, Genotyping and Mapping
3. Results
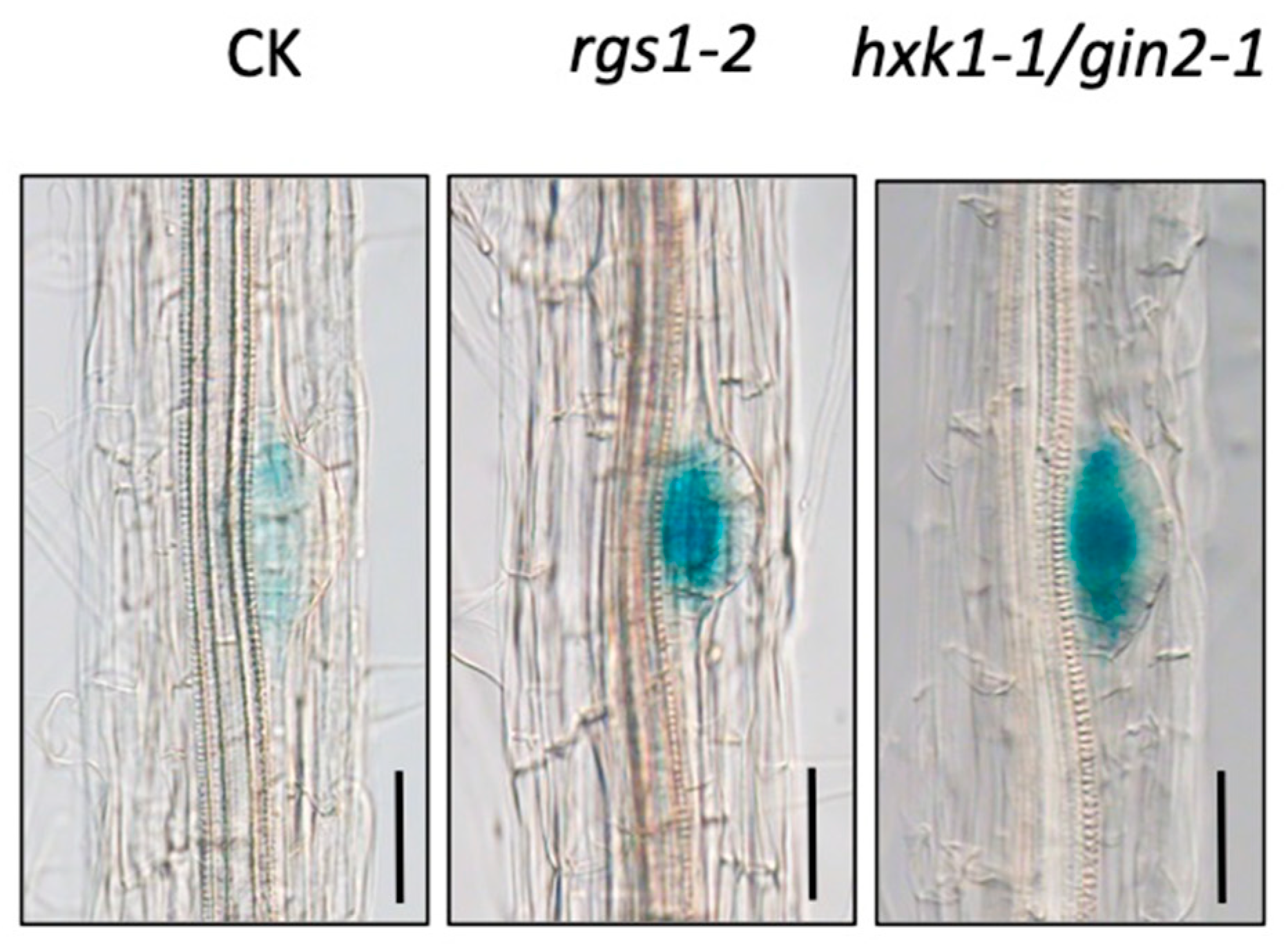
3.1. WOX7 Acts Downstream of HXK1 and RGS1
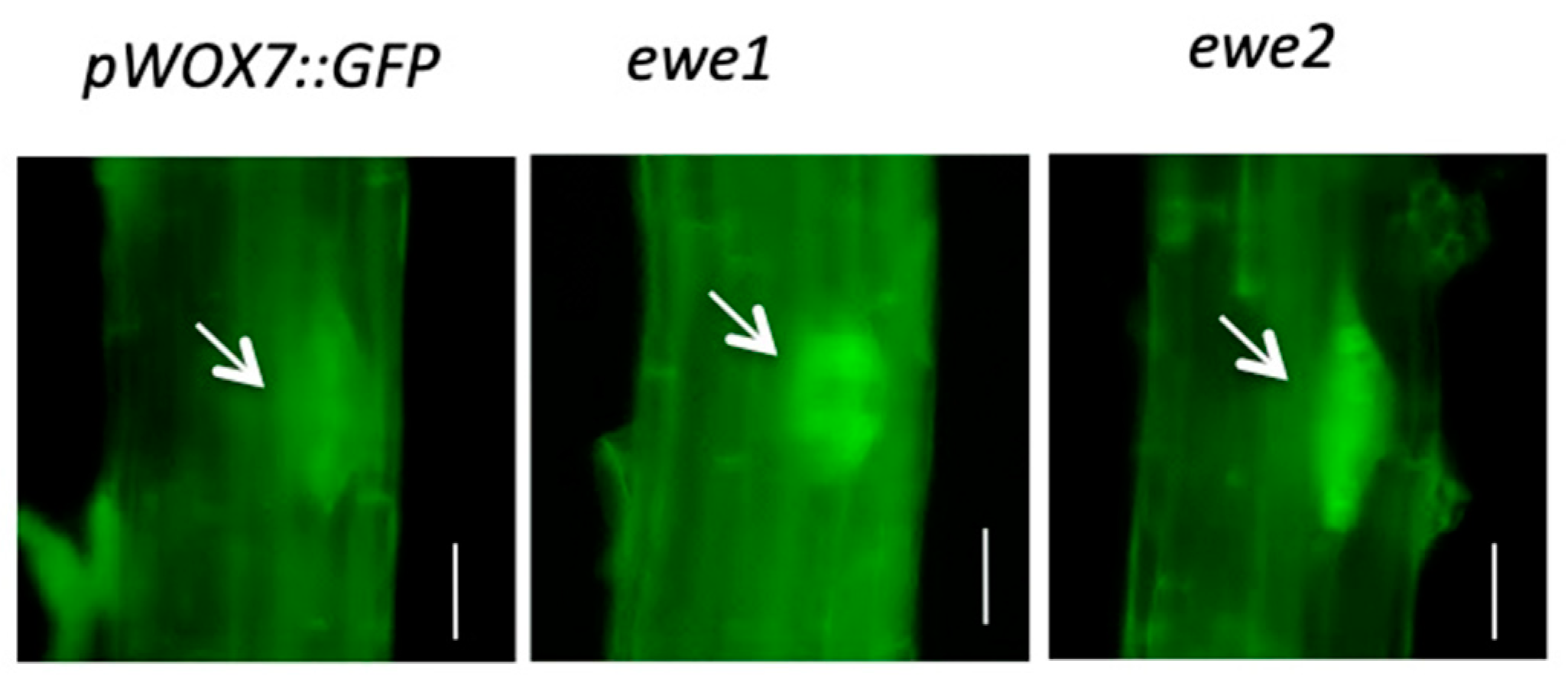
3.2. A Genetic Screen for Mutants with Altered WOX7 Expression
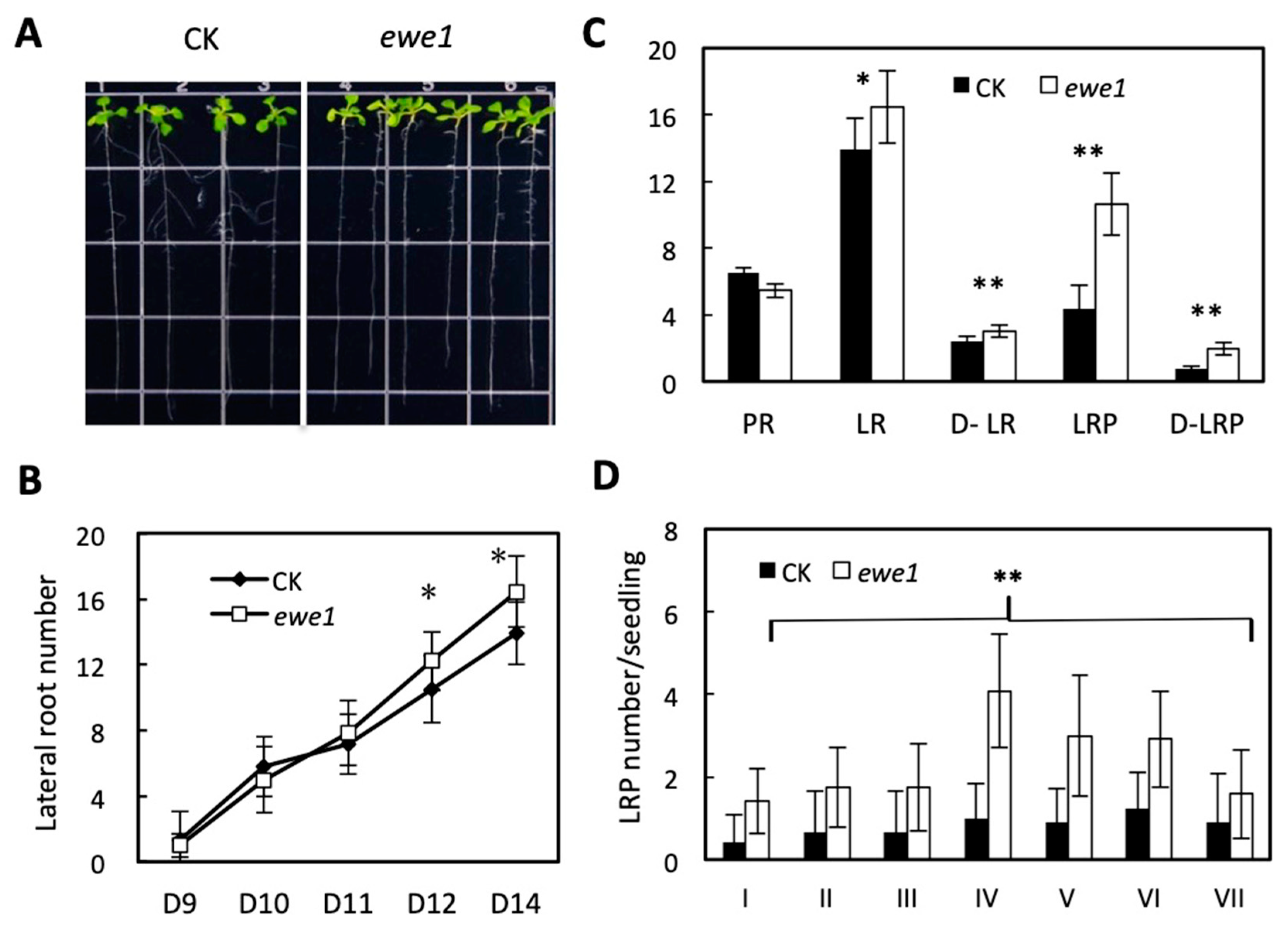
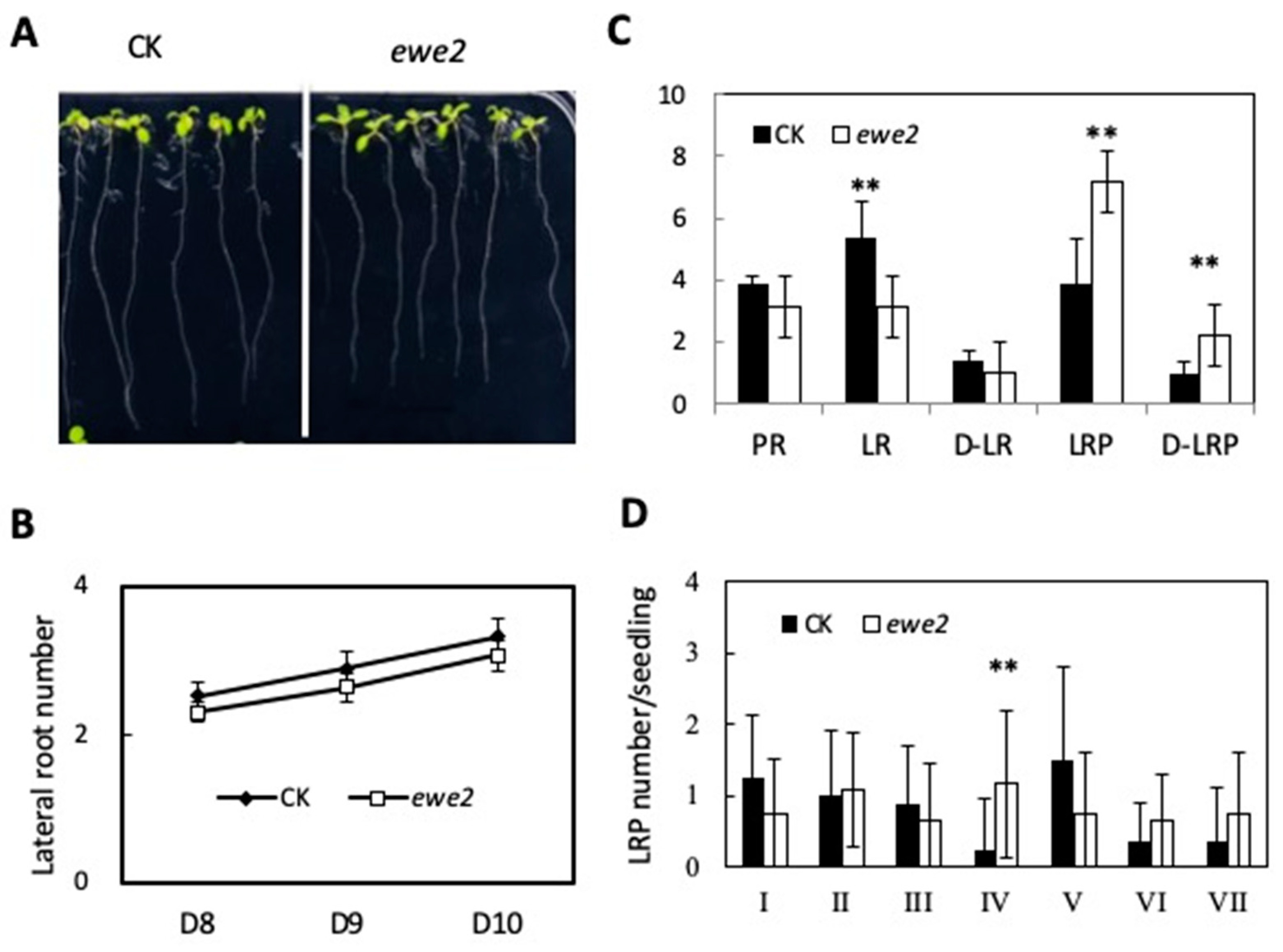
3.3. Characteristics of the ewe1 and ewe2 Mutants
3.4. Mapping the Causal Mutations in the ewe1 and ewe2 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Negi, S.; Ivanchenko, M.G.; Muday, G.K. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 2008, 55, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Delaux, P.M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattachaya, C.; Sejalon-Delmas, N.; Combier, J.P.; Becard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; De Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef]
- Linkohr, B.I.; Williamson, L.C.; Fitter, A.H.; Leyser, H.M. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002, 29, 751–760. [Google Scholar] [CrossRef]
- Malamy, J.E.; Benfey, P.N. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 1997, 124, 33–44. [Google Scholar]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Banda, J.; Bellande, K.; von Wangenheim, D.; Goh, T.; Guyomarc’h, S.; Laplaze, L.; Bennett, M.J. Lateral Root Formation in Arabidopsis: A Well-Ordered LRexit. Trends Plant Sci. 2019, 24, 826–839. [Google Scholar] [CrossRef]
- Malamy, J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Roycewicz, P.; Malamy, J.E. Dissecting the effects of nitrate, sucrose and osmotic potential on Arabidopsis root and shoot system growth in laboratory assays. Phil. Trans. R. Soc. Lond. Ser. B 2012, 367, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E.; Ryan, K.S. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001, 127, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, M.; Laxmi, A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, Y.; Karve, A.; Teixeira, P.J.; Jiang, K.; Tunc-Ozdemir, M.; Jones, A.M. Photosynthate Regulation of the Root System Architecture Mediated by the Heterotrimeric G Protein Complex in Arabidopsis. Front. Plant Sci. 2016, 7, 1255. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef]
- Karve, A.; Rauh, B.L.; Xia, X.; Kandasamy, M.; Meagher, R.B.; Sheen, J.; Moore, B.D. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 2008, 228, 411–425. [Google Scholar] [CrossRef]
- Karve, A.; Moore, B.D. Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J. Exp. Bot. 2009, 60, 4137–4149. [Google Scholar] [CrossRef]
- Chen, Y.; Ji, F.; Xie, H.; Liang, J.; Zhang, J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006, 140, 302–310. [Google Scholar] [CrossRef]
- Booker, K.S.; Schwarz, J.; Garrett, M.B.; Jones, A.M. Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS ONE 2010, 5, e12833. [Google Scholar] [CrossRef]
- Huang, J.P.; Tunc-Ozdemir, M.; Chang, Y.; Jones, A.M. Cooperative control between AtRGS1 and AtHXK1 in a WD40-repeat protein pathway in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 851. [Google Scholar] [CrossRef] [PubMed]
- Deak, K.I.; Malamy, J. Osmotic regulation of root system architecture. Plant J. 2005, 43, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, D.R.; Deak, K.I.; Ingram, P.A.; Malamy, J.E. Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 2008, 20, 2643–2660. [Google Scholar] [CrossRef] [PubMed]
- Ingram, P.; Dettmer, J.; Helariutta, Y.; Malamy, J.E. Arabidopsis Lateral Root Development 3 is essential for early phloem development and function, and hence for normal root system development. Plant J. 2011, 68, 455–467. [Google Scholar] [CrossRef]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Arabidopsis: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2002. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010, 38, e164. [Google Scholar] [CrossRef]
- NGS Raw Data for Mapping the Causal Genes in ewe1 and ewe2. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA669402 (accessed on 24 October 2020).
- Austin, R.S.; Vidaurre, D.; Stamatiou, G.; Breit, R.; Provart, N.J.; Bonetta, D.; Zhang, J.; Fung, P.; Gong, Y.; Wang, P.W.; et al. Next-generation mapping of Arabidopsis genes. Plant J. 2011, 67, 715–725. [Google Scholar] [CrossRef]
- The BAR tool. Available online: http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi (accessed on 24 October 2020).
- James, G.V.; Patel, V.; Nordstrom, K.J.V.; Klasen, J.R.; Salome, P.A.; Weigel, D.; Schneeberger, K. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013, 14, R61. [Google Scholar] [CrossRef]
- Li, L.; He, Z.; Pandy, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 2002, 277, 5260–5268. [Google Scholar] [CrossRef] [PubMed]
| Cross | Generation | Total Seedlings Analyzed | Mutant Phenotype | Wild Type | ᵪ2 (3:1) |
|---|---|---|---|---|---|
| ewe 1 × Col | F2 | 78 | 18 | 60 | 0.155 a |
| ewe2 × Col | F2 | 64 | 16 | 48 | 0.004 a |
| ewe1 × ewe2 | F1 | 10 | — | 10 |
| Chromosome | SSLP Marker | Distance to Mutation (cM) |
|---|---|---|
| 1 | YUP8H12 | 64.3 |
| 1 | T17H3 | 54.8 |
| 1 | NGA128 | 42.9 |
| 2 | CIW2 | 19 |
| 2 | NGA168 | 31 |
| 3 | CIW11 | 52.4 |
| 3 | T32N15A | 50 |
| 3 | NGA6 | 38.1 |
| 4 | CIW5 | 42.9 |
| 4 | FCA8 | 33.3 |
| 4 | F17I5A | 38.1 |
| 4 | NGA1107 | 40.5 |
| 5 | NGA151 | 40.5 |
| 5 | CIW9 | 40.5 |
| Chromosome | SSLP Marker | Distance to Mutation (cM) |
|---|---|---|
| 2 | T18E12 | 16.67 |
| 2 | F15L11 | 11.11 |
| 2 | F7E22 | 6.94 |
| 2 | T4E5 | 6.94 |
| 2 | F13J11 | 6.94 |
| 2 | F26C24 | 9.72 |
| 2 | NGA168 | 34.72 |
| Chromosome | SSLP Marker | Distance to Mutation (cM) |
|---|---|---|
| 1 | YUP8H12 | 40 |
| 1 | T17H3 | 54.8 |
| 1 | NGA128 | 54.8 |
| 2 | F504A | 25.8 |
| 2 | CIW2 | 25.8 |
| 2 | CIW3 | 29 |
| 2 | NGA1126 | 45.1 |
| 2 | NGA168 | 40.3 |
| 3 | F14P3 | 45.1 |
| 3 | NGA162 | 54.8 |
| 3 | CIW11 | 48.3 |
| 3 | NGA6 | 58 |
| 4 | CIW5 | 40.9 |
| 4 | T32N4A | 50 |
| 4 | CIW6 | 54.8 |
| 4 | FCA8A | 59.6 |
| 5 | NGA151 | 50 |
| 5 | CIW9 | 53.2 |
| Chromosome | SSLP Marker | Distance to Mutation (cM) |
|---|---|---|
| 2 | F15L11 | 5.21 |
| 2 | T12H3 | 4.17 |
| 2 | F7E22 | 4.17 |
| 2 | T4E5 | 3.13 |
| 2 | F10C8 | 3.13 |
| 2 | T17A11 | 4.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wang, B.; Zhu, X.; Li, R.; Fu, J.; Cui, H. Novel Regulators of Sugar-Mediated Lateral Root Development in Arabidopsis thaliana. Genes 2020, 11, 1257. https://doi.org/10.3390/genes11111257
Li J, Wang B, Zhu X, Li R, Fu J, Cui H. Novel Regulators of Sugar-Mediated Lateral Root Development in Arabidopsis thaliana. Genes. 2020; 11(11):1257. https://doi.org/10.3390/genes11111257
Chicago/Turabian StyleLi, Jinzhu, Bingxin Wang, Xinxing Zhu, Rong Li, Jing Fu, and Hongchang Cui. 2020. "Novel Regulators of Sugar-Mediated Lateral Root Development in Arabidopsis thaliana" Genes 11, no. 11: 1257. https://doi.org/10.3390/genes11111257
APA StyleLi, J., Wang, B., Zhu, X., Li, R., Fu, J., & Cui, H. (2020). Novel Regulators of Sugar-Mediated Lateral Root Development in Arabidopsis thaliana. Genes, 11(11), 1257. https://doi.org/10.3390/genes11111257






